Abstract
Legumes play a vital role in maintaining the nitrogen cycle of the biosphere. They conduct symbiotic nitrogen fixation through endosymbiotic relationships with bacteria in root nodules. However, this and other characteristics of legumes, including mycorrhization, compound leaf development and profuse secondary metabolism, are absent in the typical model plant Arabidopsis thaliana. We present LegumeIP (http://plantgrn.noble.org/LegumeIP/), an integrative database for comparative genomics and transcriptomics of model legumes, for studying gene function and genome evolution in legumes. LegumeIP compiles gene and gene family information, syntenic and phylogenetic context and tissue-specific transcriptomic profiles. The database holds the genomic sequences of three model legumes, Medicago truncatula, Glycine max and Lotus japonicus plus two reference plant species, A. thaliana and Populus trichocarpa, with annotations based on UniProt, InterProScan, Gene Ontology and the Kyoto Encyclopedia of Genes and Genomes databases. LegumeIP also contains large-scale microarray and RNA-Seq-based gene expression data. Our new database is capable of systematic synteny analysis across M. truncatula, G. max, L. japonicas and A. thaliana, as well as construction and phylogenetic analysis of gene families across the five hosted species. Finally, LegumeIP provides comprehensive search and visualization tools that enable flexible queries based on gene annotation, gene family, synteny and relative gene expression.
INTRODUCTION
Legumes have the ability to conduct symbiotic nitrogen fixation through endosymbiotic interactions with bacteria residing in root nodules. Thus, these plants play a vital role in maintaining the nitrogen cycle of the biosphere. Some legume species are also important resources for oils, fiber, fuel, lumber, medicine, chemicals and horticultural varieties.
In addition to root nodulation and nitrogen fixation symbiosis with rhizobia, legumes possess many unique features that are not found in the typical plant model Arabidopsis thaliana, including mycorrhization, compound leaf development, a protein-rich physiology, profuse secondary metabolism, secondary compounds with valuable health-promoting properties, glandular trichome development and border cells within roots. Therefore, legumes are considered as another important plant model for studying physiology, genomics, plant–microbe interaction, sustainable agriculture, food production, security and renewable bioenergy generation.
Legumes have traditionally been divided into three main subfamilies: caesalpinioids, mimosoids and papilionoids (1), which all derived from a common ancestor around 60 million years ago (2). The papilionoids mainly include two sister clades, phaseoleae and trifolieae, which constitute >60% of all papilionoids. Of the two sister clades, the former mostly consists mostly of tropical, herbaceous species such as the soybean Glycine max. The latter consists primarily of temperate species such as Medicago sativa, a species that is closely related to two model legumes, M. truncatula and L. japonicus.
Utilizing bacterial artificial chromosome (BAC)-by-BAC, whole-genome shotgun and second-generation sequencing approaches, the genomes of three legume species G. max (http://www.phytozome.net/soybean), L. japonicus (http://www.kazusa.or.jp/lotus) and M. truncatula (http://www.medicago.org/genome) have been sequenced recently (3–5). The results of these sequencing projects have become invaluable resources for legume research. Furthermore, large-scale gene expression profiling of L. japonicus (http://www.brics.dk/cgi-compbio/Niels/index.cgi), G. max (http://digbio.missouri.edu/soybean_atlas/) and M. truncatula (http://mtgea.noble.org/) have been also performed to characterize tens of thousands of genes in these species (6–8).
Comparative genomics and transcriptomics approaches have empowered gene discovery and gene functional characterization. For example, Libault et al. (9) systematically reviewed the identified transcription factors through comparative sequence analysis and expression data. Comparative genomics and transcriptomics, coupled with comprehensive gene annotation, gene family classification and phylogenetic analysis have also been used successfully to decipher biological processes that are unique to legumes, such as nodulation in response to rhizobial infection. For example, the MtHAP2.1, MtERN and LjNIN genes, which control nodule development, were identified by analysis of collinear relationships and gene expression profiles (10–12).
