Abstract
The channel proteins belonging to the major intrinsic proteins (MIP) superfamily are diverse and are found in all forms of life. Water-transporting aquaporin and glycerol-specific aquaglyceroporin are the prototype members of the MIP superfamily. MIPs have also been shown to transport other neutral molecules and gases across the membrane. They have internal homology and possess conserved sequence motifs. By analyzing a large number of publicly available genome sequences, we have identified more than 1000 MIPs from diverse organisms. We have developed a database MIPModDB which will be a unified resource for all MIPs. For each MIP entry, this database contains information about the source, gene structure, sequence features, substitutions in the conserved NPA motifs, structural model, the residues forming the selectivity filter and channel radius profile. For selected set of MIPs, it is possible to derive structure-based sequence alignment and evolutionary relationship. Sequences and structures of selected MIPs can be downloaded from MIPModDB database which is freely available at http://bioinfo.iitk.ac.in/MIPModDB.
INTRODUCTION
Major intrinsic proteins (MIPs) form one of the largest superfamily of channel proteins (1,2). They transport water, neutral solutes such as glycerol and urea, metalloids including antimonite and arsenite and gases like CO2, nitric oxide and ammonia across the membranes (3–5). Water-transporting aquaporin and glycerol-specific aquaglyceroporin are the prominent members of this family. Members of MIP superfamily are involved in vital physiological processes such as skin moisture, gastrointestinal fluid transport, fat metabolism, epidermal proliferation, maintaining corneal and lens transparency in eyes and water homeostasis in kidney and central nervous system (6–11). MIPs are found from bacteria to humans and a large number of diverse MIP members have been identified especially in plants (12–17). The abundant MIPs identified in higher plants can be classified into at least five major subfamilies (15). In humans, they are implicated in several diseases such as nephrogenic diabetes insipidous, acute and chronic renal failure, brain edema, cataract and arsenic toxicity (18–24). Sequence analysis of MIP members clearly revealed the presence of highly conserved Asn-Pro-Ala (NPA) signature sequence motif (25,26). MIP sequences also possess internal homology in which the N- and C-terminal halves have significant sequence similarity (25). At the structural level, the N-terminal half is related to the C-terminal half by a pseudo-2-fold symmetry (27,28). Three-dimensional structures of more than 10 MIPs from different organisms such as mammalian (29–31), plant (32), Escherichia coli (33), yeast (34) and archaea (35) have been determined. They all adopt a unique hourglass fold even when the sequence identity among them is very low. Two regions of constriction have been identified within the channel. The two NPA motifs form the central constriction and the outer constriction is toward the extracellular side known as aromatic/arginine (ar/R) selectivity filter formed by four residues. Both regions are known to play important role in the solute transport and selectivity (36–39). With their involvement in human physiology and pathophysiology and with the available structural knowledge, members of MIP family are being considered as attractive drug targets (40,41). For example, aquaglyceroporin in intracellular parasite Plasmodium falciparum and its structural knowledge (42) has opened new options for novel malaria therapies (43). However, for the majority of MIPs from large number of organisms, the tissue localization, their functional properties and the biological significance are either not known or they are not clearly understood. With the advent of new generation of sequencing technologies (44,45), genome sequences of large number of organisms are available. Using conserved sequence motifs and the internal sequence similarity as constraints, we have previously searched genome sequences of plants to identify and characterize the MIP proteins (15,16). We have extended this approach to genomes of other organism groups and identified more than 1000 MIP sequences in diverse organisms. The wealth of information on MIP sequences is now stored in a database called MIPModDB and various details about the substitutions in the conserved NPA motif, gene structure and the features obtained from structural models are available for each MIP sequence. With diverse MIP sequences, one can also obtain structure-based sequence alignment and evolutionary relationship for a given set of MIP sequences.
