Abstract
The BioSample Database (http://www.ebi.ac.uk/biosamples) is a new database at EBI that stores information about biological samples used in molecular experiments, such as sequencing, gene expression or proteomics. The goals of the BioSample Database include: (i) recording and linking of sample information consistently within EBI databases such as ENA, ArrayExpress and PRIDE; (ii) minimizing data entry efforts for EBI database submitters by enabling submitting sample descriptions once and referencing them later in data submissions to assay databases and (iii) supporting cross database queries by sample characteristics. Each sample in the database is assigned an accession number. The database includes a growing set of reference samples, such as cell lines, which are repeatedly used in experiments and can be easily referenced from any database by their accession numbers. Accession numbers for the reference samples will be exchanged with a similar database at NCBI. The samples in the database can be queried by their attributes, such as sample types, disease names or sample providers. A simple tab-delimited format facilitates submissions of sample information to the database, initially via email to biosamples@ebi.ac.uk
INTRODUCTION
Biological samples are now routinely assayed by various high-throughput molecular technologies, such as microarrays, new generation sequencing or mass spectroscopy. Many data resources at the European Bioinformatics Institute (EBI), such as the archive of functional genomics data ArrayExpress (1), the European Nucleotide Archive (ENA) (2), the Proteomics Identification Database PRIDE (3) and the European Genome-phenome Archive (EGA) capture and represent information about samples linked to the (molecular) data they store. The same sample can be assayed by several technologies; for instance, cancer samples are often genotyped and profiled for DNA methylation and gene expression. Samples may have a relationship between them, for instance in cancer profiling the DNA of a tumour sample is sometimes compared to the DNA obtained from the tumour periphery or blood of the same individual. To interpret data from such experiments, it is important to know the essential sample attributes as well as the relationship between different samples and their sources. The attributes may specify the material sampled, the site—organs, tissues and phenotypic information, including disease states. We refer to all such metadata as sample data (or sample information).
Most bioinformatics resources will record sample data in the future, as molecular profiling has now moved from creating reference datasets to profiling individuals and specific conditions. Samples are often collected at one site and then distributed to several remote sites, each for a specific type of analysis. Some reference samples, such as standard cell lines, are distributed commercially and reused widely. Therefore it is becoming advantageous to record sample information in a separate dedicated database, which then can link out to the assay data stored about a specific sample in the appropriate assay databases.
This has led the EBI to establish a new database: the BioSamples Database (BioSD). The main goals of this database are:
to record and manage sample information consistently within the EBI and to link sample information to assay data across multiple resources;
to minimize data entry efforts for the user, in particular, to enable submissions of sample descriptions only once and reference them later from other databases and submissions;
to support cross database sample queries by sample description; and
to build a continuously growing set of consistently annotated samples that are repeatedly used in experiments and can be easily referenced from any of the databases within the EBI and externally. These are termed reference layer samples.
To achieve these goals, we assign stable accession numbers to samples in BioSD. Moreover, we have agreed on a common accessioning system and sample data exchange with the National Center for Biotechnology Information (NCBI), which is developing a similar database (http://www.ebi.ac.uk/biosamples/documents/BioSampleDB_EBI_NCBI.pdf). All the reference layer samples will be exchanged, accessioned in a coordinated way and made accessible from both NCBI and EBI databases.
The BioSample Database is the culmination of previous experience in recording and storing project and domain specific sample data EBI in the last 10 years. ArrayExpress (4) was the first database at EBI to deal with complex sample annotation for high-throughput gene expression data sets from 2002, other EBI molecular assay databases have made similar efforts since. Jointly with collaborators, we also developed the sample information management system PASSIM (5) for pre-registering and annotating samples collected for specific bio-medical genomics projects where the sample annotation was standardized, known in advance and tightly controlled. This system is still used for sample annotation and tracking in a number of projects including the International Cancer Genomics Project on kidney cancer CAGEKID. More recently, projects such as ISA Infrastructure (6), have developed resources that can be used to store data and sample annotations for multi-omics projects in a study centric manner. The Sample Availability System (SAIL) (7) was developed with the purpose of integrating sample information across BioBank collections.