The Legume Information System (LIS) is a community portal that hosts a vast quantity of legume-related data, including gene sequences, transcript sequences such as expressed sequence tags (ESTs), genetic markers, literature and external links to multiple legume species (13). The LIS also provides useful tools such as orthologous gene and evolutionary event analysis, a chromosome visualization tool and synteny-view in a genome browser. However, due to the complexity of tandem duplication and genomic divergence, analysis of synteny or comparison of chromosome position only may overlook a large number of orthologous genes. A typical example is the flavonoid gene family (14). Isoflavonoids, a subset of this family (15), are unique to legume species, and most are involved in mediating host specificity within this plant. Nevertheless, only a few of these genes could be identified by collinear analysis since the tandem duplications that produced this family of flavonoids most likely occurred after the large-scale genome duplication events.
Combining the analysis of gene families, phylogenetic context and tissue-specific transcriptomic profiles is a powerful method for studying complex biological events (16). The online database PLAZA (17) integrates large-scale genomics data from several plant species for plant evolution research; however, a lack of gene expression data make it less effective in inferring species-specific gene function and evolutionary history, especially in legume species.
The recent publication of legume genomics and transcriptomics data has necessitated the development of a comprehensive genomics and transcriptomics database of model legumes. Here, we present LegumeIP, an integrative database for comparative genomics and transcriptomics of model legumes, for use in studying gene function and genome evolution in this important plant family. LegumeIP currently hosts large-scale genomics and transcriptomics data including the genome sequences of three model legumes (i.e. M. truncatula, G. max and L. japonicas) and two reference plant species (i.e. A. thaliana and Populus trichocarpa) with annotation based on Uniprot (18), InterProScan (19,20), Gene Ontology (GO) (21) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) (22) and encompassing 222 217 protein-coding gene sequences. LegumeIP also contains large-scale gene expression data compiled from 104 L. japonicas microarrays, 156 microarrays from the M. truncatula gene atlas database and 14 RNA-Seq-based gene expression data from G. max gene atlas database, with profiles on time-course experiments and different tissues including four common tissues namely nodule, flower, root and leaf. In addition, LegumeIP can perform systematic synteny analysis across M. truncatula, G. max, L. japonicas and A. thaliana, as well as construct the gene family and perform gene family-wide phylogenetic analysis across the five hosted plant species. Finally, LegumeIP can perform comprehensive search and visual representation to enable flexible queries based on gene annotation, gene family, synteny and relative gene expression.
LegumeIP is freely available at http://plantgrn.noble.org/LegumeIP/
DATABASE PRODUCTION
Data source and process
The protein-coding and amino acid sequences for M. truncatula, L. japonicas and G. max were obtained from http://www.medicago.org/genome/download/, ftp://ftp.kazusa.or.jp/pub/lotus/lotus_r2.5/ and ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v7.0/Gmax/annotation/, respectively. Data for the two outgroup reference species, A. thaliana and P. trichocarpa, were acquired from ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR10_genome_release/ and ftp://ftp.jgi-psf.org/pub/JGI_data/phytozome/v7.0/Ptrichocarpa/annotation/, respectively. In total, LegumeIP integrates 222 217 protein-coding gene sequences and 221 706 amino acid sequences.
Large-scale microarray and RNA-Seq-based gene expression data for M. truncatula, L. japonicas and G. max were obtained from http://mtgea.noble.org/, http://cgi-www.cs.au.dk/cgi-compbio/Niels/index.cgi and http://digbio.missouri.edu/soybean_atlas/, respectively (6–8,23). Gene expression data for L. japonicas are available only for version 1.0 genome sequences in the atlas database. Therefore, we remapped the Affymetrix L. japonicas GeneChip probesets using BLAST to align CDS sequences (Version 2.5) against the GeneChip probe set target sequences downloaded from the ArrayExpress (http://www.ebi.ac.uk/arrayexpress/) database (E-value ≤1e-10). Only the highest-scoring hit was selected. As the result, LegumeIP integrates data from 104 L. japonicas microarrays, 156 M. truncatula microarrays and 14 RNA-Seq-based gene expression profiles for G. max, with profiles on different tissues including four common tissues (nodule, flower, root and leaf) for all three model legume species. LegumeIP also integrates data from additional tissues and time-course experiments for individual species.
