Abstract
PhosphoSitePlus (http://www.phosphosite.org) is an open, comprehensive, manually curated and interactive resource for studying experimentally observed post-translational modifications, primarily of human and mouse proteins. It encompasses 1 30 000 non-redundant modification sites, primarily phosphorylation, ubiquitinylation and acetylation. The interface is designed for clarity and ease of navigation. From the home page, users can launch simple or complex searches and browse high-throughput data sets by disease, tissue or cell line. Searches can be restricted by specific treatments, protein types, domains, cellular components, disease, cell types, cell lines, tissue and sequences or motifs. A few clicks of the mouse will take users to substrate pages or protein pages with sites, sequences, domain diagrams and molecular visualization of side-chains known to be modified; to site pages with information about how the modified site relates to the functions of specific proteins and cellular processes and to curated information pages summarizing the details from one record. PyMOL and Chimera scripts that colorize reactive groups on residues that are modified can be downloaded. Features designed to facilitate proteomic analyses include downloads of modification sites, kinase–substrate data sets, sequence logo generators, a Cytoscape plugin and BioPAX download to enable pathway visualization of the kinase–substrate interactions in PhosphoSitePlus®.
INTRODUCTION
The cellular regulation of post-translational modification [PTMs; (1)] is complex and plays a fundamental role at all levels of biological regulation. Protein phosphorylation, partly because of its early links to metabolic regulation (2) and cancer (3,4) has been the most widely studied PTM. Throughout the 1990s, the number of physiological substrates discovered grew significantly; there was a recognized need for an archive that systematically collected protein kinase phosphorylation sites (5). Such an archive, in order to be broadly useful to biologists and biomedical researchers, needs to minimally include not only the modified residue and surrounding sequence, but also its regulation by treatments and ligands, associated biological processes, upstream and downstream interactions and location relative to protein domains and other functional regions of the protein.
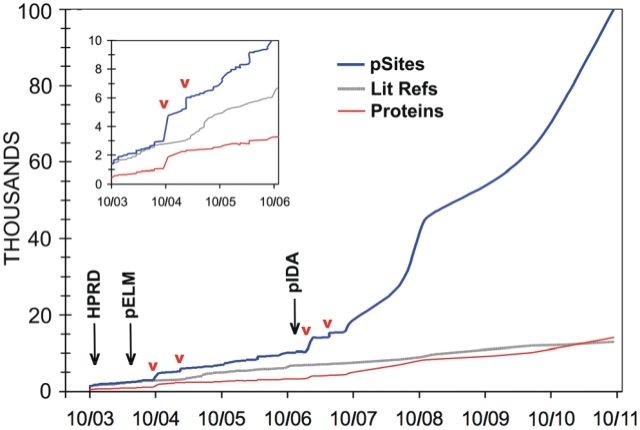
PhosphoSite® (6), launched in 2003, was designed as a resource that would comprehensively aggregate information about the structure and regulatory interactions of phosphorylation sites. At its launch it incorporated over 1200 journal articles identifying over 1200 non-redundant sites on over 500 human and mouse proteins (Figure 1). A total 12 000 references have been incorporated since then, with curated information including the regulation by treatments and of protein interactions, roles in biological processes, disease relevance, kinase–substrate interactions and relevant structural files and molecular rendering tools. Initially, nearly all curated information was derived from low-throughput (LTP) experimental methods and so curation seemed eminently tractable. This changed in the beginning of 2004 with high-throughput (HTP) tandem mass spectrometry [MS; (7)], when articles reporting many hundreds to thousands of phosphorylation sites began appearing (8–10). This technology has similarly transformed the identification of other modification types including ubiquitination (11–13), acetylation (14,15) and O-GlcNAcylation (16).
Figure 1.
Informational content of PhosphoSite® curated from the literature from its launch in 2003 through 2011. Phosphorylation sites (blue), proteins (red) and references (gray). The approximate dates that descriptions of three other resources were published are indicated by black arrows: HPRD (19), phospho.ELM (20) and PHOSIDA (21). Inflection points indicate the publication of large MS data sets; a few are marked with red arrowheads.
PhosphoSitePlus® (PSP), reengineered from PhosphoSite® and launched in Feb 2008, is comprised of 1 29 082 non-redundant sites on 14 256 non-redundant proteins (Table 1). Over 90% of these sites are from human and mouse. Phosphorylation, acetylation and ubiquitination are included, enabling the study of cross-regulation between PTMs (16,17). Recent improvements allow users to access content and tools not previously accessible including: (i) queries for extracting targeted sets of modification sites, such as those responsive to particular treatments, or found in particular types of cancer or specific cellular compartments; (ii) downloads of user-defined interactive data sets and monthly updated static data sets; (iii) the generation of sequence logos to visualize specificity profiles; (iv) downloads of Chimera (18) and (v) PyMol scripts that colorize and label known modification sites on molecular models; and (vi) pathway visualizations of kinase-substrate interactions.
Table 1.