A centralized sample repository such as BioSD must be flexible enough to capture any biological sample description for samples of a priori unknown types. Samples can have complex links and may be grouped in many ways; critically these links and groups may be unknown in advance and may not be related to any particular project or study at the point of submission. Existing samples may be combined into new studies, and various meta-analyses may be performed. The data resulting from such studies need to be linked to samples when the data is submitted at a later date. It must be made easy for biologists and project owners to pre-register samples, enter available sample information and obtain accession numbers. Information about samples may be incomplete at the time of submission and expanded or corrected incrementally. Since the assay databases at the EBI already hold large numbers of sample records, BioSD has to be able to deal with this data and scale in the future to many more.
The BioSD implementation supports these requirements. Samples can be submitted in a simple tabular format, called SampleTab, and the database allows for user-driven sample registration and submission as well as incorporating data both from external reference collections (e.g. cell lines) and from a number of assay databases at the EBI including ENA, ArrayExpress and PRIDE; it already contains sample data from over a million samples. BioSD can also act as the principal repository of sample data for future assay databases which may prefer not to store sample information locally, e.g. the database of Genetic Variation DGVa at the EBI (8). Centralization of sample information in this way allows consistency checking of annotations, encourages use of common terminologies, e.g. Experimental Factor Ontology (9) and provides a single query portal for sample related data. A simple query interface allows the user to query all samples in BioSD by various properties or attributes and to navigate to assay databases.
BioSD DATA MODEL AND IMPLEMENTATION
Sample data are represented as two types of objects: samples and sample groups. Rather than develop multiple different types of object to represent all possible sample types (individual, blood, biopsy, cell line, mouse strain, etc.), we model a generic sample and use attributes to define types. A sample is an identifiable object to which annotation such as species, disease information or cell type is attached. Samples can be derived-from other samples. For example, an individual can be represented as a typed sample with sex, age and ethnicity information. A blood sample obtained at a specific time from that individual would be represented as a separate sample linked to the individual through the derived-from relationship. Other types of relationships, such as pedigree relationships between individuals, may also be recorded. Multiple samples can be asserted to refer to aliquots of the same physical material. These are modelled as multiple sample objects in the database, and we establish equivalence relationships between them. This allows provenance information to be recorded about the equivalence relationships, as well as incremental addition of information to a particular sample without creating information ownership conflicts.
In most cases samples are naturally grouped, for instance, cell lines of a specific collection, e.g. the National Institute of Aging, or the samples related to a publication or project. Samples in the same group are typically annotated consistently, i.e. the same attributes are provided for all (or at least most samples), and the same terms are used to annotate all the samples in the group. This is not necessarily true for samples that belong to different groups—samples representing human subjects or bacterial cell cultures will have largely different attributes, and we cannot automatically assume that attributes of the same name in different sample groups has exactly the same meaning or use the same terminology. Logically, a sample may belong to more than one group.
Use of groups enables batch submissions. Assay databases already commonly group samples, and queries can return individual samples and related samples within a group.
Data to populate the BioSD comes from three different types of sources:
Samples submitted directly to BioSD for referencing in later data submissions to assay databases. For example, commercial cell lines, or samples used in large analysis projects such as ENCODE. We propose a format called SampleTab to be used for this route (see below).
Sample data imported from assay databases (termed assay samples). For existing assay databases, the sample information is usually also retained in the respective assay database, however new assay databases at EBI may store sample information only in the BioSD.
Data exchange of reference samples submitted to NCBI.
In many cases, there is a one-to-one relationship between a submission and a sample group for samples submitted to BioSD directly. A curated subset of samples acquired this way (alongside with those exchanged with the NCBI) constitutes the reference sample set. We are also actively working with standard sample collections to populate the database. Some samples acquired through route 2 may also be included in the reference set after curation. Note that route 1 supports the submission of samples belonging to coordinated multi-omics studies—the samples are submitted to BioSD and then referenced from the respective assay databases.