Comprehensive gene annotation
Genome sequences were annotated using a series of manually curated standard databases. The protein sequences were first searched against UniProt using BLASTP with a cutoff E-value ≤1e-04. The top five most meaningful query results were considered valid. This method was also used to annotate protein sequences against GO, KEGG, Transporter Classification Database (TCDB) (24) and Plant Transcription Factor Database (PlantTFDB) (25). Conserved domains between protein sequences were identified with InterProScan software using its default E-value cutoff thresholds (20).
Systematic synteny identification
Alignment of syntenic regions between non-legume and legume species is an effective approach for identifying patterns of evolutionary conservation and divergence across genomes. LegumeIP employs the DAGchainer program (26) to identify syntenic regions between M. truncatula, L. japonicas, G. max and A. thaliana. First, we performed an all-by-all BLASTN search with an E-value cutoff ≤1e-10 to identify intraspecies paralogous pairs and interspecies homologous pairs. Then, we applied DAGchainer (parameters Z = 12, D = 10, g = 1 and a = 5) to discover orthologous pairs of interspecies collinear regions. Parameters with -s and -i in DAGchainer were applied to identify collinear paralogous pairs in the same species. To all the identified homologous pairs within syntenic regions, we applied the F3x4 model from the PAML4.0 software package (27) to estimate the ratio of the number of non-synonymous substitutions/non-synonymous site (Ka) to the number of synonymous substitutions/synonymous site (Ks) (i.e. Ka/Ks).
Cross-species gene family and phylogenetic analysis
Due to the frequent occurrence of tandem duplication or sequence diversity, the order of orthologous genes within a collinear/syntenic region may have become disrupted during evolutionary development (28). To better study gene function and genome evolution, two groups of putative gene families were constructed in the five species based on protein-coding sequence similarity. This was accomplished by the TribeMCL (29) and OrthoMCL (30) clustering algorithms that are complementary to each other. Although TribeMCL outputted fewer gene families, each resultant family consisted of more member genes but with a higher false-positive rate that was likely due to the inherent nature of BLAST hits within the TribeMCL algorithm. In contrast, OrthoMCL yielded a large number of smaller gene families with a lower false-positive rate. To construct the TribeMCL gene families, we first performed an all-by-all BLASTP search of the protein-coding nucleotide sequences from the five plant species using an E-value ≤1e-10 as the cutoff threshold. TribeMCL (with default option I = 2.0) was then employed to delineate large gene families based on the BLAST results. We applied the OrthoMCL method to construct gene families with fewer member genes and a lower false-positive rate based on the same blast result. Application of both algorithms resulted in the grouping of 95.70% of 212 653 protein-coding genes into 12 166 TribeMCL gene families and 70.40% of all protein-coding genes grouped into 19 315 OrthoMCL gene families.
To construct phylogenetic trees for the gene families, multiple sequence alignment was first performed using the MUSCLE software (31). Unrooted trees were then created using PHYLIP software (including the seqboot, proml and consensus programs) with 100 bootstrap replications.
Analysis of large-scale gene expression data
To enable comparative transcriptomics analysis across the three model legume species, LegumeIP integrates microarray-based and RNA-Seq-based gene expression profiling data from four common tissues, namely nodule, root, flower and leaf, in its core expression table. Furthermore, large-scale gene expression profiles for additional tissues and time-course experiments were also included for individual species. The hosted gene expression profiling experiments are described in the Supplementary Materials and Methods. We used a rank-based method to normalize the gene expression data. Briefly, the genes identified by each microarray or RNA-Seq data set were first ranked from lowest to highest in terms of expression. The ranks were subsequently divided by the total number of expressed genes. Furthermore, the expression values from individual tissues were calculated by averaging the ranks of different experimental conditions applied to the same tissue.
Clustering of gene families from the three legume species was estimated based on the Pearson's correlation coefficient of normalized expression in the four tissues using the hierarchical clustering algorithm (32).