Number of post-translationally modified proteins and sites in PSP
| Modification type | Proteins | Sites |
|---|---|---|
| Serine phosphorylation | 10 704 | 65 511 |
| Threonine phosphorylation | 7134 | 19 609 |
| Tyrosine phosphorylation | 6460 | 15 053 |
| Ubiquitinylation | 5139 | 18 996 |
| Acetylation | 3287 | 7869 |
| Sumoylation | 356 | 622 |
| O-Glycosylation | 177 | 602 |
| Di-methylation | 167 | 378 |
| Mono-methylation | 153 | 303 |
| Methylation | 56 | 139 |
Two resources in addition to PSP were launched in 2003–2004 that aggregated experimentally observed phosphorylation sites: HPRD (19) and phospho.ELM (20) (Figure 1). A fourth, PHOSIDA (10), was launched in the fall of 2006. HPRD aggregates information about multiple types of PTMs, comprising 93 710 instances, as well as protein–protein interactions, subcellular localization and tissue expression. Phospho.ELM is a database of experimentally verified phosphorylation sites in eukaryotic proteins. Its last release contains more than 42 500 phosphorylation sites on 8718 proteins from different species. PHOSIDA (21), first described in November 2006 (10), archives only MS2 assignments, all of which are derived from the same research group. It is comprised of more than 80 000 PTM sites from nine different species. These include 24 262 sites on 8283 human proteins and 25 085 sites on 9234 mouse proteins. Modification sites in addition to phosphorylation, including O-glycosylation and acetylation, have recently been included. SysPTM (22) contains information curated from literature about nearly 50 PTM types, and is comprised of 1 17 349 PTM sites on 33 421 proteins. CPLA (23) has aggregated 7151 experimentally identified acetylation sites in 3311 proteins.
DATA MANAGEMENT OF SEQUENCES, STRUCTURES AND ASSOCIATED DATA
Protein sequences
Parent proteins are usually defined as the longest of alternatively spliced isoforms. Protein sequences are imported hierarchically: first, from the reviewed, manually annotated entries of UniProtKB/Swiss-Prot (24); secondly, from NCBI/RefSeq (25) entries with accession IDs beginning with NP; and thirdly, from unreviewed, automatically annotated entries from UniProtKB/TrEMBL or from RefSeq entries with accession IDs beginning with XP. If no matches are found, then Ensembl (26) entries are searched. When a new protein sequence is imported, its human, mouse or rat orthologs are also loaded. Isoform sequences other than that of the parent protein are imported only when the only evidence for the existence of the modification site is present in an isoform. Approximately 93% of sequences in PSP are from UniPROT KB, 6% from NCBI and <0.5% from Ensembl. The sequences and accession numbers of parent proteins are synchronized with the UniPROT KB every 6 months. All other sequences are updated as needed during curation in order to accommodate experimental results.
Additional protein data
Accession numbers from UniPROT KB, NCBI and Ensembl (24–26), as well as gene symbols from HGNC (27), are curated for all proteins when possible. Basic protein descriptions include information parsed from UniPROT KB (24), and may include additional information from the literature. Descriptions are updated in bulk occasionally. Gene Ontology (28) annotations are parsed from NCBI (25). Editors assign protein types.
Structural files
A total of 22 958 PDB (29) files corresponding to proteins in PSP are stored locally for rendering molecular structures opened by the default Astex Java viewer. PDB (29) is queried monthly for new structures associated with proteins in PSP. 4500 of the 14 300 proteins in PSP, or 31%, have associated PDB files corresponding to some segment of the protein's sequence. 15 307 sites, 12% of all sites in PSP, are located within the resolved regions of solved structures or within relatively unstructured regions in which the modified residue is resolved, e.g. Akt1 Lys14 in PDB_1H10: F..K*N..TFI..KER (30).
Protein and site groups
Individual modification sites and proteins are organized by orthologous (31) groups. Protein and site groups include all members of the orthologous group. Unless explicitly stated otherwise, the terms protein and site shall apply to their respective groups throughout this article. The associated numbers of proteins and sites are thus non-redundant by definition, unless otherwise stated.
PSP ontology
Most curated information is parsed into the 27 controlled vocabularies (CVs) that constitute the PSP ontology. 10 of these CVs provide terms used in the search interfaces.
Database and application software
Data are stored in an Oracle 10 relational database with a schema consisting of about 100 different tables. The curatorial interface is a custom-designed Java/Swing application that enables curators to enter new data or modify existing data if needed. The web interface is based on Java/Struts/Hibernate running on a Tomcat 5 sever.
Links to antibody and siRNA products from cell signaling technology
Proteins and modification sites for which cell signaling technology (CST) sells products are marked with links to the appropriate product pages in the online catalog. This arrangement for a comprehensive scientific resource with commercial links was a part of research proposals from CST that were funded by National Institutes of Health Small Business Innovation Research grants.
CURATED MODIFICATION SITE CONTENT
Modification sites
The total number of modification sites is 1 29 082 on 14 256 proteins (Table 1). Seventy-eight percent are phosphorylation, 15% ubiquitination and 6% acetylation. The numbers of discovered phospho-Tyr, ubiquitinyl-Lys and acetyl-Lys have been greatly amplified by the availability of appropriate affinity reagents (9,13–15). For example, pY constitutes only a few percent of total cellular protein phosphorylation, but the systematic use of anti-pY antibodies in MS2 protocols has increased its representation in the known repertoire to 27%, 10-fold higher than that expected based on its natural frequency. Note that methylation here represents instances in which the records did not distinguish between mono-, bi- or tri-methylation.
Species
Although PTMs from 18 organisms are included in PSP, 99.7% are mammalian. The five organisms with the highest numbers of PTMs are human (70 338), mouse (57 862), rat (7665), cow (553) and chicken (366).