DATABASE CONSTRUCTION
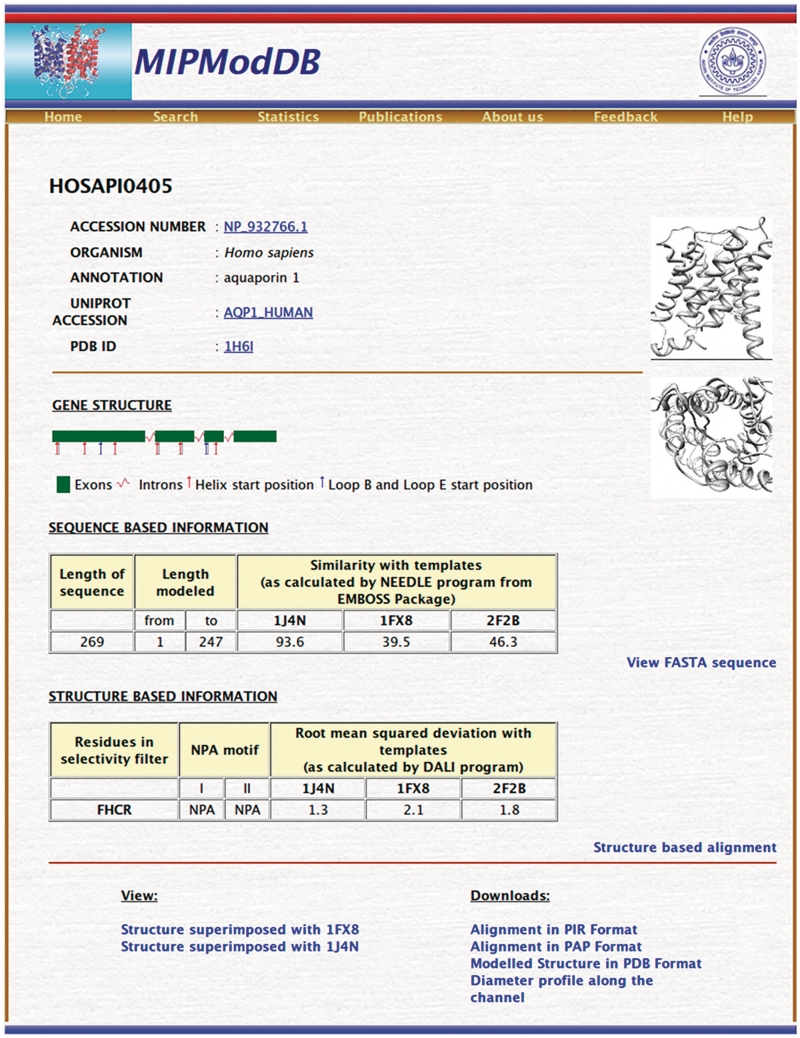
MIP genes were identified using BLAST (46) from completed and partial genomes available in NCBI database and the protocol used for this purpose is the same as described in our previous studies (15,16). Thirteen human aquaporins and five plant MIPs belonging to the five major plant subfamilies were used as query sequences. Sequences of short length and those sequences with missing transmembrane segments and important loop regions were discarded. We also found that some MIPs have been wrongly annotated. For example, an MIP from Salinispora tropica is annotated as ‘low molecular weight phosphotyrosine protein phosphatase’ (NCBI accession: YP_001157491). MIPs from plants identified from previous work (13–16,47) were also included. We have also searched the motif-oriented database, MIPDB (48), and considered those MIP sequences not identified in the above BLAST search. The final data set contained 1008 MIP sequences from 341 different organisms. For each MIP sequence, presence of conserved NPA sequence motif and internal sequence similarity were examined and the sequences were also confirmed by pattern databases like Pfam (49) and PROSITE (50). The three-dimensional structure of each MIP sequence was modeled by homology modeling procedure using the same protocol that was applied earlier for plant MIPs (15,16). Structures of bovine AQP1, E. coli GlpF and archeal AQPM were used as template structures and their PDB (51) IDs are 1J4N, 1FX8 and 2F2B, respectively. The channel radius profile was calculated using the HOLE program (52) as described previously (16). Thus the contents of this database can be largely categorized into sequence and structural data and are explained in more details in the following sections. The important statistics related to sequence and structure data of MIPModDB is given in Table 1. Both the sequence and structure data for a representative MIP sequence is shown in Figure 1.
Table 1.
Important statistics of MIPModDB
| Number of MIP sequences | 1008 |
| Number of organisms | 341 |
| Substitutions in the NPA motifs | 219 |
| Only in the first NPA motif (loop B) | 74 |
| Only in the second NPA motif (loop E) | 82 |
| Both NPA motifs | 63 |
| MIPs with selectivity filter similar to water-channelsa (FHTR + FHAR + FHCR + FHSR) | 349 |
| MIPs with selectivity filter similar to glycerol channelsa (WGYR + WGFR + WGWR) | 170 |
| Experimentally determined MIP structures | 38 |
aThe selectivity filter is formed by four residues and the corresponding amino acids are given in one letter codes. The first and second residues come from the second and the fifth transmembrane segments, respectively. The other two residues are contributed by the loop E. See text for details.
Figure 1.
Screenshot of a representative MIP protein, human aquaporin 1. Information about gene structure, substitutions in NPA motif, residues forming the ar/R selectivity filter, sequence similarity with the templates, RMSD calculated for the modeled structure with the three template structures are some of the features reported for a given MIP in the protein page.
MIP SEQUENCE DATA
In general, only a single MIP has been identified in most of the microbial genomes. Plants have large number of MIPs in comparison to animals. For each MIP, its source and the NCBI accession ID are given. Each MIP sequence is also given a unique identifier derived from its scientific name. The first two characters are taken from its genus and the next four characters are from its species name followed by a four digit unique number. Wherever it is available, UNIPROT (53) accession ID is also provided. Apart from the primary structure information, sequence data includes exon–intron organization of the gene, substitution (if any) in the conserved NPA motif and percentage sequence similarity with the template sequences that are used in the homology modeling procedure to build three-dimensional models. Sequence similarity between a given MIP sequence and the template sequences was calculated using the program NEEDLE as available in the EMBOSS suite of programs (54). Only the modeled part of the target MIP sequence was considered for this purpose.
