Abstract
The Aspergillus Genome Database (AspGD; http://www.aspgd.org) is a freely available, web-based resource for researchers studying fungi of the genus Aspergillus, which includes organisms of clinical, agricultural and industrial importance. AspGD curators have now completed comprehensive review of the entire published literature about Aspergillus nidulans and Aspergillus fumigatus, and this annotation is provided with streamlined, ortholog-based navigation of the multispecies information. AspGD facilitates comparative genomics by providing a full-featured genomics viewer, as well as matched and standardized sets of genomic information for the sequenced aspergilli. AspGD also provides resources to foster interaction and dissemination of community information and resources. We welcome and encourage feedback at aspergillus-curator@lists.stanford.edu.
INTRODUCTION
The Aspergillus Genome Database (AspGD) is a unique web-based resource focusing on a single genus with a dense representation of sequenced and publicly available whole-genome sequences. AspGD’s mission is to serve the scientific research community by improving the structural and functional annotation associated with the Aspergillus genomes and providing a web-based platform for interrogating the similarities and differences between strains and species. The database currently houses 10 genomes representing eight distinct species and, with third-generation sequencing technologies making sequencing more accessible, we expect that number to increase markedly over the next year. As more genomic data become available, there is a concomitant increase in the power of the comparative platform for exploring the diversity of the aspergilli, which inhabit diverse environmental niches and include a pathogen of humans, agriculturally devastating toxin-producers and species with utility in industry and in the laboratory.
A hallmark of AspGD is the expert literature-based curation of the gene, protein and sequence data for key Aspergillus species. Species were prioritized for curation in consultation with the Aspergillus Genome Research Policy Committee (AGRPC), as well as the larger Aspergillus research community at their annual meetings. The first species targeted for curation was Aspergillus nidulans, an important model organism and the best characterized species with the largest body of literature (1). The next species chosen was Aspergillus fumigatus, the main cause of aspergillosis, an invasive fungal infection affecting immune-compromised patients, due to its clinical relevance (2,3). We are currently curating Aspergillus niger, which is of great importance to industrial applications involving enzyme production (4,5).
Comprehensive curation and annotation of A. nidulans and A. fumigatus genes
AspGD now features comprehensive curation and annotation of the genomic catalog of two entire Aspergillus species, A. nidulans and A. fumigatus. AspGD curators have read the entire available published scientific literature describing genes in both organisms. From this corpus, we have collected gene names, including all of the alternate names (aliases) that have been published for each gene; annotations describing the gene product function, cellular role and localization, with the controlled vocabularies provided by Gene Ontology (GO) (6); mutant phenotypes and lists of all published references concerning each gene. To augment the literature-based annotation, we have inferred additional GO annotations using orthology to characterized genes in other aspergilli (at AspGD) and in Saccharomyces cerevisiae (at the Saccharomyces Genome Database, SGD, www.yeastgenome.org), as well as the presence of characterized InterPro protein domains and motifs (7). We also use tRNAscan software to generate consistent catalogs of tRNA gene model annotations for each genome in AspGD (8). Current AspGD curation statistics are summarized in Table 1. We are now in the process of adding A. niger information into AspGD, and will be expanding the scope of AspGD literature curation to cover additional species in the future, including Aspergillus flavus, Aspergillus oryzae, Aspergillus clavatus, Aspergillus terreus and Neosartorya fischeri/Aspergillus fischerianus. The A. fumigatus genome statistics, like those for the A. nidulans genome, are now summarized in a Genome Snapshot page on the AspGD web site, which displays tabular and graphical summaries of gene statistics and annotation and is updated daily to reflect the most up-to-date state of the characterization of the genome and gene catalog.
Table 1.