LTP content
Information from nearly 13 000 papers and 600 different journals characterizing modification sites with LTP methods has been curated into PSP. The top journals and associated numbers of curated articles are: J Biol Chem, 3665; Mol Cell Biol, 734; Oncogene, 546; Proc Natl Acad Sci USA, 530; and EMBO J, 375. 10 700 of the sites in PSP have been observed with LTP methods. 7168 (67%) of these LTP sites are associated with only 1 record, 2745 (26%) with 2–5 records, 615 (6%) with 6–25 records and 173 (2%) with >25 records.
HTP content
Sites based on MS assignments can come either from journal publications or from the CST research group (9,32,33). Over 110 journal publications and 2258 curation sets from CST have been curated. Over 2 60 000 (redundant) site assignments have been curated from journal publications and over 65 000 from CST curation sets. A majority of sites in PSP have only been reported using MS. Of 1 29 668 sites, 1 16 660 (90%) have been seen only with MS. 72 362 (62%) of these MS’ only sites are associated with only 1 record, 30 886 (26%) with 2–5 records, 11 245 (10%) with 6–25 records, and 2116 (2%) with >25 records.
Data quality issues
If a recently curated paper includes site assignment scores, only sites with scores of P ≥ 0.95 or an Ascore ≥13 (34) have been curated. Previously curated data sets that contain numerical scores are being reanalyzed to exclude sites that do not meet these standards to filter out as much unreliable data as possible. Since most MS2 peptide and site assignments are published en masse in unreviewed Supplementary Tables and come from labs with different experimental practices and levels of expertise, it is impossible to implement a common acceptability standard on MS2 data curated into PSP. [For a discussion of these issues, see Ref. (35).] Users must use caution and good judgment when evaluating site assignments based upon MS2 methods alone.
USING PSP
This section outlines the navigational structure of PSP, with a few examples that will help the user to effectively navigate the site. Detailed navigational help is available on the site itself. First, an online Flash tutorial (www.phosphosite.org/staticTrainingTutorial.do) under the Using PhosphoSite tab provides instructions for use; its index allows users to jump to a specific topic. Second, links to help pages, indicated by question marks in blue, are available in all search interfaces next to the red SEARCH buttons.
Navigating PSP
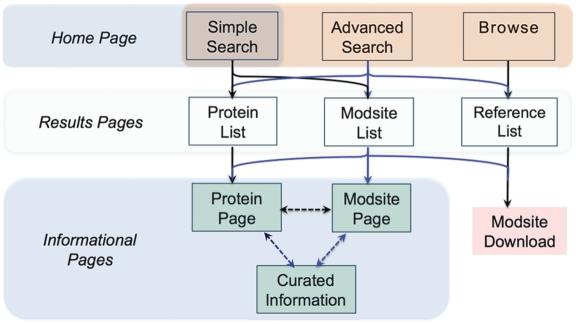
The structure and navigation of PSP is outlined in Figure 2 The site is organized around three types of pages: search, results and informational. Searches are initiated directly from the Homepage, advanced search and browse pages. ‘Results’ pages are lists of items matching the search string and are linked to Protein, Site, or Reference Pages, depending upon the nature of the query. Selection of an item from a ‘Results’ page opens up the corresponding informational page. The three informational pages (Protein, Modification Site and Curated Information) are all interlinked (Figure 2), allowing for ease of navigation between levels. The Homepage can be accessed from any page in the site by clicking on the word Home in the upper left-hand corner. Table 2 summarizes the modes for submitting searches and the pages from which they are launched.
Figure 2.
Overview of the navigational flow and content of PSP.
Table 2.
Four search pages and types of queries launched from them: T, text; L, text list, C, controlled vocabulary
| Query term | Homepage | Advanced search pages |
||
|---|---|---|---|---|
| Protein, sequence, site search comparative | Reference search | Site search | ||
| Kinase | T | |||
| Protein | T | T,L | ||
| Accession ID | T,L | |||
| PubMed ID | T,L | |||
| Author | T | |||
| Sequence or motif | T,L | T | ||
| Molecular weight | T | T | ||
| Protein type | C | C | C | |
| Cellular component | C | C | C | |
| Domain | C | C | C | |
| Treatment | C | C | ||
| Cell line | C | C | ||
| Cell type | C | C | ||
| Tissue | C | C | ||
| Disease | C | C | ||
| Biological process | C | |||
| Molecular function | C | |||
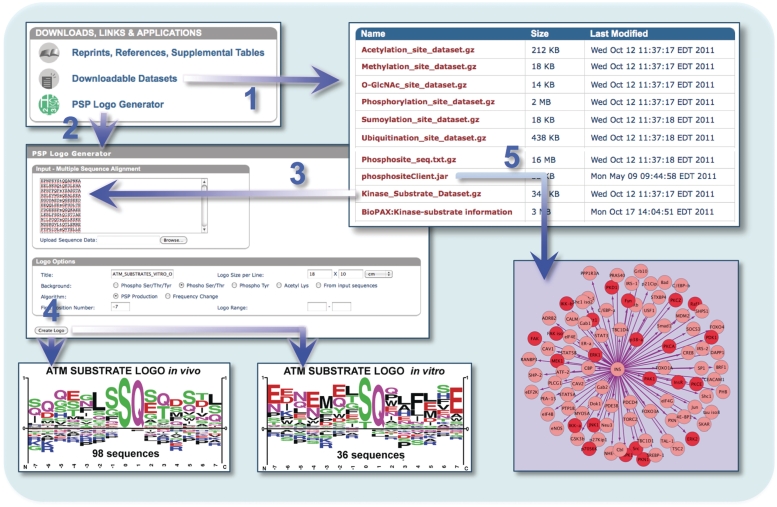
The Homepage is the hub of the site, the page from which all functionalities and actions are initiated. These include two types of simple searches and links to three advanced search and three browse pages. The Downloads, Links & Applications section on the lower left part of the Homepage (Figure. 3) provides access to multiple downloads and applications. The Downloadable Data set section provides downloads of six different modification type data sets, an archive of all protein sequences in PSP and the PSP Kinase-Substrate data set in three formats: a tab-delimited text file, a Cytoscape Plugin (36) and a file in BioPAX (37) format. The PSP Logo Generator tool at this location accepts lists of aligned sequences of identical length for sequence logo analysis. Two algorithms are available for logo generation: Frequency Change and PSP Production. Both differ from the original algorithm (38) by graphing under- and over-represented amino acids as negative and positive values, respectively. The Frequency Change method is as described (39). The PSP Production algorithm keeps the sum of absolute values of the negative and positive numbers equal to one, permitting a better view of amino acids that are represented at frequencies closer to the expected. In the example in Figure 3, ATM substrate sequences were separated into in vivo and in vitro groups and analyzed with the PSP Logo Generator.