Gene structure
For each MIP, gene structure is represented in the form of a graphical diagram. It gives the length and the positions of the introns with respect to the secondary structures of the corresponding MIP. The red and blue vertical bars indicate the starting positions of helices and loops B/E, respectively. The information regarding the positions of each individual introns was extracted from the NCBI database annotations. Transmembrane segments are marked based on the modeled structures (see below). Knowledge of intron–exon organization helps to understand the evolution of MIPs across different organisms and MIP subfamilies within a same species. For example, in plants it has been shown that the number and positions of introns are conserved within a given MIP subfamily (14–16).
Substitutions in NPA motif
In addition to its role in solute transport and selectivity (36,55–57), substitutions in the highly conserved NPA motifs seem to be important in other functional roles such as protein targeting (58) and full expression of the protein (59). In our data set, substitutions in only the first NPA motif occur in 74 examples. In 82 cases, substitutions are found only in the second NPA. Both NPA motifs are substituted in 63 MIPs (Table 1). In total, substitutions in at least one of the NPA motifs are found in about 22% of the total MIPs in our data set. Majority of the substitutions involve mutation of either Pro or Ala of NPA motif. Only a handful of examples (less than 16) are found in which Asn is mutated indicating its important role in both in structure as a helix capping residue and function as a residue responsible for cation exclusion as demonstrated in recent studies (60).
DATA FROM MIP STRUCTURAL MODELS
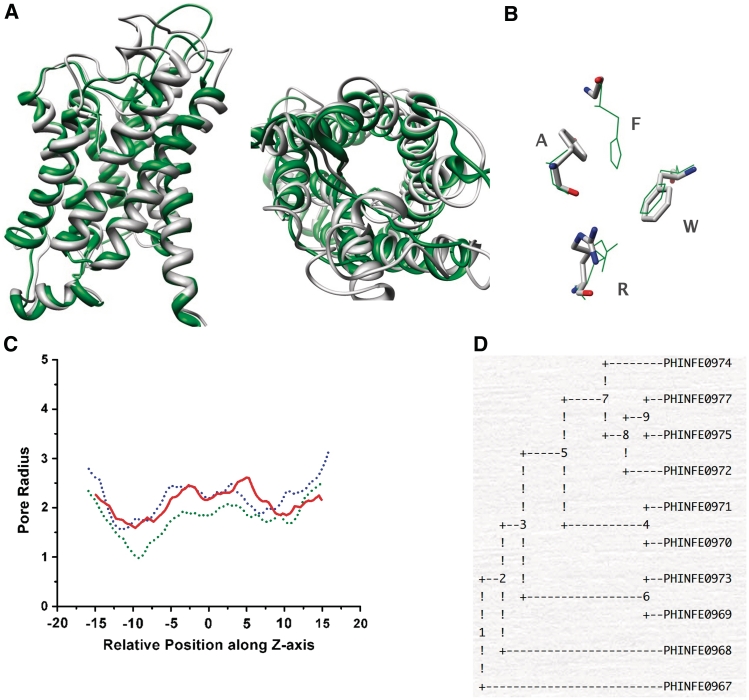
Structure-based data includes the atomic model obtained using the homology modeling procedure, residues that form the ar/R selectivity filter, structure-based sequence alignment, conservation of residues at the helix–helix interface and the HOLE radius profile. The structure-based details also include the root mean square deviation (RMSD) calculated for the modeled MIP and each of the template structure using the program DALI (61). The superposed figures are available in two different orientations (Figure 2A). Many MIPs have long N- and C-terminal extensions and hence the start and end positions of the polypeptide segment used in the homology modeling method are given.
Figure 2.
(A) Superposition of the modeled MIP structure with 1J4N, the structure of bovine aquporin shown in two different orientations, namely, parallel (left) and perpendicular (right) to the channel axis. (B) The four residues of the ar/R selectivity filter superposed on that of 1J4N structure. (C) Comparison of HOLE radius profiles plotted for the water channel (green), glycerol facilitator (blue) and the modeled MIP structure (red). The ar/R selectivity filter is approximately located at −10 Å. (D) Phylogenetic tree calculated for all MIPs of a representative organism Phytophthora infestans using parsimony method.
Aromatic/arginine selectivity filter
Four residues form the outer constriction nearly 8 Å from the conserved NPA motif toward the extracellular side. These residues are contributed by second and fifth TM segments and loop E. This aromatic/arginine (ar/R) selectivity filter has been implicated in obstructing the proton conduction (62) and efficient solute transport (37,63,64). The four ar/R selectivity filter residues, represented by their one letter codes, are given for each MIP. For example, ‘FIIR’ indicates that the first and second residues Phe and Ile are from TM2 and TM5, respectively, and the last two are the loop E residues. Analysis of selectivity filter residues indicates that there are 349 MIPs, which have selectivity filter (FHTR, FHAR, FHCR or FHSR) similar to that found in water-selective aquaporin channels (FHTR or FHCR). The number of MIPs having selectivity filter (WGYR, WGFR or WGWR) similar to the glycerol-specific aquaglyceroporin (WGFR) is 170. Thus, about 50% of the total MIPs in the database have selectivity filter typical of aquaporin or aquaglyceroporin. The remaining half has substitutions that can alter the size and chemical nature of the outer constriction. This will have major influence in the nature of solute that is being transported by the channel. The channel diameters of water channel and the aquaglyceroporin at the ar/R constriction are 2.0 and 3.5 Å, respectively. The selectivity filter residues of the predicted MIP model superposed on that of the pure water channel from bovine aquaporin and that of aquaglyceroporin from E. coli are available (Figure 2B). The HOLE radius profiles of all three MIPs, water channel, glycerol channel and the modeled MIP channel, can be compared (Figure 2C). This will give an idea about the size of the region around the ar/R selectivity filter region with respect to both water channel and aquaglyceroporin and it will help the user to predict the possible size of a solute that can pass through this constriction.