AspGD curation statistics
| Aspergillus nidulans | Aspergillus fumigatus | Aspergillus niger | |
|---|---|---|---|
| Number of ORFs | 11 286 | 9887 | 14 071 |
| Number of tRNAs | 222 | 178 | 264 |
| Manual GO annotations | 20 233 | 16 994 | N/A |
| Features with manual GO annotations | 8781 | 7786 | N/A |
| Orthology-based GO annotations | 8295 | 9220 | 9637 |
| Features with orthology-based GO annotations | 1973 | 2229 | 2342 |
| Protein-domain (InterPro)-based GO annotations | 14 505 | 13 068 | 16 614 |
| Features with protein-domain (InterPro)-based GO annotations | 5748 | 5226 | 6348 |
| Features with orthology-based description lines | 5016 | 5430 | N/A |
AspGD curation statistics were determined as of 1 September 2011. The number of Open Reading Frames (ORFs) includes characterized (verified) and uncharacterized genes that are predicted to encode protein products. The tRNA complement was predicted at AspGD using tRNAscan-SE (8). GO annotations have been assigned based on manual curation of the scientific literature, predicted based on orthology to characterized gene products from S. cerevisiae or other aspergilli, and predicted based on characterized InterPro protein domains (7). Manual curation of the scientific literature about A. nidulans and A. fumigatus is complete and up-to-date; curation of A. niger is commencing in September 2011. In species for which manual curation is complete, predicted gene products that are uncharacterized, but which have orthologs, are given descriptions containing orthology-based information.
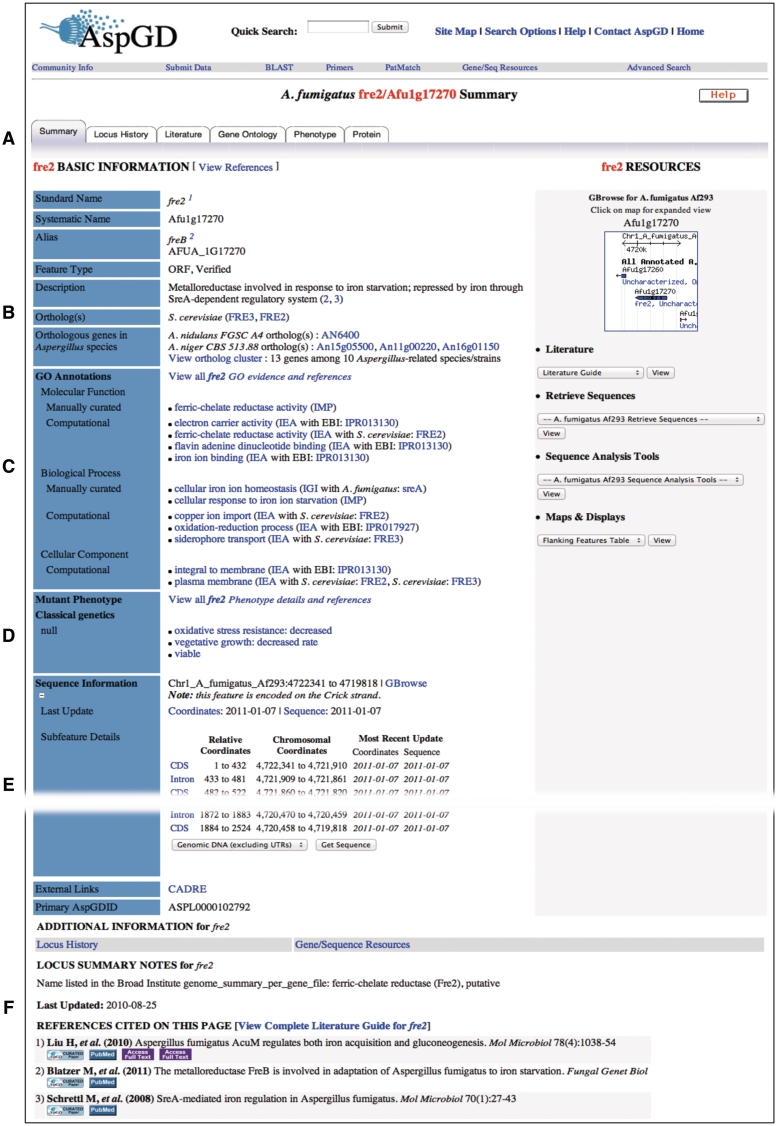
The information for each gene is displayed on the AspGD web site and also available in downloadable files. We can provide custom-format data files upon request; to request a file, contact us at http://www.aspergillusgenome.org/cgi-bin/suggestion. The web site is organized around Locus Summary pages (Figure 1) that list the basic information about each gene and link to additional tabbed pages that provide more extensive information, such as the history of sequence and major annotation changes (Locus History), the full citation list (Literature Guide), the gene product annotation and supporting evidence (GO) and specifics of each mutant phenotype with details including specifics of the experimental conditions, observations and strain background. Recently, we have implemented an additional Protein Details page for each protein-coding gene. The protein details shown include a graphical display of protein domain organization, the molecular structure of the top hit in the Protein Data Bank (PDB) (9), the protein sequence, the set of predicted physicochemical properties and links to tabular enumerations of all unique domains and domains shared with other proteins in the genome, transmembrane segments and signal peptides, plus other domain exploration tools.