Sample groups are also used to provide information in scenarios where it is not possible to release detailed information about specific samples. For example, it may be known that a group of human samples have age, sex and birth date available, but for ethical reasons these details cannot be provided. However, summary information at the group level can be provided within ethical guidelines, e.g. age is between 18 and 30 years. Similarly, for some toxicology data pooling of individual samples has the consequence that only mean data per group of samples is available.
Finally, using the group concept allows us for provision of sample group context to support queries. For instance, HeLa derived samples are used in assays and reference collections. Provision of group level information such as ‘Coriell catalogue’ or ‘ArrayExpress experiment’ gives context to query results when many hits appear.
The BioSample Database implementation was designed to accommodate highly variable sample descriptions and to be flexible enough to support future changes without large system modifications such as schema changes in RDBMS. The core of BioSD is a custom graph-based data engine that manages information as objects with arbitrary sets of attributes attached, and which can be linked by defined relationships such as derived-from or equivalent-to. The data engine includes a semantic description of loaded data such as types of objects, possible attributes and relationships, and rules that allow objects to have associations to attributes or relationships. Therefore, the data model is flexible, easy to extend and edit, enabling us to focus on optimal data organization for our query and data representation use cases, rather than conforming to existing data constraints from multiple external databases.
Using a semantically annotated graph for data description allows us to enrich information by inferring new relationships between objects, e.g. sample equivalence, pedigree relations and sample similarity. Data indexing and search services are implemented that select and process information from the object graph. The most important is a full text index that allows users to find samples and groups according to their annotations. BioSD also supports tag-based search when user can select information according to some pre-defined tags used to group samples by criteria such as data source or related projects.
BioSD FILE FORMAT: SampleTab
We have developed a file format termed SampleTab to represent information about BioSamples. This is aimed primarily for use by biologists, is human readable, suitable for data exchange, and was inspired by spreadsheet-like tab-separated formats such as MAGE-TAB (10) and ISA-TAB (11). Each SampleTab file describes samples as a collection of attribute-value pairs. In addition, each file contains information about the provenance of both the sample material and the data describing the samples. A full description of the SampleTab file and examples for different sample types are available (http://www.ebi.ac.uk/microarray-srv/biosd/static/st.html), therefore only a brief summary is provided here.
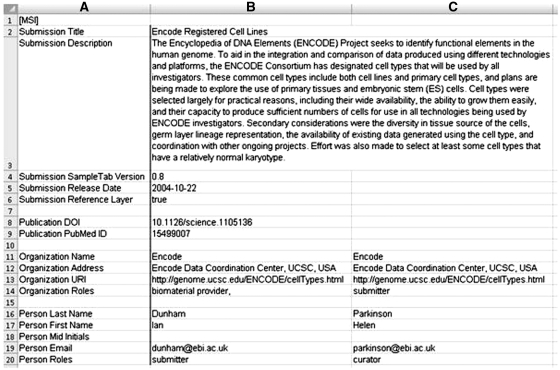
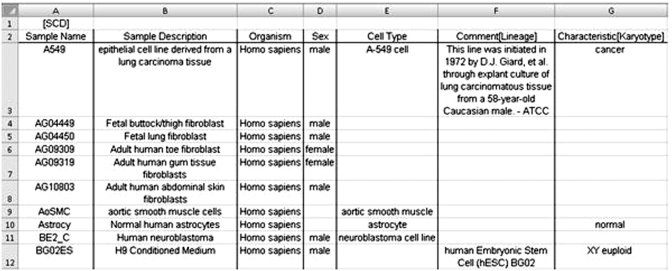
A SampleTab file is composed of two parts—a Meta-Sample Information (MSI) section and a Sample Characteristics Description (SCD) section. In a completed SampleTab file, the start of these sections are indicated by lines ‘[MSI]’ and ‘[SCD]’ respectively, but in a working copy they may be stored as separate spreadsheets in a workbook. An example of MSI and SCD sections is given in Figures 1 and 2, respectively.
Figure 1.
Example of the MSI section of a SampleTab file.
Figure 2.
Example of the SCD section of a SampleTab file.