Figure 3.
The Downloads, Links and Applications section provides (1) multiple modification site data sets from PhosphoSitePlus, the complete archive of current reference proteins (Phosphosite_seq), and the extensive Kinase–Substrate data set in three formats: the Kinase_Substrate_Data set is a tab-delimited txt file, a Cytoscape Plugin (phosphositeClient_jar), and BioPAX format (37); (2) a logo generator with which users can analyze uploaded data sets; (3) ATM substrate sequences extracted from the kinase-substrate data set were segregated into in vitro and in vivo reactions and (4) analyzed using the PSP Logo Generator and (5) a subset of the data extracted from PSP and visualized using Cytoscape (36).
Homepage searches
Protein searches are initiated by submitting the name of a protein in the Protein Name query field. The results page shows a list of all proteins whose primary or alternative names matches the search string. Selecting a protein opens up the corresponding Protein Page.
Substrate searches are initiated by entering the name of a kinase in the Substrates Of query field. The results page will show a list of all kinases that match the query string. Selecting a kinase opens up a downloadable list of experimentally determined substrate sequences. The substrate sequence preference of the kinase can be profiled by clicking on Show Sequence Logo link at the top of the page. This version of the sequence logo generator has more options that the stand-alone version described above: the modification type, organism associated with the kinase and modified amino acid(s) can all be selected. The data set used to generate the logo includes only one representative for each site group, eliminated bias due to over-representation of multiple instances of orthologous sites.
‘Homepage informational content’ includes the What's New section that describes new features that are not in the tutorial and site statistics that are updated daily. Four tabs in the upper right-hand corner open up informational pages describing the function, content and contact information for PSP.
Advanced searches
There are three search interfaces: (i) Protein, Sequence or Reference Search; (ii) Site Search and (iii) Comparative Site Search. Sixteen search types launch from the Advanced Search Pages (Table 2). Users can search by one or multiple parameters connected by the AND operator.
Seven search fields accept free text. Ten fields, marked with a blue SELECT button on the search interfaces, only accept pre-curated terms that need to be selected from the CVs.
Searching from pre-curated terms in the advanced search pages
Successful searches using terms from a CV requires five steps: (i) click on the blue SELECT button to open the Selection Window. Wait for the list of terms to appear in the middle window; (ii) begin typing letters in the search field at the top of the page, reducing the available terms shown in the search window; (iii) when the intended search term is found, highlight it and click the ADD button. The selected term will appear in the query frame at the bottom of the page; (iv) when all desired search terms have been selected and in the lower frame, click the SUBMIT button at the bottom of the Selection Window. This will populate the search field in the Search Page with the selected terms; and (v) initiate the search (finally) by clicking the red SEARCH button at the bottom of the frame.
Protein, sequence or reference searches
Protein Searches retrieve lists of proteins based on seven query types (Table 2). The sequence/motif search in this frame is case-insensitive, unlike that in the following Sequence Search. It retrieves the names of all proteins in PSP that contain the specified sequence.
‘Sequence searches’ retrieve lists of proteins and sequences containing specified sequences, degenerate motifs and domains. The powerful and versatile search syntax, patterned after the Swiss Institute of Bioinformatics’ ScanProsite tool (40), has been modified in this interface to interpret lower case letters as modified residues when the check box is turned on. For example, a search for [VIL]SXXSR returns 2136 hits without the box checked. With the box checked, a search for [VIL]sXXSR returns 60 hits and [VIL]sXXsR returns 26 hits. A unique search from the Sequence Search section allows users to submit a list of peptides and organisms to search for known PTMs on the peptide. When a match is found, the sequence is added to the output with the known sites marked.
‘Reference searches’ retrieve lists of literature references specified by authors, proteins or PubMedIDs. Author searches only accept one last name. Reference Search Results link to the PubMed citation and to Curated Information pages.
Site Searches and comparative site searches
Site Searches retrieve lists of modified sites matching the selected criteria. Comparative Site Searches retrieve sites that possess specified attributes and exclude others. Both can be specified by eight CV terms; Site Searches can be restricted by an additional four parameters (Table 2).