Figure 1.
Locus Summary page for A. fumigatus fre2. (A) Navigation tabs lead to expanded reports on Locus History, relevant literature, GO and phenotype annotations and protein details. (B) Basic Information section provides gene names and aliases, a headline-like description of the gene product, links to orthologous genes in other Aspergillus species in AspGD and links to orthologs in S. cerevisiae at SGD. (C) GO section lists functional annotations derived from manual curation of available data and from orthology-based and protein domain-based computational analysis. (D) Mutant Phenotype section outlines literature-derived experimental data for mutant phenotypes. (E) Sequence Information section gives an overview of the gene model and provides access to sequences (parts of this section have been removed from the figure to conserve space). (F) Additional Information section includes other notes and the list of references cited on the Locus Summary page. The comprehensive citation list for the gene is found on the gene’s Literature Guide, accessible from the Literature tab near the top of the Locus Summary page.
Accommodation of the new A. fumigatus information in AspGD alongside the A. nidulans data required numerous extensive updates to the web site and the database structure. In the new, multispecies format, we have interconnected the Locus Summary pages between species via orthology relationships. The Locus Summary page for a A. nidulans gene is connected to the Locus Summary page for its ortholog in A. fumigatus, and vice versa, by hyperlinks displayed in the section labeled ‘Orthologous genes in Aspergillus species’, located near the top of the page, making it easy to navigate between orthologs (Figure 1). The link labeled ‘View ortholog cluster’ displays the entire gene cluster in the Sybil comparative genomics browser, which is described in more detail below.
All of the tools on the AspGD web site have been updated to handle multispecies information. Tools such as Quick Search, which queries data from multiple species, feature a redesigned results page (Figure 2A) with sections on the page for results that are specific to each species and a separate section for results that are not species specific (e.g. GO terms, authors and reference information, colleagues). Tools that query sequence-based data now provide a species selection option (Figure 2B), whereby the scope of the search is limited to the species of interest.
Figure 2.
Multispecies search tools. (A) Quick Search Results page is divided into sections for general results, such as GO terms or authors and for species-specific results, such as gene names, names of orthologous genes, as well as words found in descriptions, notes and phenotypes. (B) Gene and Sequence Resources tool retrieves DNA sequences for genes or chromosomal regions; the arrow points to the new pull-down menu for species selection. (C) Advanced Search tool allows retrieval of genes based on their properties; arrows indicate options for selection of a species, types of features, other properties and GO terms (lower part of the page is removed to conserve space).
To accommodate new data, we also have implemented new search capabilities within AspGD. The Advanced Search tool, which searches the gene catalog based on gene properties, rather than gene names, has several additional options (Figure 2C). Users may now choose to search for pseudogenes, repeat regions, as well as for genes with or without annotated introns, untranslated regions (UTRs) and introns in UTRs. All of the previously provided options remain available, including searches for protein-coding, tRNA, rRNA, experimentally characterized, uncharacterized, deleted or unmapped genes or for genes with annotations to particular functional or localization (GO) classifications.