Database development
LegumeIP was developed using the Java and Groovy languages. The system runs on a Linux-based RESIN J2EE web server architecture using MySQL as its database management system. Circos software (33) and Gbrowse_syn, which is a Gbrowse-based synteny browser (34), were adopted for visualization of macro- and micro-syntenic relationships, respectively. The interactive phylogenetic tree is rendered by Archaeopteryx (35) and gene expression profiles are plotted using the OpenFlashChart package (http://teethgrinder.co.uk/open-flash-chart/). Gene cluster is depicted as heatmap in the format of a background-colored HTML table.
User-friendly web interface for data access
LegumeIP provides a comprehensive web interface for searching and exploring genes, gene families, syntenic regions and gene expression patterns. For example, through a simplified Keyword Gene Search interface, users can search LegumeIP by gene name, description, family and specific biological classification system, such as a GO term, InterProScan domain name, transporter family, transcription factor family, KEGG pathway or compound name. In the Advanced Gene Search page, more complex searching criteria can be used, including combinations of keywords for querying quantitative tissue-specific gene expression patterns. The search results are listed in a table with links for batch downloading, a detailed page of comprehensive gene annotations, plots depicting the transcriptomics profile if applicable and sequence information. Users can also navigate to corresponding synteny and gene family pages using the ‘TribeGroup’, ‘OrthoGroup’ and ‘Included gene in synteny’ links. The phylogenetic tree and gene cluster heatmap are provided in detail on the gene family page.
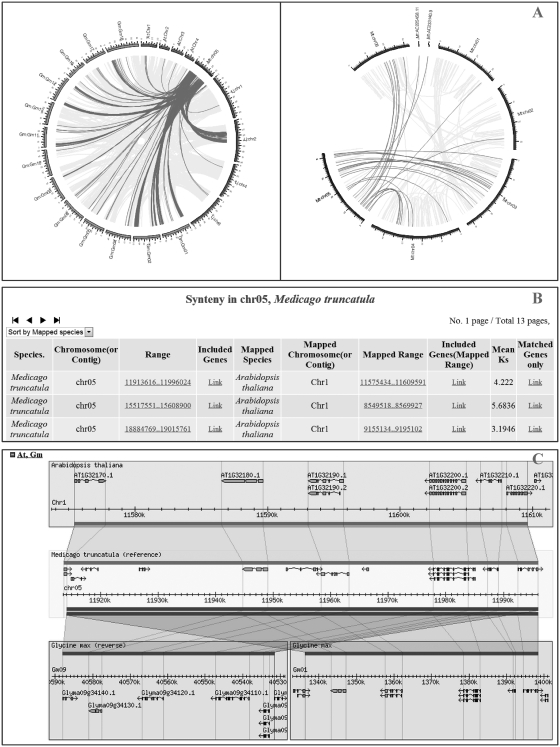
LegumeIP provides a simplified page to allow users to explore syntenic regions by chromosome or Contig ID. The macrosynteny outputs, including interspecies and intraspecies syntenic regions are represented as Circos maps (Figure 1A) and a summary table (Figure 1B) with links to detail pages, such as visualization of microsynteny in the GBrowse_syn module (Figure 1C) and a list of genes within the syntenic region.
Figure 1.
Web interfaces for (A) searching, (B) exploring and (C) visualizing macrosyntenys and microsyntenys.
LegumeIP also integrates BLAST search interfaces, allowing users to search homologous genes or protein sequences based on sequence similarity.
DEMONSTRATION OF THE UTILITY OF LEGUMEIP
Below we demonstrate the utility of LegumeIP in discovering and characterizing gene candidates in legumes. Additional examples are available in the Help page of the LegumeIP web server.
Mining UGT and p450 genes in M. truncatula with gene search tools
The UDP-glycosyltransferase (UGT) gene family (36), which is reportedly involved in the biogenesis of important secondary metabolites such as flavonoids, is highly enriched in the legume M. truncatula compared to A. thaliana. Therefore, much research on plant secondary metabolism has been focused on this family. Since these enzymes feature a conserved InterProScan domain, IPR002213, we used this domain ID as a keyword to search for UGT genes in the M. truncatula genome. This search yielded 351 genes as potential UGT candidates for further study.