The MSI section of a SampleTab file has row-based formatting where the first column consists of attributes describing four categories of information. These are: the BioSD submission, any associated publications, organizations and contacts. At a minimum, the following must be included: Submission Title, Submission SampleTab Version and either an organization or individual email address for contact.
In the SCD section, there is one header row containing attribute names. Each subsequent row represents a sample (or several samples derived from each other). Not every sample has to have a value for each attribute, for example, where no data are available (e.g. Sex of AsSMC and Astrocy samples in Figure 2). As a minimum, each sample must have a ‘Sample Name’. It is expected that almost all submitted samples will contain an ‘Organism’ attribute specifying the species, though for some data this may not be applicable (e.g. meta-genomic samples). Most samples will also contain a ‘MaterialType’ attribute—e.g. purified DNA, cell line, blood sample. We encourage the submitters to provide additional information, such as collection location, genetic modifications. It is also possible to encode relationships between the samples, such as derived from relationship between individuals and blood samples taken from them.
We do not seek to specify what information must be provided in SampleTab files based on a priori assumptions of the data; the format and the process of submission remain flexible to accept the data as it exists, as it may change in the future as standards are developed and applied. Therefore, submissions in SampleTab format may provide any number of additional columns labelled as either characteristics of the sample or comments about the sample. This can be seen in Figure 2 in the ‘Characteristic[Karyotype]’ column and ‘Comment[Lineage]’columns. Through this mechanism, submitters can capture information relevant to them in terminology they are familiar with, without being required to understand lengthy and technical specification documents.
Submission to BioSD is via email to biosamples@ebi.ac.uk using submission templates. Additional submission tools and routes are in development and will be released as open source applications. Pre-submission enquiries and data retrieval queries can also be directed to that address. Managed format extensions and subsequent versions of SampleTab will be available to support the future needs of submitters and for data exchange. We welcome comments and feedback on the SampleTab format.
BioSD QUERY INTERFACE, APIs AND CONTENT
BioSD contents can be browsed or queried by sample or sample group attributes, such as ‘blood’, ‘human’, ‘cancer’, ‘ENCODE’. The user interface follows the group/sample concept and represents search results as a list of groups that match the query criteria. For querying a common search engine-like syntax is used; users can enter a combination of keywords of interest. Logical expressions with operations like AND, NOT are also supported. Search results can be restricted by hits in groups, samples, attributes names, attribute values and any combination of the above, and by source: assay databases, or reference layer samples.
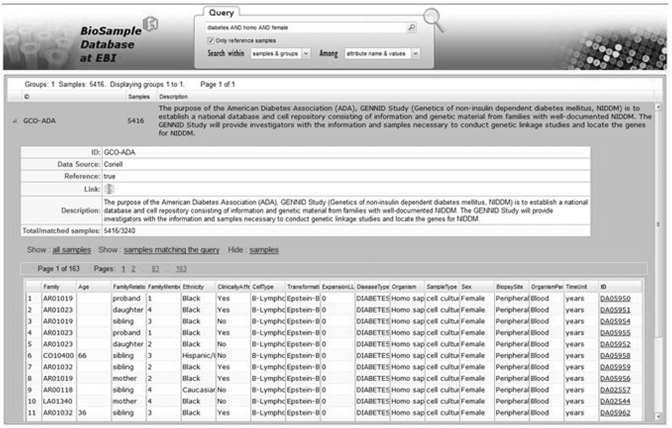
Every group record in the search result list can be expanded to provide more detailed information including: contacts, publications, affiliations. In addition to the group description, the list of samples in the group is shown (see Figure 3). Each row corresponds to one sample, and each column denotes one sample attribute. Users can choose to view the complete sample group, or the subset of samples matching the input query.
Figure 3.
Example of search results in BioSD web interface.
By the end of 2011, the BioSD will contain over 1 million samples from reference collections and EBI assay databases including ArrayExpress [including GEO database exchanged data (12)] and the SRA component of the European Nucleotide Archive (2). An automated pipeline system was constructed to extract, parse and load data from each source via existing database APIs, or from file downloads where no suitable API was available. For example, International Mouse Strain Repository (IMSR) data was obtained from the tab-separated files available through http://www.findmice.org/reportlist.jsp. For each of these diverse sources, custom format conversion software was developed to generated SampleTab format. Further processing steps assign accessions to samples and to groups, combine samples into submissions, ensure controlled vocabulary and literature references are valid.