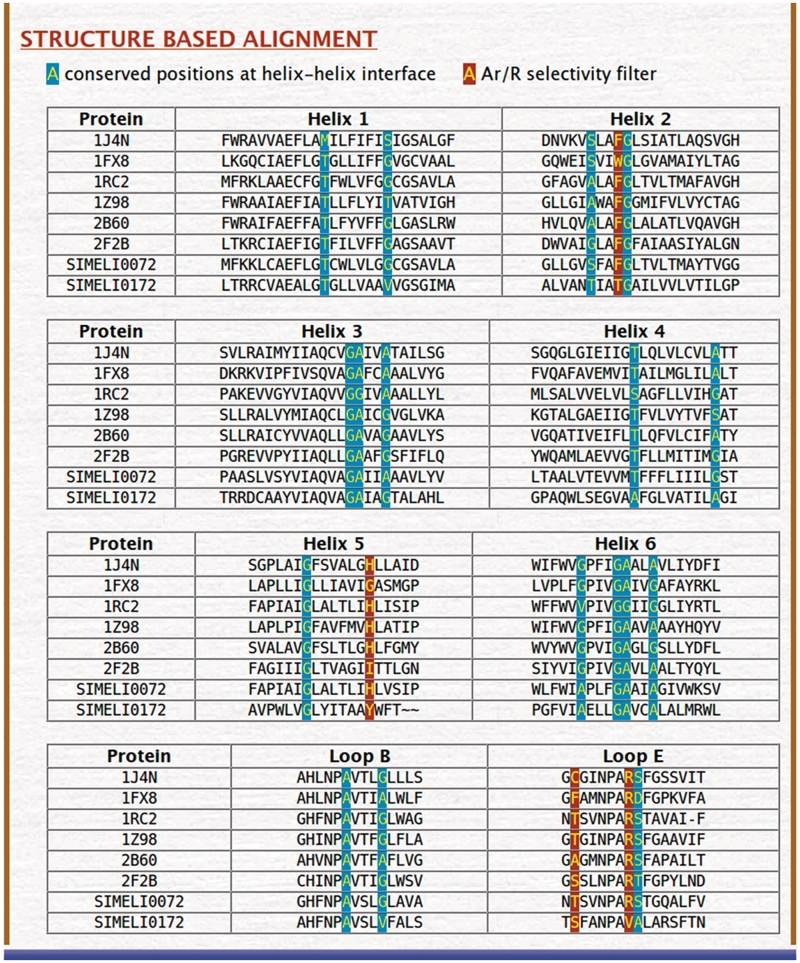
Structure-based sequence alignment
As mentioned earlier, MIP sequences are diverse. For example, the sequence identities between some of the plant MIP subfamilies are as low as 20% (13,15). In such cases, programs such as ClustalW (65) that are used to generate multiple sequence alignment of a given set of sequences are unlikely to produce meaningful alignment for a diverse set of MIP sequences. Instead, if the structurally equivalent positions belonging to the TM helical segments are aligned, they are likely to result in high conservation and indicate the importance of residues in certain positions. Structure-based sequence alignment for all the TM segments and the functionally important loops B and E is provided for all the MIPs present in the same organism along with the template sequences. We have also previously identified 17 positions that occur in the helix–helix interface. They are small and weakly polar residues and have been shown to be highly conserved if the amino acids Ala, Thr, Ser, Cys and Gly are considered as a group (15,16). These positions along with the residues that form the ar/R selectivity filter are highlighted in the structure-based sequence alignment. An example of structure-based sequence alignment as obtained from the MIPModDB database is shown in Figure 3.
Figure 3.
Structure-based sequence alignment for a selected set of MIPs. This alignment is always produced with the six high-resolution MIP structures and their PDB IDs are also shown. This alignment is produced for the six transmembrane segments and the two functionally important loops B and E. The residues forming the ar/R selectivity filter are shown in the dark brown background. Seventeen positions previously identified to occur in the helix–helix interface (16) are highly group conserved when small and weakly polar residues are considered together as a group. They are displayed in cyan background.