CYP84 constitutes a family of cytochrome P450-dependent mono-oxygenases defined by ferulate 5-hydroxylase activity, which is reportedly mediates a plant defense mechanism (37). Thus, we searched for CYP84 genes in the M. truncatula genome using the keywords ‘ferulate’ and ‘5-hydroxylase’ and found 10 candidate genes. Microarray data showing detectable expression levels (i.e. higher than the 10th percentile in at least one of the four common tissues analyzed) were available. These genes likely function as P450-dependent mono-oxygenases. However, experimental validation is still required as many of the P450 subfamilies are quite similar in terms of both sequence and structure.
In both cases, users can batch download all of the candidate sequences by clicking on links located on the upper right-hand corner of the result pages.
Mining SymRK genes for symbiosis analysis in legumes
Symbiosis with rhizobia in the nodule is the source of nitrogen fixation in legumes. The leucine-rich repeat receptor kinases [also known as symbiosis receptor-like kinase (SymRK)] are reportedly involved in the signaling pathway that mediates early root response to bacterial and fungi infection in epidermal tissues of root nodules. These kinases also mediate the uptake of symbiotic bacteria and fungi into plant cells (38,39). In addition, SymRKs play an essential role in nodulation initiation. For example, SymRK in Lotus (previous gene name, CM0177.3340.r2.m), together with other Nod-factor genes, was reportedly involved in signaling that leads to epidermal calcium spikes (40).
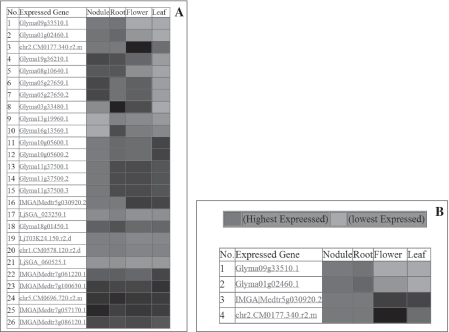
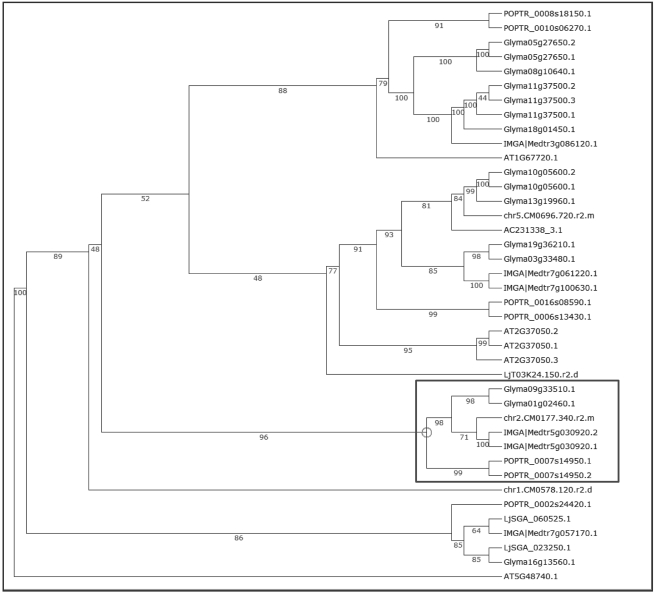
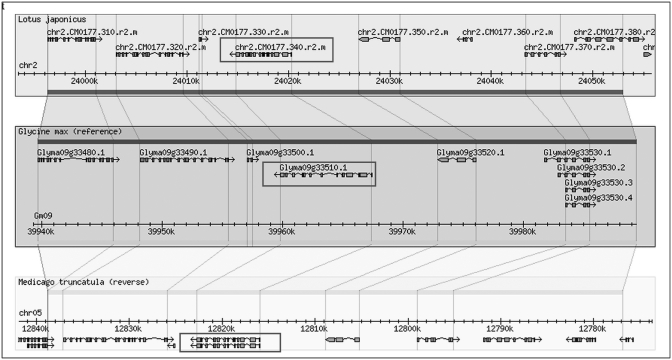
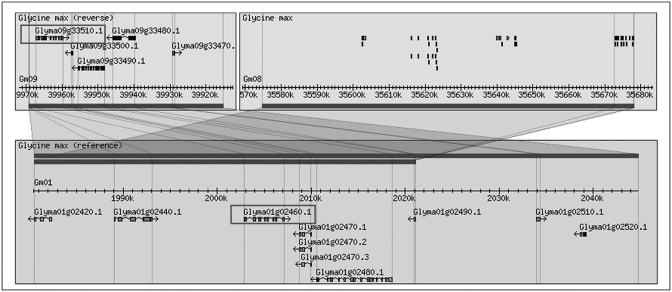
Using the keyword ‘SymRK’, we retrieved seven SymRK genes using LegumeIP. The results further indicated that all of these genes were assigned into one family, the TribeMCL00867 group, or according to the OrthoMCL method, the OrthoMCL07722 group. Corresponding probesets were available on the Medicago GeneChip for four of the seven genes, and each of these was highly expressed in nodule and root (Figure 2), suggesting their possible roles in nodule initiation. The reconstructed phylogenetic tree demonstrated that these genes most likely also belong to the same clade (Figure 3) and gene family, OrthoMCL07722. Synteny analysis further indicated their derivation from common ancestral genes (Figures 4 and 5). Analysis of the phylogenetic tree enabled us to identify two P. trichocarpa genes located in the same clade (Figure 3), suggesting that the function of SymRK genes may not be specific to only legume species and may be related to ectomycorrhiza symbiosis in P. trichocarpa. Altogether the results produced from LegumeIP provide additional support for the SymRK gene family as the common genetic basis of root nodule symbiosis (38,39).