FUTURE
BioSD already contains information about substantial number of reference samples that are routinely used in functional genomics experiments. We encourage the scientific community to reference these samples by their accession numbers, in particular, when data obtained by assaying them are submitted to any of the EBI assay databases. If necessary or desired, additional information about the samples can be added. We will work with all the EBI assay databases to make sure that accessing and referencing existing samples in BioSD is simple. As some assay databases also hold sample information locally, we will establish a system that automatically pushes requested sample information from BioSD into the respective assay database. There will also be a mechanism for handling coordinated multi-omics submissions across assay databases at the EBI.
One of the tasks for BioSD is to establish submission modification tools that allow the submitters to add or edit information about existing samples easily. Ensuring that the sample information in BioSD is consistent and updated is a non-trivial issue. Many of the sources used do not expose an API with updates by type. Instead, we periodically re-parse all the source information, compare with information previously loaded into the BioSample Database, and update where appropriate. Improvement of existing APIs will make this process considerably easier.
We will continue to work with the reference sample collection owners to populate the BioSD with sample information. Online submission tools will be developed to make SampleTab submissions easier for direct submitters. The reference layer will also be gradually expanded through the curation of sample information present in the EBI assay databases where these samples fulfil the reference layer criteria developed jointly with the NCBI. All the reference samples will be exchanged with the BioSample Database at NCBI, and the information about these will be held in both databases.
It is possible to navigate from samples and groups in BioSD to relevant assays in assay databases to retrieve the assay data by following the hyperlink. Some databases (including ArrayExpress) currently do not accession individual samples, which makes it non-straight forward to create and maintain links to individual assays from individual samples in BioSD. Sample links have been currently implemented for ENA and PRIDE with group links for ENA and ArrayExpress. To make the BioSD database more useful, in future hyperlinks from individual samples in BioSD to assay data in all assay databases will be provided.
A controlled access mechanism allowing the users to keep their sample descriptions private either for later release (e.g. after a publication), or to enable restricted access compliant with ethical requirements is under test.
The GUI will be further developed to enable more sophisticated queries, filtering of existing search results, improved layout and information download. Query power will be improved by using the Experimental Factor Ontology (EFO) (9) based query expansion; for instance, a search for ‘cancer’ would match all the subtypes and synonyms of cancer, such as ‘carcinoma’ and ‘malignant neoplasia’. In addition, as the user-base of the BioSamples Database expands and diversifies, we will conduct user experience studies to determine other areas for improvement.
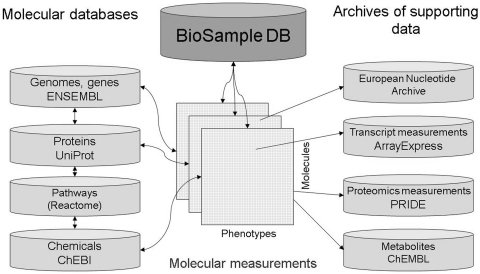
In future, BioSD will become the central location where all information about biological samples at the EBI (see Figure 4) are stored and referenced from other relevant databases within the EBI, as well as externally, and where such information can be easily queried and discovered.
Figure 4.
Overview of future integration between BioSD and other EBI databases.