Comparative genomics resources in AspGD
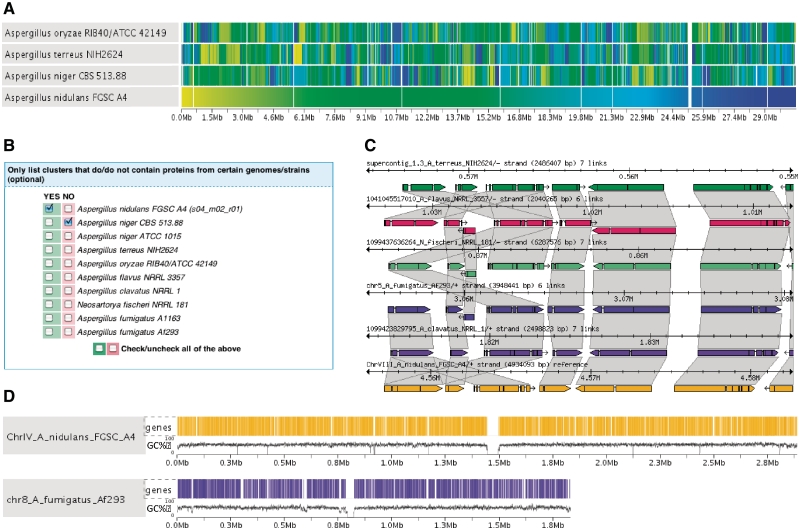
AspGD provides a rich and user-friendly interface for the comparison of all genomes currently deposited in our database. With the integration of Sybil comparative genomics software into AspGD (http://sybil.sourceforge.net/), researchers can visualize synteny and gene structure across species, and list and retrieve the sequence of homologous genes present in any genome subset defined by the user, including genes present in all species, those specific to a single genome and genes occurring in one clade but absent in another (Figure 3). These capabilities enable researchers to detect gene structure and gene content differences associated with a clade, species or strain containing a particular phenotype of interest. In addition, multiple pre-computed sequence alignments of nucleotide and amino acid sequence among the members of each cluster can be visualized through the AspGD web interface.
Figure 3.
Search and visualization tools in the Sybil comparative genomics module. (A) Synteny gradient indicating the location of homologous blocks relative to a reference sequence, in this example A. nidulans FGSC A4. Vertical bars with the same shade of color represent orthologous genes across genomes. (B) Ortholog/Paralog cluster search tool, that allows researchers to retrieve homolog clusters meeting user-defined criteria regarding strain or species distribution. (C) Genomic region comparison depicting the structural annotation of syntenic and non-syntenic genes in a homologous genomic region. The trapezoids connect orthologous genes. (D) Whole-genome display showing the gene density and GC content along the sequence of A. nidulans chromosome IV and A. fumigatus chromosome 8. Colored vertical bars indicate gene locations.
The Sybil module also provides users with an overview of syntenic blocks shared among genome sequences stored in AspGD (Figure 3A), as well as the gene density of protein coding genes, tRNA, rRNA, guanine-cytosine (GC) content and skew along each chromosomal sequence, thus providing a tool for visualization of genomic translocations, inversions and deletions (Figure 3).
AspGD as a repository of consistently formatted genomic sequences and features
Genome sequencing and structural annotation of the aspergilli has been performed at various sequencing centers, using different platforms, across a time span of multiple years (1–5,10,11), (http://www.aspergillusflavus.org/genomics/, http;//www.broadinstitute.org/annotation/genome/aspergillus_group/GenomeDescriptions.html#AT1). A significant challenge in comparative genomics analysis thus becomes the generation of a set of data with sufficient consistency to serve as a reasonable initial starting point. To begin to address these issues, we have generated, and posted for download, comprehensive sets of sequence files in consistent formats for A. nidulans FGSC A4 and nine additional genomes: A. clavatus NRLL 1, A. flavus NRRL 3357, A. fumigatus A1163, A. fumigatus Af293, A. niger ATCC 1015, A. niger CBS 513.88, A. oryzae RIB40/ATCC 42149, A. terreus NIH2624 and N. fischeri NRLL 181.
Improvement of gene structure
As transcriptome data for the species and strains with genomic sequences stored in AspGD become available, we are mapping and incorporating these data into the structural annotation of those genomes. This effort will progressively refine the current gene models and their products, further facilitating comparative analyses and molecular or biochemical experiments that depend on this information.