‘Browse interfaces’ retrieve lists of sites associated with diseases, cell lines or tissues. Results are returned in a table with a different column for each associated record. Selecting one of the three types opens a listing of all instances of each type. These include 48 disease listings, 244 cell lines and 24 tissues. Note that for diseases, both parent and children terms are listed, resulting in listings for leukemia as well as for T-cell leukemia, chronic myelogenous leukemia, etc. Clicking on one of the diseases, cell lines or tissues opens a list of all associated records. Upon selecting and submitting the desired records, a table will be generated and downloaded to your desktop containing a separate column for each record and if there are multiple samples in each record, separate columns for them.
Informational pages
Protein Pages aggregate a variety of structural and functional information about the protein, as well as organizing all modification sites. Site Pages present detailed information about that site, its biological characteristics (including regulation, effects, disease and references). Curated Information Pages, the most granular in PSP, outline the experimental details curated for all sites reported in the article. Only Protein Pages, the most widely visited informational page, will be considered in greater detail below.
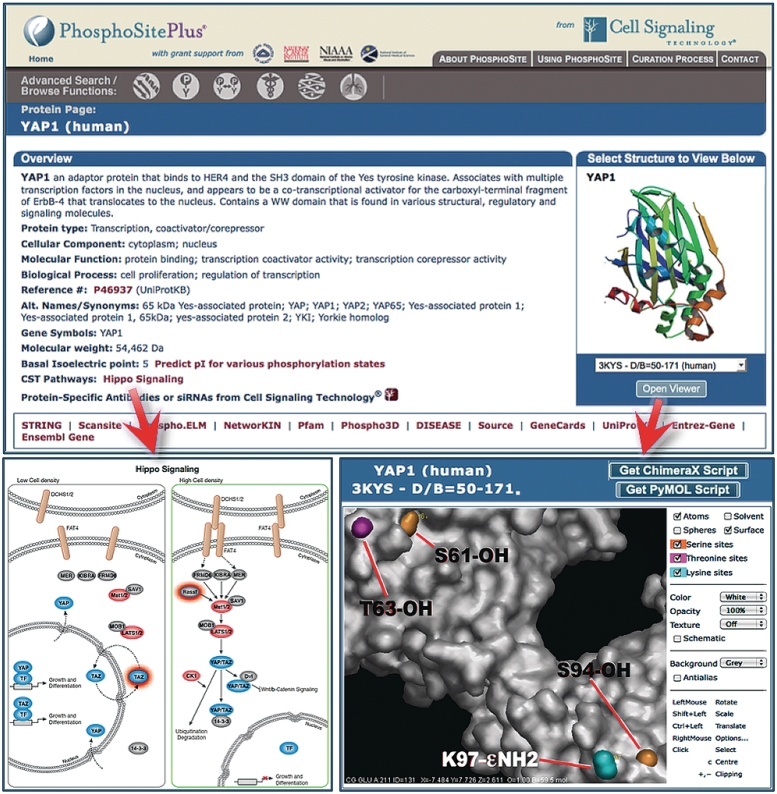
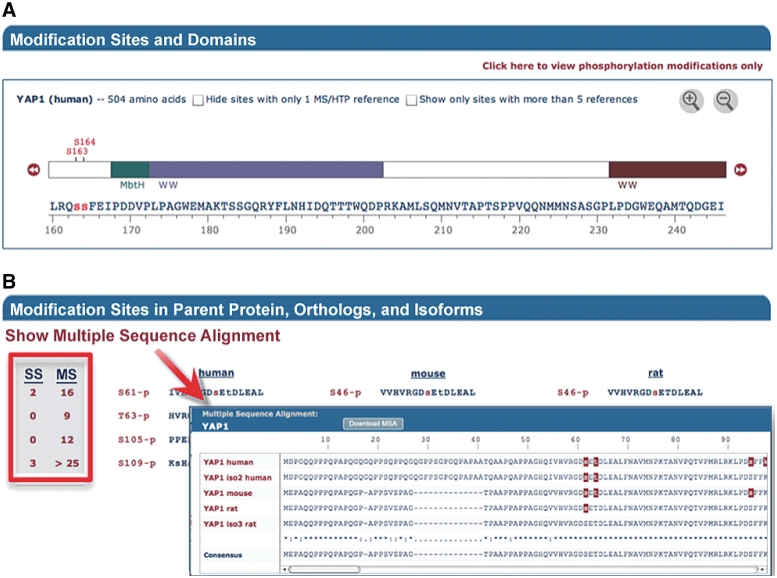
‘Protein Pages’ are divided into four sections. The topmost is the ‘Overview Section’, an example of which is shown in Figure 4 for the protein YAP1. It provides key information about the protein including a brief description, GO terms and links related to the specific protein. If the protein is included in a CST pathway, a link is provided to its online version (Figure 4 lower left). Clicking on the Open Viewer button will open the chosen PDB file in the Viewer window with the OpenAstexViewer (41) (Figure 4, lower right). The rendered structure includes labels on the modifiable amino acids that have been identified in PSP. The reactive side-chain groups (hydroxyl and ε-amino) are color coded by modification type. Scripts for rendering the structure with colored and labeled modifiable side chain groups in PyMOL and Chimera (18) can be downloaded from the upper right-hand corner of the Viewer window. The second section of the Protein Page (data not shown) lists modified residues and briefly summarizes the consequences of the modification on the protein's function or biological processes, with hyperlinks to the relevant Site Pages where more detail is available. The lower two sections of the YAP1 protein page are shown in Figure 5. The ‘Modification Sites and Domains’ section (Figure 5A) of the Protein Page provides a zoomable linear representation of the protein sequence, modification sites and Pfam-A domains. Purported transmembrane segments, extracted from UniPROT KB, are labeled with TM. Lower case letters represent modified residues in all sequences. The ‘Modification sites in parent protein, ortholog and isoforms’ section (Figure. 5B) provides a table of modification sites and flanking sequences. The two left most columns outlined in red show the total number of times the site was characterized using low-throughput site-specific methods (SS) or mass spectrometry (MS) across all species and isoforms; these numbers are linked to a Site Group page summarizing the references.