FUNDING
All the authors and the majority of the BioSD development were primarily funded from the EMBL core budget provided by the EMBL member countries with contributions from the European Commission grants CAGEKID (HEALTH-F4-2010-241669) and ENGAGE (HEALTH-F4-2007-201413 from the European Commission FP7 program). Funding for open access charge: EMBL Core Funds.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank our NBCI colleagues: Tanya Barrett, Steve Sherry and Jim Ostell for fruitful discussions and for sharing their XML schema with us. We benefitted greatly from experience with, and discussions on PASSIM, BII/ISA projects and SAIL, in particular, with Juris Viksna, Maria Krestyaninova, Susanna Assunta Sansone and Philippe Rocca-Serra. We also had fruitful discussions with Gramene, 1000 Genomes and the Encode projects about optimal access models and data representation needs for their respective communities and species. We thank many of the EBI staff, in particular, James Malone, Tony Burdett, Chao Pang, Ilkka Lappalainen, Lisa Skipper, Attila Csordas, Chris Hunter, Phil Jones, Paula de Matos, Henning Hermjakob, Sarah Hunter, John Overington, Christoph Steinbeck, Paul Flicek, Ewan Birney and Graham Cameron for discussion on BioSD applications, use cases and implementation.
REFERENCES
- 1.Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Holloway E, et al. ArrayExpress update–an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;39(Database issue):D1002–D1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leinonen R, Akhtar R, Birney E, Bower L, Cerdeno-Tárraga A, Cheng Y, Cleland I, Faruque N, Goodgame N, Gibson R, et al. The European nucleotide archive. Nucleic Acids Res. 2011;39(Database issue):D28–D31. doi: 10.1093/nar/gkq967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vizcaíno JA, Côté R, Reisinger F, Barsnes H, Foster JM, Rameseder J, Hermjakob H, Martens L. The proteomics identifications database: 2010 update. Nucleic Acids Res. 2010;38(Database issue):D736–D742. doi: 10.1093/nar/gkp964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brazma A, Parkinson H, Sarkans U, Shojatalab M, Vilo J, Abeygunawardena N, Holloway E, Kapushesky M, Kemmeren P, Lara GG, et al. ArrayExpress–a public repository for microarray gene expression data at the EBI. Nucleic Acids Res. 2003;31:68–71. doi: 10.1093/nar/gkg091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Viksna J, Celms E, Opmanis M, Podnieks K, Rucevskis P, Zarins A, Barrett A, Neogi SG, Krestyaninova M, McCarthy MI, et al. PASSIM–an open source software system for managing information in biomedical studies. BMC Bioinformatics. 2007;8:52. doi: 10.1186/1471-2105-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rocca-Serra P, Brandizi M, Maguire E, Sklyar N, Taylor C, Begley K, Field D, Harris S, Hide W, Hofmann O, et al. ISA software suite: supporting standards-compliant experimental annotation and enabling curation at the community level. Bioinformatics. 2010;26:2354–2356. doi: 10.1093/bioinformatics/btq415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gostev M, Fernandez-Banet J, Rung J, Dietrich J, Prokopenko I, Ripatti S, McCarthy MI, Brazma A, Krestyaninova M. SAIL - a software system for sample and phenotype availability across biobanks and cohorts. Bioinformatics. 2011;27:589–591. doi: 10.1093/bioinformatics/btq693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Church DM, Lappalainen I, Sneddon TP, Hinton J, Maguire M, Lopez J, Garner J, Paschall J, DiCuccio M, Yaschenko E, et al. Public data archives for genomic structural variation. Nat. Genet. 2010;42:813–814. doi: 10.1038/ng1010-813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malone J, Holloway E, Adamusiak T, Kapushesky M, Zheng J, Kolesnikov N, Zhukova A, Brazma A, Parkinson H. Modeling sample variables with an experimental factor ontology. Bioinformatics. 2010;26:1112–1118. doi: 10.1093/bioinformatics/btq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rayner TF, Rocca-Serra P, Spellman PT, Causton HC, Farne A, Holloway E, Irizarry RA, Liu J, Maier DS, Miller M, et al. A simple spreadsheet-based, MIAME-supportive format for microarray data: MAGE-TAB. BMC Bioinformatics. 2006;7:489. doi: 10.1186/1471-2105-7-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sansone SA, Rocca-Serra P, Brandizi M, Brazma A, Field D, Fostel J, Garrow AG, Gilbert J, Goodsaid F, Hardy N, et al. The first RSBI (ISA-TAB) workshop: “can a simple format work for complex studies?”. OMICS. 2008;12:143–149. doi: 10.1089/omi.2008.0019. [DOI] [PubMed] [Google Scholar]
- 12.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]