Construction of phylogenetic tree for selected MIPs
The database provides an interface whereby the user will be able to analyze the evolutionary relationship among the selected group of MIPs. For constructing a phylogenetic tree, the user needs to select at least three sequences. The user will have the option to choose one of the three different methods, namely, neighbor-joining method, maximum likelihood method and maximum parsimony method to construct a phylogenetic tree. In addition to the complication in generating a multiple sequence alignment of diverse MIP sequences, many MIPs have long N- and C-termini as well as long loops connecting the TM segments. Hence to avoid errors, while constructing the evolutionary tree, the input used for this purpose is the structure-based sequence alignment of TM helical regions and the loops B and E. The program PHYLIP which is part of the EMBOSS suite of programs (54) is being utilized by the server to create the phylogenetic trees (Figure 2D).
IMPLEMENTATION
The information content on MIPs is maintained as a relational database using MySQL (http://www.mysql.com). This allows easy access and storage. The database is hosted on a web server running apache (http://www.apache.org/) on Fedora Core Linux platform and can be queried through the web interface which is implemented in PHP (v5.2) scripting language (http://www.php.net/).
ACCESS TO MIP DATA
MIPModDB allows the users to browse, retrieve and query the database. The Statistics page lists all the MIPs in three different categories: (i) based on the NPA sequence motif, (ii) selectivity filter residues and (iii) organism-wise grouping. Users can browse MIPs according to the conservation or substitutions that are found in the NPA boxes. One can also look for MIPs with particular residues that form the selectivity filter. MIPs are also organized as per the organism in which they occur. They are arranged in the descending order in which the model tree Populus tricocarpa is present on the top with the maximum number of 55 MIPs.
The database interface allows the user to retrieve and identify MIPs using various features of MIPs as query. It can be searched by unique accession and complete or partial amino acid sequence. The search facility also allows the user to select MIP(s) with particular residues in the selectivity filter. Alternatively, MIPs with specific substitutions in the NPA motif also can be queried. A query based on an organism name will retrieve all MIP sequences from a particular species. More than one MIP features can also be used to narrow down the search. More detailed information can be retrieved by following the associated links. In addition to searching specific MIPs, the database enables users to download all the MIP sequences from a given organism or all the sequences that have the same ar/R selectivity filter residues in FASTA format. MIP sequences with specific substitutions in the NPA motif can also be downloaded. Sequence alignments used to generate the three-dimensional models can be downloaded in PIR or PAP format. For each MIP, the coordinates of the model are available in the PDB format. Similarly, the phylogenetic tree for a selected set of MIPs can be downloaded.
COMPARISON WITH OTHER TRANSPORTER DATABASES
There are databases which are developed specifically for membrane proteins that are involved in transporting the solutes across the membranes. The database TransportDB (66) provides information about the complete list of transporters for a given organism. For example, in the case of humans, the type of transporters listed include those that are ATP-dependent, ion channels, secondary transporters and unclassified. It also lists all outer membrane porins and channels from different organisms. The other database TCDB is a Transporter Classification Database (67) and is a classification system for membrane transporters. The superfamilies of transporters in this database include channel-forming toxins and peptides, transporters, symporters, antiporters, porins, carriers and ion channels. Both the above databases do not include all the MIP superfamily members whose members are known to predominantly transport neutral solutes. For example, while TransportDB lists human MIPs, MIP members in other organisms are not found. TCDB does not seem to include MIP superfamily although some MIP sequences are found. Moreover, while the above two databases largely contain sequence information and known PDB structures, MIPModDB provides structural models and associated information for more than 1000 MIP sequences.
FUTURE DIRECTIONS
Few MIPs have been functionally very well characterized and experimental studies are being carried out to determine the solute transport properties of many more MIPs. In the next version of MIPModDB database, whenever it is available, functional properties of MIPs, post-translational modification and cellular localization will be annotated and the related literature will be linked through PUBMED. As new MIPs are being recognized, the users will have the option to upload the sequences in the future version. Ultimately, our software will follow a pipeline procedure that will take an MIP sequence to series of steps which will include extracting the sequence features, building a homology model, identifying the selectivity filter residues, generating the HOLE radius profile and a possible prediction of the solutes that are likely to be transported.
FUNDING
The Department of Biotechnology, Government of India (BT/HRD/34/17/2008, partial); CSIR (senior research fellowship to A.B.G. and R.K.V.). Funding for open access charge: Indian Institute of Technology Kanpur.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Vivek Modi for his help and all our lab members for the useful input. A.B.G. and R.K.V. would like to thank CSIR for a senior research fellowship.