Figure 2.
(A) Expression cluster displayed as heatmap for all expressed member genes in TribeMCL00867 and (B) OrthoMCL07722, which include SymRK genes.
Figure 3.
Phylogenetic tree of the TribeMCL00867 gene family.
Figure 4.
SymRK orthologous genes within a syntenic region common to L. japonicus, G. max and M. truncatula.
Figure 5.
SymRK paralogous genes within a syntenic region in G. max.
CONCLUSIONS AND FUTURE PERSPECTIVES
The rapid growth of legume-related genomics and transcriptomics data demands development of integrated databases and advanced bioinformatics analysis tools for not only efficiently managing, storing, retrieving and sharing data, but also for effectively integrating, analyzing and mining large volumes of highly complex information.
LegumeIP compiles data and related analytical tools such as comprehensive Uniprot-/InterProScan-/GO-/KEGG-based gene annotations, relative gene expression data, gene family classification and macrosynteny analysis. The data and analytical tools are thoughtfully organized to enable quick searches for genes of interest through user-friendly web interfaces. Transcriptomics profiling, synteny and phylogenetic analysis are powerful tools that can be used to discover gene function, including identifying genes associated with the symbiotic nitrogen fixation process in legumes. Moreover, synteny and phylogenetic analysis are helpful in better understanding the evolution of terrestrial plants such as legumes. All of these features demonstrate the enormous potential of LegumeIP as a vital tool in studying fundamental biological questions.
We are committed to continually improving LegumeIP. Additional microarray- and RNA-Seq-based gene expression data will be populated into the LegumeIP database as it is made available in public repositories. In addition, we will integrate large-scale genomic and EST sequences from different sources, such as the Medicago Hapmap project (http://www.medicagohapmap.org/).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Material and Methods.
FUNDING
National Science Foundation (Grant ABI-0960897 to P.X.Z.) and Samuel Roberts Noble Foundation. Funding for open access charge: National Science Foundation; Samuel Roberts Noble Foundation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to Dr Firoz Ahmed for the critical reading of this article and for providing valuable comments.