Figure 4.
Protein Page overview section for YAP1. (lower left) The linked Hippo Pathway from CST, with links to PSP proteins, can be opened and downloaded from this page. (lower right) Selected PDF files, in this case 3KYS (46), can be opened in the Viewer window with the OpenAstexViewer (41).
Figure 5.
Protein Page sections 3 and 4 for YAP1. (A) The Modification Sites and Domains section. (B) Modification Sites in Parent Protein, Orthologs, and Isoforms section. The two left most columns outlined in red show the total number of references using SS or MS to characterize the site group.
RESULTS AND DISCUSSION
Overlap of MS and LTP phosphorylation sites
The overlap of LTP with MS sites in PSP is 48% (5229 out of 10 727 LTP sites), considerably higher than the 17% (846 out of 5095 LTP sites) previously reported (20). The 2.6-fold higher overlap rate may reflect the larger sample size in the present article, which is based on 1 00 249 phosphorylation sites compared to 38 259 (20).
Location of various modification types within protein structures
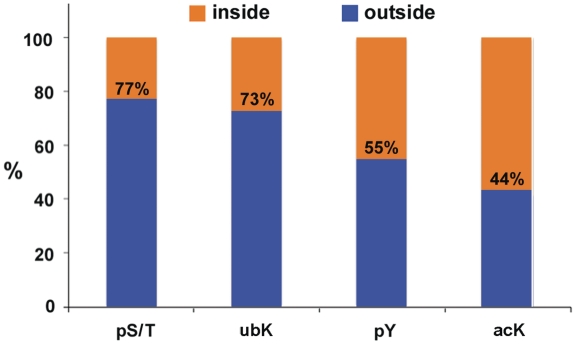
It has been observed that protein Ser/Thr phosphorylation occurs predominantly within intrinsically disordered protein regions (42,43), which are mainly outside of structural domains. In order to determine if other modification types are similarly situated, the location of pS/T modifications and three other modification types were scored as either inside or outside of Pfam-A domains (44). The results (Figure 6) show that 77% of phospho-Ser/Thr (pS/T) sites occur outside structured domains, a result consistent with previous observations (42,43). Two of three other modification types do not follow this pattern. A 23% of the pS/T sites occur within domains, while 45% of the phospho-Tyr sites and 56% of the acetyl-Lys (acK) sites fall within domains, a 2.0- and 2.4-fold increase, respectively. The distribution of ubiquitinyl-Lys (ubK) is similar to that of pS/T. The relative increase of the pY content within domains may reflect the conservation of these sites, many of which are regulatory, compared to pY sites located between domains, which may reflect the disproportionate loss of tyrosines that has occurred during metazoan evolution (45).
Figure 6.
The locations of four protein posttranslational modifications relative to Pfam-A domains. The locations of 4 modification types were scored as either inside or outside of Pfam-A domains. pS/T, phospho-Ser/-Thr; pY, phospho-Tyr; acK, acetyl-Lys; ubK, ubiquitinyl-Lys.
Using structural information to identify potentially important regulatory PTMs
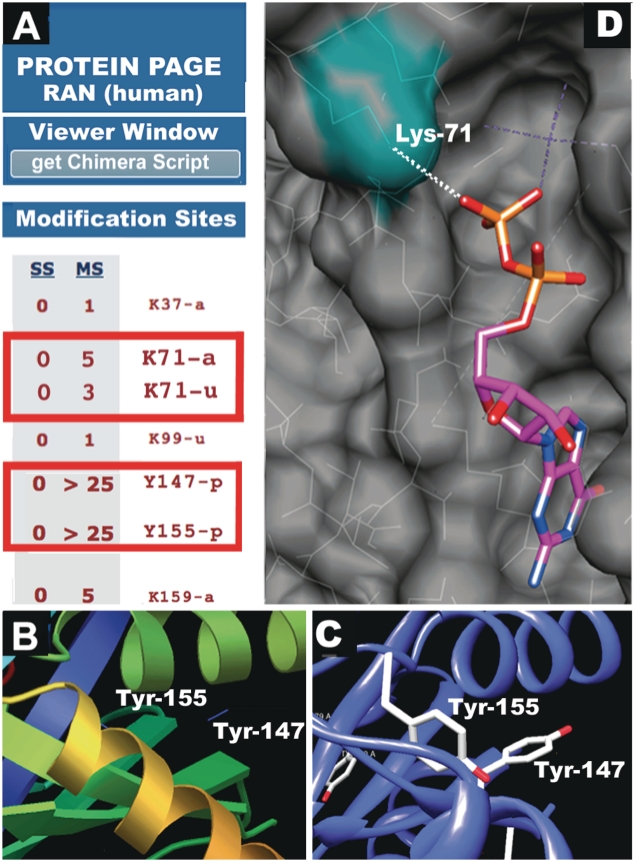
The molecular visualization capabilities of PSP, recently improved with the addition of downloadable Chimera and PyMOL scripts, are designed to enable researchers to quickly identify modification sites that lie within domains and to visualize the topology of the modified amino acid. The exact position of the modified site may change from that of the unmodified site in the structure but the first-order approximation afforded by this view allows for rapid identification of sites that may be functionally significant. The following examples using the small G protein RAN show how these tools might be useful when evaluating previously unreported sites that are found in PSP. The table of modification sites at the bottom of the RAN Protein Page (Figure 7A) shows two regions in the Ras domain that have elevated levels of PTMs: Lys-71 is acetylated and ubiquitinylated and Tyr-147 and -155 are phosphorylated The RAN PDB file 3CH5 was opened from the drop down menu in the molecular viewer section of the h. Figure 7B shows Tyr-147 and -155 in the default viewer. The ChimeraX script was downloaded from the upper right-hand side of the default viewer window and opened in Chimera (Figure 7C). Examining the same section as shown in Figure 7B, Tyr-147 and -155 appear to be interposed in a hydrophobic region between two α-helices, suggesting that the phosphorylation of these residues has the potential to open this region up and affect the function of RAN. Figure 7D shows that the ε-amino group of Lys-71 (green) is very close (1.29 Å) from O2B of the GDP molecule, suggesting that modification of this lysine may affect the binding of RAN to GDP (and GTP). Both of these possibilities present testable hypotheses, and show that PSP might be used to quickly identify potentially interesting functional roles of PTMs that lie within regions of a protein for which there are solved structures. This sort of analysis is limited to those 15 307 site groups that are located within the resolved regions of solved structures.