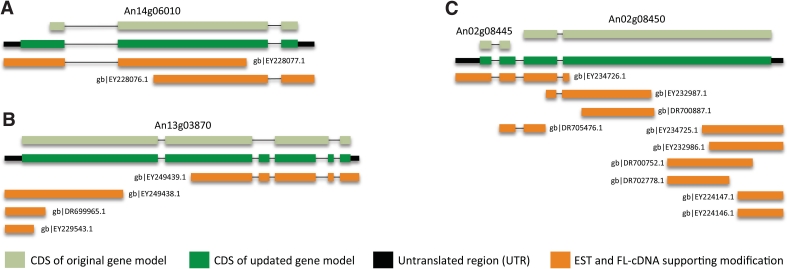
To begin, we have performed a set of gene structure modifications on the genome of A. niger CBS 513.88 through incorporation of publicly available experimental data comprising A. niger expressed sequence tags (ESTs) and full-length cDNAs (12–17). The analysis was performed by PASA (Program to Assemble Spliced Alignments) (18), which automatically redefines intron–exon boundaries, extends UTRs and identifies possible novel isoforms. The modifications performed by PASA are supported by mapping and clustering of transcriptome data, assembly of the aligned sequence data and comparison with the current structural annotation of the genomes analyzed (Figure 4). The PASA analysis performed on A. niger CBS 513.88 resulted in the modification of 4230 genes (30% of the total gene set): the UTRs of 3452 genes were extended, coding sequence alterations were made for 308 genes, the internal exon structure of 719 genes was modified and 15 pairs of genes were merged into single loci. Some genes were subject to multiple types of modification. As more species are added to AspGD, and more experimental transcriptome data become available for Aspergillus species, including RNA-Seq, this PASA pipeline will be used for iterative improvements of all of these reference genome annotations.
Figure 4.
Examples of modifications to A. niger gene models based on analysis with PASA. The changes include (A) extension of coding sequence (CDS), (B) update of internal exon structure and (C) gene merging. Rectangles represent gene exons and are colored according to the legend. Displayed with each structure diagram is the name of the gene modified by the PASA analysis or the GenBank accession number of each EST and full-length cDNA sequence supporting the modifications.
AspGD and community interconnections
As a community-focused database, AspGD integrates and assembles information of various types in addition to genomic sequence and literature curation to improve and facilitate access to, and sharing of community resources. A recent and significant challenge for the A. nidulans research community was the simultaneous existence of two different versions of the genome annotation, both with their individual strengths. A major collaborative community effort produced the Eurofungbase annotation, in which individual experts conducted manual review and updating of genes with which they were familiar (10). In a parallel effort, researchers at the Broad Institute produced a genome-wide reannotation using modern computational tools. At AspGD, we made resolution of this situation a priority. We analyzed the differences and performed a systematic merge of the two sets, first retaining the careful manual efforts of the Eurofung team, and then integrating the updated computationally based gene model changes where they did not introduce a conflict with the manual curation. The Locus History that we provide for each gene provides details on all updates made, or information conflicts, which pertain to the annotation of the gene model.
A particularly valuable experimental resource for the A. nidulans community is the systematic A. nidulans gene knockout collection. Knockout cassettes are now available from the Fungal Genetics Stock Center (FGSC, http://www.fgsc.net/Aspergillus/KO_Cassettes.htm). The community asks that mutant strains generated with the cassettes be deposited with the FGSC as soon as possible after construction. At AspGD, we have added a ‘KO Cassette Available’ link to the External Resources section on the Locus Summary page of each gene for which this is the case, to facilitate utilization of this resource and generation of the strain collection.
New web-based community resources in AspGD include a page for sharing of laboratory methods and protocols, and a page for downloading of community-provided images and movies (http://www.aspgd.org/Methods.shtml and http://www.aspgd.org/Images.shtml, respectively). To contribute to either of these resources, or to suggest other pages that might be useful, please contact the AspGD curators at aspergillus-curator@lists.stanford.edu.
SUMMARY/FUTURE DIRECTIONS
Going forward, we will curate the experimental data from the scientific literature on A. niger and then A. oryzae, as decided by community consensus at the Asperfest meeting at the 10th European Conference on Fungal Genetics, in 2010. Subsequently, we will curate A. flavus, A. clavatus, A. terreus and N. fischeri/A. fischerianus. Each additional species will be curated and fully incorporated into AspGD alongside A. nidulans and A. fumigatus. We will also be further enhancing the web site and tools to be able to store and display sequence data for multiple strains of any given Aspergillus species.