REFERENCES
- 1.Benga G. Water channel proteins (later called aquaporins) and relatives: past, present and future. IUBMB Life. 2009;61:112–133. doi: 10.1002/iub.156. [DOI] [PubMed] [Google Scholar]
- 2.Danielson JAH, Johanson U. Phylogeny of major intrinsic proteins. In: Jahn TP, Bienert GP, editors. MIPs and Their Role in the Exchange of Metalloids. Vol. 679. Springer-Verlag, Berlin: 2010. pp. 19–32. [Google Scholar]
- 3.Hachez C, Chaumont F. Aquaporins: A family of highly regulated multifunctional channels. In: Jahn TP, Bienert GP, editors. MIPs and their Role in the Exchange of Metalloids. Vol. 679. Springer-Verlag, Berlin: 2010. pp. 1–18. [DOI] [PubMed] [Google Scholar]
- 4.Wu B, Beitz E. Aquaporins with selectivity for unconventional permeants. Cell. Mol. Life Sci. 2007;64:2413–2421. doi: 10.1007/s00018-007-7163-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bienert GP, Bienert MD, Jahn TP, Boutry M, Chaumont F. Solanaceae XIPs are plasma membrane aquaporins that facilitate the transport of many uncharged substrates. Plant J. 2011;66:306–317. doi: 10.1111/j.1365-313X.2011.04496.x. [DOI] [PubMed] [Google Scholar]
- 6.Ishibashi K, Kondo S, Hara S, Morishita Y. The evolutionary aspects of aquaporin family. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2011;300:R566–R576. doi: 10.1152/ajpregu.90464.2008. [DOI] [PubMed] [Google Scholar]
- 7.Ding T, Gu F, Fu L, Ma YJ. Aquaporin-4 in glioma invasion and an analysis of molecular mechanisms. J. Clin. Neurosci. 2010;17:1359–1361. doi: 10.1016/j.jocn.2010.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Noda Y, Sohara E, Ohta E, Sasaki S. Aquaporins in kidney pathophysiology. Nat. Rev. Nephrol. 2010;6:168–178. doi: 10.1038/nrneph.2009.231. [DOI] [PubMed] [Google Scholar]
- 9.Hara-Chikuma M, Verkman AS. Roles of aquaporin-3 in the epidermis. J. Invest. Dermatol. 2008;128:2145–2151. doi: 10.1038/jid.2008.70. [DOI] [PubMed] [Google Scholar]
- 10.Verkman AS, Ruiz-Ederra J, Levin MH. Functions of aquaporins in eye. Prog. Retin. Eye Res. 2008;27:420–433. doi: 10.1016/j.preteyeres.2008.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ma TH, Verkman AS. Aquaporin water channels in gastrointestinal physiology. J. Physiol. London. 1999;517:317–326. doi: 10.1111/j.1469-7793.1999.0317t.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zardoya R. Phylogeny and evolution of the major intrinsic protein family. Biol. Cell. 2005;97:397–414. doi: 10.1042/BC20040134. [DOI] [PubMed] [Google Scholar]
- 13.Chaumont F, Barrieu F, Wojcik E, Chrispeels MJ, Jung R. Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 2001;125:1206–1215. doi: 10.1104/pp.125.3.1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johanson U, Karlsson M, Johansson I, Gustavsson S, Sjovall S, Fraysse L, Weig AR, Kjellbom P. The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 2001;126:1358–1369. doi: 10.1104/pp.126.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gupta AB, Sankararamakrishnan R. Genome-wide analysis of major intrinsic proteins in the tree plant Populus trichocarpa: characterization of XIP subfamily of aquaporins from evolutionary perspective. BMC Plant Biol. 2009;9 doi: 10.1186/1471-2229-9-134. Art. No. 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bansal A, Sankararamakrishnan R. Homology modeling of major intrinsic proteins in rice, maize and Arabidopsis: comparative analysis of transmembrane helix association and aromatic/arginine selectivity filters. BMC Struct. Biol. 2007;7:Art. No. 27. doi: 10.1186/1472-6807-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park W, Scheffler BE, Bauer PJ, Campbell BT. Identification of the family of aquaporin genes and their expression in upland cotton (Gossypium hirsutum L.) BMC Plant Biol. 2010;10 doi: 10.1186/1471-2229-10-142. Art. No. 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loreto C, Reggio E. Aquaporin and vascular diseases. Curr. Neuropharmacol. 2010;8:105–111. doi: 10.2174/157015910791233196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verkman AS. Aquaporins: translating bench research to human disease. J. Exp. Biol. 2009;212:1707–1715. doi: 10.1242/jeb.024125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morishita Y, Sakube Y, Sasaki S, Ishibashi K. Molecular mechanisms and drug development in aquaporin water channel diseases. Aquaporin superfamily (superaquaporins): expansion of aquaporins restricted to multicellular organisms. J. Pharmacol. Sci. 2004;96:276–279. doi: 10.1254/jphs.fmj04004x7. [DOI] [PubMed] [Google Scholar]
- 21.King LS, Kozono D, Agre P. From structure to disease: the evolving tale of aquaporin biology. Nat. Rev. Mol. Cell Biol. 2004;5:687–698. doi: 10.1038/nrm1469. [DOI] [PubMed] [Google Scholar]