REFERENCES
- 1.Doyle JJ, Luckow MA. The rest of the iceberg. Legume diversity and evolution in a phylogenetic context. Plant Physiol. 2003;131:900–910. doi: 10.1104/pp.102.018150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the tertiary. Syst. Biol. 2005;54:575–594. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- 3.Schmutz J, Cannon SB, Schlueter J, Ma J, Mitros T, Nelson W, Hyten DL, Song Q, Thelen JJ, Cheng J, et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 4.Sato S, Nakamura Y, Kaneko T, Asamizu E, Kato T, Nakao M, Sasamoto S, Watanabe A, Ono A, Kawashima K, et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008;15:227–239. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young ND, Debellé F, Oldroyd GED, Geurts R, Cannon SB, Udvardi MK, Benedito VA, Mayer KFX, Gouzy J, Schoof H, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011 doi: 10.1038/nature10625. doi:10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hogslund N, Radutoiu S, Krusell L, Voroshilova V, Hannah MA, Goffard N, Sanchez DH, Lippold F, Ott T, Sato S, et al. Dissection of symbiosis and organ development by integrated transcriptome analysis of lotus japonicus mutant and wild-type plants. PLoS One. 2009;4:e6556. doi: 10.1371/journal.pone.0006556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Libault M, Farmer A, Joshi T, Takahashi K, Langley RJ, Franklin LD, He J, Xu D, May G, Stacey G. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010;63:86–99. doi: 10.1111/j.1365-313X.2010.04222.x. [DOI] [PubMed] [Google Scholar]
- 8.Benedito VA, Torres-Jerez I, Murray JD, Andriankaja A, Allen S, Kakar K, Wandrey M, Verdier J, Zuber H, Ott T, et al. A gene expression atlas of the model legume Medicago truncatula. Plant J. 2008;55:504–513. doi: 10.1111/j.1365-313X.2008.03519.x. [DOI] [PubMed] [Google Scholar]
- 9.Libault M, Joshi T, Benedito VA, Xu D, Udvardi MK, Stacey G. Legume transcription factor genes: what makes legumes so special? Plant Physiol. 2009;151:991–1001. doi: 10.1104/pp.109.144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schauser L, Roussis A, Stiller J, Stougaard J. A plant regulator controlling development of symbiotic root nodules. Nature. 1999;402:191–195. doi: 10.1038/46058. [DOI] [PubMed] [Google Scholar]
- 11.Combier JP, Frugier F, de Billy F, Boualem A, El-Yahyaoui F, Moreau S, Vernie T, Ott T, Gamas P, Crespi M, et al. MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes. Dev. 2006;20:3084–3088. doi: 10.1101/gad.402806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Middleton PH, Jakab J, Penmetsa RV, Starker CG, Doll J, Kalo P, Prabhu R, Marsh JF, Mitra RM, Kereszt A, et al. An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell. 2007;19:1221–1234. doi: 10.1105/tpc.106.048264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gonzales MD, Archuleta E, Farmer A, Gajendran K, Grant D, Shoemaker R, Beavis WD, Waugh ME. The Legume Information System (LIS): an integrated information resource for comparative legume biology. Nucleic Acids Res. 2005;33:D660–D665. doi: 10.1093/nar/gki128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winkel-Shirley B. Flavonoid Biosynthesis: a colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol. 2001;126:485–493. doi: 10.1104/pp.126.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaufman PB, Duke JA, Brielmann H, Boik J, Hoyt JE. A comparative survey of leguminous plants as sources of the isoflavones, genistein and daidzein: implications for human nutrition and health. J Altern. Complement. Med. 1997;3:7–12. doi: 10.1089/acm.1997.3.7. [DOI] [PubMed] [Google Scholar]