Figure 7.
The topology of modified residues from PSP on the small G protein RAN visualized with the PDB file for 3CH4. (A) The table of modification sites on the Ran (human) Protein Page. (B) PDB 3CH5 was opened in the Astex viewer. Tyr-147 and -155 are marked. (C) The PSP ChimeraX script was downloaded and opened in Chimera. Tyr-147 and -155 are marked, and the tyrosyl hydroxyls that can be phosphorylated are colored red. (D) Chimera model of 3CH4 in space-filling mode. The acetylated ε-amino group of Lys-71 is colored green.
CONCLUSION
As seen in Figure 1, we are in the middle of a period of rapid expansion of information about post-translationally modified sites discovered using both LTP and HTP technologies. Trying to keep up and to figure out how it all fits together is a fantastic challenge. LTP literature is regularly scanned for reports of new sites and for new information about established sites that contribute to our understanding of the site. The natural language processing software I2E from Linguamatics is helping us identify and preprocess the growing volume of LTP literature. Curation, however, is still done manually in-house. Curation of HTP literature possess the challenges of throughput and quality. We periodically upgrade the processing software to improve throughput and reevaluate the quality standards of sites that are already in PSP and those yet to be entered. PSP will continue to aggregate both LTP and HTP information in order to remain a comprehensive resource about PTMs for biomedical researchers. Hopefully PSP will prove useful to researchers trying to figure out how it all fits together.
FUNDING
National Institutes of Health (grant numbers R43GM065768 R44AA014848, R44CA126080). Funding for open access charge: Departmental budget.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization and Informatics at the University of California, San Francisco (supported by NIH P41 RR001081). Thanks to: Dr. Michael Comb for support of this project from its inception; Drs. Lew Cantley, Michael Yaffe and Tony Hunter for advice on its structure and content; Dr. Gael McGill for help and guidance with early web design and Chris Roberts and Chris Gang for their excellent computational contributions.
REFERENCES
- 1.Walsh CT, Garneau-Tsodikova S, Gatto GJ., Jr Protein posttranslational modifications: the chemistry of proteome diversifications. Angew Chem. Int. Ed. Engl. 2005;44:7342–7372. doi: 10.1002/anie.200501023. [DOI] [PubMed] [Google Scholar]
- 2.Mayer SE, Krebs EG. Studies on the phosphorylation and activation of skeletal muscle phosphorylase and phosphorylase kinase in vivo. J. Biol. Chem. 1970;245:3153–3160. [PubMed] [Google Scholar]
- 3.Varmus H, Hirai H, Morgan D, Kaplan J, Bishop JM. Function, location, and regulation of the src protein-tyrosine kinase. Princess Takamatsu Symp. 1989;20:63–70. [PubMed] [Google Scholar]
- 4.Sefton BM, Hunter T, Beemon K, Eckhart W. Evidence that the phosphorylation of tyrosine is essential for cellular transformation by Rous sarcoma virus. Cell. 1980;20:807–816. doi: 10.1016/0092-8674(80)90327-x. [DOI] [PubMed] [Google Scholar]
- 5.Pearson RB, Kemp BE. Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol. 1991;200:62–81. doi: 10.1016/0076-6879(91)00127-i. [DOI] [PubMed] [Google Scholar]
- 6.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: A bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 7.MacCoss MJ, Wu CC, Yates JR., III Probability-based validation of protein identifications using a modified SEQUEST algorithm. Anal. Chem. 2002;74:5593–5599. doi: 10.1021/ac025826t. [DOI] [PubMed] [Google Scholar]
- 8.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, Cohn MA, Cantley LC, Gygi SP. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl Acad. Sci. USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rush J, Moritz A, Lee KA, Guo A, Goss VL, Spek EJ, Zhang H, Zha XM, Polakiewicz RD, Comb MJ. Immunoaffinity profiling of tyrosine phosphorylation in cancer cells. Nat. Biotechnol. 2005;23:94–101. doi: 10.1038/nbt1046. [DOI] [PubMed] [Google Scholar]
- 10.Olsen JV, Blagoev B, Gnad F, Macek B, Kumar C, Mortensen P, Mann M. Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 2006;127:635–648. doi: 10.1016/j.cell.2006.09.026. [DOI] [PubMed] [Google Scholar]