- 22.Agre P, Kozono D. Aquaporin water channels: molecular mechanisms for human diseases. FEBS Lett. 2003;555:72–78. doi: 10.1016/s0014-5793(03)01083-4. [DOI] [PubMed] [Google Scholar]
- 23.Bedford JJ, Leader JP, Walker RJ. Aquaporin expression in normal human kidney and in renal disease. J. Am. Soc. Nephrol. 2003;14:2581–2587. doi: 10.1097/01.asn.0000089566.28106.f6. [DOI] [PubMed] [Google Scholar]
- 24.Loonen AJM, Knoers NVAM, van Os CH, Deen PMT. Aquaporin 2 mutations in nephrogenic diabetes insipidus. Semin. Nephrol. 2008;28:252–265. doi: 10.1016/j.semnephrol.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 25.Heymann JB, Engel A. Structural clues in the sequences of the aquaporins. J. Mol. Biol. 2000;295:1039–1053. doi: 10.1006/jmbi.1999.3413. [DOI] [PubMed] [Google Scholar]
- 26.Kruse E, Uehlein N, Kaldenhoff R. The aquaporins. Genome Biol. 2006;7 doi: 10.1186/gb-2006-7-2-206. Art. no. 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gonen T, Walz T. The structure of aquaporins. Quart. Rev. Biophys. 2006;39:361–396. doi: 10.1017/S0033583506004458. [DOI] [PubMed] [Google Scholar]
- 28.Fujiyoshi Y, Mitsuoka K, de Groot BL, Phillippsen A, Grubmuller H, Agre P, Engel A. Structure and function of water channels. Curr. Opin. Struct. Biol. 2002;12:509–515. doi: 10.1016/s0959-440x(02)00355-x. [DOI] [PubMed] [Google Scholar]
- 29.Ho JD, Yeh R, Sandstrom A, Chorny I, Harries WE, Robbins RA, Miercke LJ, Stroud RM. Crystal structure of human aquaporin 4 at 1.8 A and its mechanism of conductance. Proc. Natl Acad. Sci. USA. 2009;106:7437–7442. doi: 10.1073/pnas.0902725106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sui H, Han B-G, Lee JK, Walian P, Jap BK. Structural basis of water-specific transport through the AQP1 water channel. Nature. 2001;414:872–878. doi: 10.1038/414872a. [DOI] [PubMed] [Google Scholar]
- 31.Horsefield R, Norden K, Fellert M, Backmark A, Tornroth-Horsefield S, van Scheltinga TAC, Kvassman J, Kjellbom P, Johanson U, Neutze R. High-resolution X-ray structure of human aquaporin 5. Proc. Natl Acad. Sci. USA. 2008;105:13327–13332. doi: 10.1073/pnas.0801466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tornroth-Horsefield S, Wang Y, Hedfalk K, Johanson U, Karlsson M, Tajkhorshid E, Neutze R, Kjellbom P. Structural mechanism of plant aquaporin gating. Nature. 2006;439:688–694. doi: 10.1038/nature04316. [DOI] [PubMed] [Google Scholar]
- 33.Savage DF, Egea PF, Robles-Colmenares Y, O'Connell JD, III, and Stroud RM. Architecture and selectivity in aquaporins: 2.5 A structure of aquaporin Z. PLoS Biol. 2003;1:334–340. doi: 10.1371/journal.pbio.0000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fischer G, Kosinska-Eriksson U, Aponte-Santamaria C, Palmgren M, Geijer C, Hedfalk K, Hohmann S, de Groot BL, Neutze R, Lindkvist-Petersson K. Crystal structure of a yeast aquaporin at 1.15 A reveals a novel gating mechanism. PLOS Biol. 2009;7 doi: 10.1371/journal.pbio.1000130. Art. No. e1000130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee JK, Kozono D, Remis J, Kitagawa Y, Agre P, Stroud RM. Structural basis for conductance by the archaeal aquaporin AqpM at 1.68 A. Proc. Natl Acad. Sci. USA. 2005;102:18932–18937. doi: 10.1073/pnas.0509469102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.de Groot BL, Frigato T, Helms V, Grubmuller H. The mechanism of proton exclusion in the aquaporin-1 water channel. J. Mol. Biol. 2003;333:279–293. doi: 10.1016/j.jmb.2003.08.003. [DOI] [PubMed] [Google Scholar]
- 37.Beitz E, Wu B, Holm LM, Schultz JE, Zeuthen T. Point mutations in the aromatic/arginine region in aquaporin 1 allow passage of urea, glycerol, ammonia and protons. Proc. Natl Acad. Sci. USA. 2006;103:269–274. doi: 10.1073/pnas.0507225103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu BH, Steinbronn C, Alsterfjord M, Zeuthen T, Beitz E. Concerted action of two cationfilters in the aquaporin water channel. EMBO J. 2009;28:2188–2194. doi: 10.1038/emboj.2009.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fu D, Libson A, Miercke LJW, Weitzman C, Nollert P, Krucinski J, Stroud RM. Structure of a glycerol-conducting channel and the basis for its selectivity. Science. 2000;290:481–486. doi: 10.1126/science.290.5491.481. [DOI] [PubMed] [Google Scholar]
- 40.Frigeri A, Nicchia GP, Svelto M. Aquaporins as targets for drug discovery. Curr. Pharm. Des. 2007;13:2421–2427. doi: 10.2174/138161207781368738. [DOI] [PubMed] [Google Scholar]
- 41.Castle NA. Aquaporins as targets for drug discovery. Drug Discov. Today. 2005;10:485–493. doi: 10.1016/S1359-6446(05)03390-8. [DOI] [PubMed] [Google Scholar]