Anticipating the wealth of large-scale data that are becoming available from the use of new high-throughput technologies, we are preparing to provide tools for interactive web-based visualization of these data. We are also incorporating RNA-Seq as an experimental input into the PASA genome annotation pipeline, to use this information alongside comparative data to improve the gene model annotations across all of the sequenced Aspergillus genomes
We welcome and encourage all suggestions, comments and questions and can be reached by email at aspergillus-curator@lists.stanford.edu.
FUNDING
Funding for open access charge: The National Institute of Allergy and Infectious Diseases at the US National Institutes of Health (grant R01 AI077599 to G.S. and J.W.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
AspGD thanks the Aspergillus research community for their enthusiastic support and very constructive suggestions and feedback. We particularly appreciate the efforts of the individuals who have taken the time to review the curation of the genes with which they are familiar, and to suggest curatorial improvements that benefit the entire community of AspGD users. We thank Jane Mabey Gilsenan and the entire CADRE project for data-sharing and collaboration. We also thank Mike Cherry and the entire Saccharomyces Genome Database project, upon which AspGD is based.
REFERENCES
- 1.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Baştürkmen M, Spevak CC, Clutterbuck J, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 2.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 3.Fedorova ND, Khaldi N, Joardar VS, Maiti R, Amedeo P, Anderson MJ, Crabtree J, Silva JC, Badger JH, Albarraq A, et al. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 2008;4:e1000046. doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 2007;2:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 5.Andersen MR, Salazar MP, Schaap PJ, van de Vondervoort PJ, Culley D, Thykaer J, Frisvad JC, Nielsen KF, Albang R, Albermann K, et al. Comparative genomics of citric-acid-producing Aspergillus niger ATCC 1015 versus enzyme-producing CBS 513.88. Genome Res. 2011;6:885–897. doi: 10.1101/gr.112169.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gene Ontology Consortium. Creating the gene ontology resource: design and implementation. Genome Res. 2001;11:1425–1433. doi: 10.1101/gr.180801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zdobnov EM, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 8.Lowe TM, Eddy SE. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997;5:955–964. doi: 10.1093/nar/25.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wortman JR, Gilsenan JM, Joardar V, Deegan J, Clutterbuck J, Andersen MR, Archer D, Bencina M, Braus G, Coutinho P, et al. The 2008 update of the Aspergillus nidulans genome annotation: a community effort. Fungal Genet. Biol. 2009;46(Suppl. 1):S2–S13. doi: 10.1016/j.fgb.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 12.Semova N, Storms R, John T, Gaudet P, Ulycznyj P, Min XJ, Sun J, Butler G, Tsang A. Generation, annotation, and analysis of an extensive Aspergillus niger EST collection. BMC Microbiol. 2006;6:6–7. doi: 10.1186/1471-2180-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.te Biesebeke R, Levin A, Sagt C, Bartels J, Goosen T, Ram A, van den Hondel C, Punt P. Identification of growth phenotype-related genes in Aspergillus oryzae by heterologous macroarray and suppression subtractive hybridization. Mol. Genet. Genomics. 2005;273:33–42. doi: 10.1007/s00438-004-1082-9. [DOI] [PubMed] [Google Scholar]
- 14.Choi JY, Lee DW, Koh JS, Kim JH, Yang MS, Chae KS. Identification of expressed sequence tags (ESTs) of the highly transcribed genes in Aspergillus niger. Biotechnol. Lett. 1999;21:381–384. [Google Scholar]
- 15.Richardson P, Lucas S, Rokhsar D, Wang M, Lindquist EA, Baker SE, Dai Z, Panther KS, Hubbard LJ. 2007 DOE Joint Genome Institute. [Google Scholar]
- 16.Tsang A, Storms RK. 2000 Department of Biology, Concordia University. [Google Scholar]
- 17.Williams BA, Tsang A, Storms RK. 1995 Deptartment of Biology, Concordia University. [Google Scholar]
- 18.Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK, Jr, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31:5654–5666. doi: 10.1093/nar/gkg770. [DOI] [PMC free article] [PubMed] [Google Scholar]