- 16.Cannon SB, Ilut D, Farmer AD, Maki SL, May GD, Singer SR, Doyle JJ. Polyploidy did not predate the evolution of nodulation in all legumes. PLoS One. 2010;5:e11630. doi: 10.1371/journal.pone.0011630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Proost S, van Bel M, Sterck L, Billiau K, Van Parys T, Van de Peer Y, Vandepoele K. PLAZA: a comparative genomics resource to study gene and genome evolution in plants. Plant Cell. 2009;21:3718–3731. doi: 10.1105/tpc.109.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O'Donovan C, Martin MJ, Gattiker A, Gasteiger E, Bairoch A, Apweiler R. High-quality protein knowledge resource: SWISS-PROT and TrEMBL. Brief Bioinform. 2002;3:275–284. doi: 10.1093/bib/3.3.275. [DOI] [PubMed] [Google Scholar]
- 19.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene Ontology: tool for the unification of biology. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He J, Benedito VA, Wang M, Murray JD, Zhao PX, Tang Y, Udvardi MK. The Medicago truncatula gene expression atlas web server. BMC Bioinformatics. 2009;10:441. doi: 10.1186/1471-2105-10-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saier MHJ, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo A-Y, Chen X, Gao G, Zhang H, Zhu Q-H, Liu X-C, Zhong Y-F, Gu X, He K, Luo J. PlantTFDB: a comprehensive plant transcription factor database. Nucleic Acids Res. 2008;36:D966–D969. doi: 10.1093/nar/gkm841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas BJ, Delcher AL, Wortman JR, Salzberg SL. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics. 2004;20:3643–3646. doi: 10.1093/bioinformatics/bth397. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 28.Zhu H, Kim DJ, Baek JM, Choi HK, Ellis LC, Kuester H, McCombie WR, Peng HM, Cook DR. Syntenic relationships between Medicago truncatula and Arabidopsis reveal extensive divergence of genome organization. Plant Physiol. 2003;131:1018–1026. doi: 10.1104/pp.102.016436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enright AJ, van Dongen S, Ouzounis CA. An efficient algorithm for large-scale detection of protein families. Nucleic Acids Res. 2002;30:1575–1584. doi: 10.1093/nar/30.7.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krzywinski M, Schein J, Birol I, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKay SJ, Vergara IA, Stajich JE. Using the Generic Synteny Browser (GBrowse_syn) Curr. Protoc. Bioinformatics. 2010 doi: 10.1002/0471250953.bi0912s31. Chapter 9, Unit 9.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han MV, Zmasek CM. phyloXML: XML for evolutionary biology and comparative genomics. BMC Bioinformatics. 2009;10:356. doi: 10.1186/1471-2105-10-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lim E-K, Bowles DJ. A class of plant glycosyltransferases involved in cellular homeostasis. EMBO J. 2004;23:2915–2922. doi: 10.1038/sj.emboj.7600295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Meyer K, Cusumano JC, Somerville C, Chapple CC. Ferulate-5-hydroxylase from Arabidopsis thaliana defines a new family of cytochrome P450-dependent monooxygenases. Proc. Natl Acad. Sci. USA. 1996;93:6869–6874. doi: 10.1073/pnas.93.14.6869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Markmann K, Giczey G, Parniske M. Functional adaptation of a plant receptor-kinase paved the way for the evolution of intracellular root symbioses with bacteria. PLoS Biol. 2008;6:e68. doi: 10.1371/journal.pbio.0060068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gherbi H, Markmann K, Svistoonoff S, Estevan J, Autran D, Giczey G, Auguy F, Peret B, Laplaze L, Franche C, et al. SymRK defines a common genetic basis for plant root endosymbioses with arbuscular mycorrhiza fungi, rhizobia, and Frankiabacteria. Proc. Natl Acad. Sci. USA. 2008;105:4928–4932. doi: 10.1073/pnas.0710618105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kosuta S, Held M, Hossain MS, Morieri G, Macgillivary A, Johansen C, Antolin-Llovera M, Parniske M, Oldroyd GE, Downie AJ, et al. Lotus japonicus symRK-14 uncouples the cortical and epidermal symbiotic program. Plant J. 2011;67:929–940. doi: 10.1111/j.1365-313X.2011.04645.x. [DOI] [PubMed] [Google Scholar]