- 42.Newby ZER, O'Connell JD, III, Robles-Colmenares Y, Khademi S, Miercke LJW, Stroud RM. Crystal structure of the aquaglyceroporin PfAQP from the malarial parasite Plasmodium falciparum. Nat. Struct. Mol. Biol. 2008;15:619–625. doi: 10.1038/nsmb.1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kun JF, de Carvalhi EG. Novel therapeutic targets in Plasmodium falciparum: aquaglyceroporins. Expert Opin. Ther. Targets. 2009;13:385–394. doi: 10.1517/14728220902817839. [DOI] [PubMed] [Google Scholar]
- 44.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 45.Lister R, Gregory BD, Ecker JR. Next is now: new technologies for sequencing genomes, transcriptomes, and beyond. Curr. Opin. Plant Biol. 2009;12:107–118. doi: 10.1016/j.pbi.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Danielson JAH, Johanson U. Unexpected complexity of the aquaporin gene family in the moss Physcomitrella patens. BMC Plant Biol. 2008;8:Art. No. 45. doi: 10.1186/1471-2229-8-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.El Karkouri K, Gueune H, Delamarche C. MIPDB: a relational database dedicated to MIP family proteins. Biol. Cell. 2005;97:535–543. doi: 10.1042/BC20040123. [DOI] [PubMed] [Google Scholar]
- 49.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunesekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sigrist CJA, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE: a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berman HM, Westbrook J, Feng Z, Gilliand G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MSP. HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J. Mol. Graph. 1996;14:354–360. doi: 10.1016/s0263-7855(97)00009-x. [DOI] [PubMed] [Google Scholar]
- 53.The UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 55.de Groot BL, Grubmuller H. Water permeation across biological membranes: mechanism and dynamics of aquaporin-1 and GlpF. Science. 2001;294:2353–2357. doi: 10.1126/science.1066115. [DOI] [PubMed] [Google Scholar]
- 56.Chakrabarti N, Tajkhorshid E, Roux B, Pomes R. Molecular basis of proton blockage in aquaporins. Structure. 2004;12:65–74. doi: 10.1016/j.str.2003.11.017. [DOI] [PubMed] [Google Scholar]
- 57.Jensen MO, Tajkhorshid E, Schulten K. The mechanism of glycerol conduction in aquaglyceroporins. Structure. 2001;9:1083–1093. doi: 10.1016/s0969-2126(01)00668-2. [DOI] [PubMed] [Google Scholar]
- 58.Guan XG, Su WH, Yi F, Zhang D, Hao F, Zhang HG, Liu YJ, Feng XC, Ma TH. NPA motifs play a key role in plasma membrane targeting of aquaporin-4. IUBMB Life. 2010;62:222–226. doi: 10.1002/iub.311. [DOI] [PubMed] [Google Scholar]
- 59.Ikeda M, Andoo A, Shimono M, Takamatsu N, Taki A, Muta K, Matsushita W, Uechi T, Matsuzaki T, Kenmochi N, et al. The NPC motif of aquaporin-11, unlike the NPA motif of known aquaporins, is essential for full expression of molecular function. J. Biol. Chem. 2011;286:3342–3350. doi: 10.1074/jbc.M110.180968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wree D, Wu BH, Zeuthen T, Beitz E. Requirement of asparagine in the aquaporin NPA sequence signature motifs for cation exclusion. FEBS J. 2011;278:740–748. doi: 10.1111/j.1742-4658.2010.07993.x. [DOI] [PubMed] [Google Scholar]
- 61.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, Chen HN, Steinbronn C, Wu BH, Beitz E, Zeuthen T, Voth GA. Enhancement of proton conductance by mutations of the selectivity filter of aquaporin-1. J. Mol. Biol. 2011;407:607–620. doi: 10.1016/j.jmb.2011.01.036. [DOI] [PubMed] [Google Scholar]
- 63.Aponte-Santamaria C, Hub JS, de Groot BL. Dynamics and energetics of solute permeation through the Plasmodium falciparum aquaglyceroporin. Phys. Chem. Chem. Phys. 2010;12:10246–10254. doi: 10.1039/c004384m. [DOI] [PubMed] [Google Scholar]
- 64.Savage DF, O'Connell JD, III, Miercke LJW, Finer-Moore J, Stroud RM. Structural context shapes the aquaporin selectivity filter. Proc. Natl Acad. Sci. USA. 2010;107:17164–17169. doi: 10.1073/pnas.1009864107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thompson JD, Higgins DG, Gibson TJ. Clustal W - improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ren Q, Chen K, Paulsen IT. TransportDB: a comprehensive database resource for cytoplasmic membrane transport systems and outer membrane channels. Nucleic Acids Res. 2007;35:D274–D279. doi: 10.1093/nar/gkl925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Saier MHJ, Yen MR, Noto K, Tamang DG, Elkan C. The transporter classification database: recent advances. Nucleic Acids Res. 2009;37:D274–D278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]