- 11.Shi Y, Chan DW, Jung SY, Malovannaya A, Wang Y, Qin J. A data set of human endogenous protein ubiquitination sites. Mol. Cell Proteomics. 2011;10 doi: 10.1074/mcp.M110.002089. M110 002089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, Sowa ME, Rad R, Rush J, Comb MJ, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 15.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Z, Udeshi ND, Slawson C, Compton PD, Sakabe K, Cheung WD, Shabanowitz J, Hunt DF, Hart GW. Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci. Signal. 2010;3:ra2. doi: 10.1126/scisignal.2000526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang XJ, Seto E. Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol. Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 19.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dinkel H, Chica C, Via A, Gould CM, Jensen LJ, Gibson TJ, Diella F. Phospho.ELM: a database of phosphorylation sites–update 2011. Nucleic Acids Res. 2011;39:D261–D267. doi: 10.1093/nar/gkq1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gnad F, Gunawardena J, Mann M. PHOSIDA 2011: the posttranslational modification database. Nucleic Acids Res. 2011;39:D253–D260. doi: 10.1093/nar/gkq1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li H, Xing X, Ding G, Li Q, Wang C, Xie L, Zeng R, Li Y. SysPTM: a systematic resource for proteomic research on post-translational modifications. Mol. Cell Proteomics. 2009;8:1839–1849. doi: 10.1074/mcp.M900030-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Z, Cao J, Gao X, Zhou Y, Wen L, Yang X, Yao X, Ren J, Xue Y. CPLA 1.0: an integrated database of protein lysine acetylation. Nucleic Acids Res. 2011;39:D1029–D1034. doi: 10.1093/nar/gkq939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Consortium U. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Seal RL, Gordon SM, Lush MJ, Wright MW, Bruford EA. genenames.org: the HGNC resources in 2011. Nucleic Acids Res. 2011;39:D514–D519. doi: 10.1093/nar/gkq892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas CC, Deak M, Alessi DR, van Aalten DM. High-resolution structure of the pleckstrin homology domain of protein kinase b/akt bound to phosphatidylinositol (3,4,5)-trisphosphate. Curr. Biol. 2002;12:1256–1262. doi: 10.1016/s0960-9822(02)00972-7. [DOI] [PubMed] [Google Scholar]
- 31.Mirny LA, Gelfand MS. Using orthologous and paralogous proteins to identify specificity determining residues. Genome Biol. 2002;3:PREPRINT0002. doi: 10.1186/gb-2002-3-3-preprint0002. [DOI] [PubMed] [Google Scholar]
- 32.Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, et al. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–1203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- 33.Gu TL, Deng X, Huang F, Tucker M, Crosby K, Rimkunas V, Wang Y, Deng G, Zhu L, Tan Z, et al. Survey of tyrosine kinase signaling reveals ROS kinase fusions in human cholangiocarcinoma. PLoS One. 2011;6:e15640. doi: 10.1371/journal.pone.0015640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat. Biotechnol. 2006;24:1285–1292. doi: 10.1038/nbt1240. [DOI] [PubMed] [Google Scholar]
- 35.Bradshaw RA, Medzihradszky KF, Chalkley RJ. Protein PTMs: post-translational modifications or pesky trouble makers? J. Mass. Spectrom. 45:1095–1097. doi: 10.1002/jms.1786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D'Eustachio P, Schaefer C, Luciano J, et al. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vacic V, Iakoucheva LM, Radivojac P. Two Sample Logo: a graphical representation of the differences between two sets of sequence alignments. Bioinformatics. 2006;22:1536–1537. doi: 10.1093/bioinformatics/btl151. [DOI] [PubMed] [Google Scholar]
- 40.de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartshorn MJ. AstexViewer: a visualisation aid for structure-based drug design. J. Comput. Aided Mol. Des. 2002;16:871–881. doi: 10.1023/a:1023813504011. [DOI] [PubMed] [Google Scholar]
- 42.Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Collins MO, Yu L, Campuzano I, Grant SG, Choudhary JS. Phosphoproteomic analysis of the mouse brain cytosol reveals a predominance of protein phosphorylation in regions of intrinsic sequence disorder. Mol. Cell Proteomics. 2008;7:1331–1348. doi: 10.1074/mcp.M700564-MCP200. [DOI] [PubMed] [Google Scholar]
- 44.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan CS, Pasculescu A, Lim WA, Pawson T, Bader GD, Linding R. Positive selection of tyrosine loss in metazoan evolution. Science. 2009;325:1686–1688. doi: 10.1126/science.1174301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li Z, Zhao B, Wang P, Chen F, Dong Z, Yang H, Guan KL, Xu Y. Structural insights into the YAP and TEAD complex. Genes Dev. 2010;24:235–240. doi: 10.1101/gad.1865810. [DOI] [PMC free article] [PubMed] [Google Scholar]