Abstract
IDEAL, Intrinsically Disordered proteins with Extensive Annotations and Literature (http://www.ideal.force.cs.is.nagoya-u.ac.jp/IDEAL/), is a collection of knowledge on experimentally verified intrinsically disordered proteins. IDEAL contains manual annotations by curators on intrinsically disordered regions, interaction regions to other molecules, post-translational modification sites, references and structural domain assignments. In particular, IDEAL explicitly describes protean segments that can be transformed from a disordered state to an ordered state. Since in most cases they can act as molecular recognition elements upon binding of partner proteins, IDEAL provides a data resource for functional regions of intrinsically disordered proteins. The information in IDEAL is provided on a user-friendly graphical view and in a computer-friendly XML format.
INTRODUCTION
The discovery of intrinsically disordered proteins (IDPs) has brought about a paradigm change in structural biology (1,2). Although proteins were believed to adopt unique 3D structures to function, IDPs do not, by themselves, assume any stable 3D structure under physiological conditions, and yet they participate in crucial biological processes such as signal transduction and transcription control (3–5). Some proteins contain long intrinsically disordered regions (IDRs) while others are fully disordered. In contrast to the long studied 3D structures of proteins, investigations on IDPs started only about 10 years ago and, as yet, knowledge of IDPs is not well collected and integrated. Although the first database of IDPs, Disprot (6), has more than 600 well-annotated entries, this number is much smaller than the over 70 thousands entries in the Protein Data Bank (PDB) (7). Considering that the protein 3D structural databases such as PDB, SCOP (Structural Classification of Proteins) (8) and CATH (9), have played important roles in deepening our understanding of the nature of protein structures and functions, the development of IDP databases are essential to the progress of IDP research.
We have developed a database, IDEAL (IDPs with Extensive Annotations and Literature) in which experimentally verified IDRs are collected. In the database construction process, we paid special attention to the functional regions in IDRs, for example, regions that interact with other molecules and post-translational modification sites. In particular, we have extensively curated IDRs that adopt unique 3D structures when they bind to other molecules by the ‘coupled folding and binding’ process (10–16). We have called these IDRs the protean segments (ProS). The information in IDEAL is provided on a user-friendly web-interface and in computer-friendly XML files.
CONTENTS OF IDEAL
Summary of the annotation process
We used the UniProt amino acid sequence (17) as the reference, and marked structural and functional features along the sequences. A unique serial identifier, IID (IDEAL Identification), was assigned to each protein in IDEAL, starting with IID0001 for human proteins, IID5001 for other eukaryotic proteins and IID9001 for all other proteins including virus proteins. Ordered and disordered regions were annotated as follows: First, ordered regions were obtained from the structural regions atomically detailed in the PDB. Then, disordered regions were located by careful assessment of PDB coordinates and by reading the literature. After identifying the ordered and disordered regions, the ProSs were manually determined. Finally, miscellaneous information, such as binding sites and post-translational modifications, was derived mainly from UniProt annotations, and structural domains were assigned by homology searches.
Proteins stored in IDEAL
As a starting point for the annotation, we chose UniProt human nuclear proteins with PDB structures (712 proteins), because eukaryotic nuclear proteins are known to contain long IDRs (18,19). We have annotated more than 120 human nuclear proteins. Out of them, the overlap with DisProt is only one-third at most, indicating IDEAL and DisProt complement each other. Most of the PDB structures for these proteins are hetero-oligomers in which the protein was associated with its binding partners. Annotations for these partner proteins are also in IDEAL, regardless of the source organism or the presence or absence of IDRs.
Ordered/disordered regions
The most important part of the IDEAL annotation is to identify the ordered and disordered amino acid segments. Ordered regions can be assigned by referring to the PDB. It is not straightforward to identify the disordered regions. In IDEAL, disordered regions are judged using several criteria; (i) missing residues in the X-ray structures, (ii) regions that interfere with protein crystallization in X-ray experiments, (iii) regions that fluctuate greatly in ensemble number of NMR model structures and (iv) regions that have been shown to be flexible in experiments using NMR, CD and other methods, and that have no corresponding structures in PDB. Of the four categories, (i) can be automatically obtained from PDB. Regions that were identified using the other three categories could only be judged manually. Although fluctuating regions in category (iii) could be found automatically by comparing the PDB coordinates of a group of models, the regions were only accepted as IDRs after curators confirmed the fluctuations by examining the corresponding literature. Category (iv) requires the most laborious procedure to be obtained, but provides variable information. Curators conduct manual literature searches to obtain such information as much as possible.
Protean segment
One of the reasons why IDPs have drawn so much attention is the discovery of the phenomenon known as coupled folding and binding in which a short flexible segment binds to its binding partner by forming a specific structure which acts as the molecular recognition element (10–16). In IDEAL, we explicitly annotated this short flexible region as ProS when both unstructured and structured information is available for the region. We defined two categories for ProS, verified ProS and possible ProS. A verified ProS is a sequence for which there is evidence of both a disordered isolated state and an ordered binding state. A possible ProS is a sequence for which there is only evidence of an ordered binding state, but circumstantial evidence suggests that the sequence is disordered in the isolated state. A possible ProS is, for example, a sequence from a protein whose homolog contains a verified ProS in the corresponding position. Another example would be the one in which the binding partner of a possible ProS binds a verified ProS using the same interface.
Sequences involved in coupled folding and binding have been addressed in several ways, for example, molecular recognition features (MoRFs) (20) and eukaryotic linear motifs (ELMs) (21) have been studied. Although ProS, MoRF and ELM are similar concepts, MoRF has a length limitation of 70 residues and an ELM should have a motif that can be described in a regular expression. On the other hand, the definition of ProS depends only on evidence of a disorder-order transition. Although most ProSs bind to a partner protein, by its definition, ProS can include IDRs whose structures are induced upon binding to small ligands. ProSs do not necessarily assume secondary structures in the binding state, and long IDRs or IDRs without a motif can also be ProSs. Some relatively long IDRs, such as p27Kip1 (PDB:1jsu) and Tcf3 (PDB:1g3j), can transform into ordered states (22). ProSs can also cover these IDRs.
MISCELLANEOUS INFORMATION
We integrate the miscellaneous information from UniProt, namely, regions interacting with other molecules, motifs and post-translational modifications. During the annotation process, the curators find interaction sites, sequence motifs or other information that has not been described in UniProt, the new information is included in IDEAL. IDEAL also provides SCOP (version 1.75) and Pfam (23) (version 24.0) domain assignments using reverse PSI-Blast (24) and HMMer (25). Note that ordered regions assigned in the order/disorder annotation process are experimentally verified ordered regions, while the structural domain assignments were done using homology searches.
USING IDEAL
Browse and search entries
‘The list’ on the top page of IDEAL provides an easy way to access any of the entries in IDEAL. The list enumerates all entries in IDEAL, where IID, protein name, organism, total sequence length and the presence/absence of ProS are tabulated. IDEAL also provides a search tool, which always appears in the blue bar at the top of each page ([1] in Figure 1). Users can choose from ‘Full text’, ‘UniProt accession’, ‘Protein Name’ and ‘PDB id’ categories, and enter some words or an ID into the input field. The BLAST search is available through the ‘BLAST search’ link button, and the user can input an amino acid sequence to find homologs in the IDEAL entries.
Figure 1.
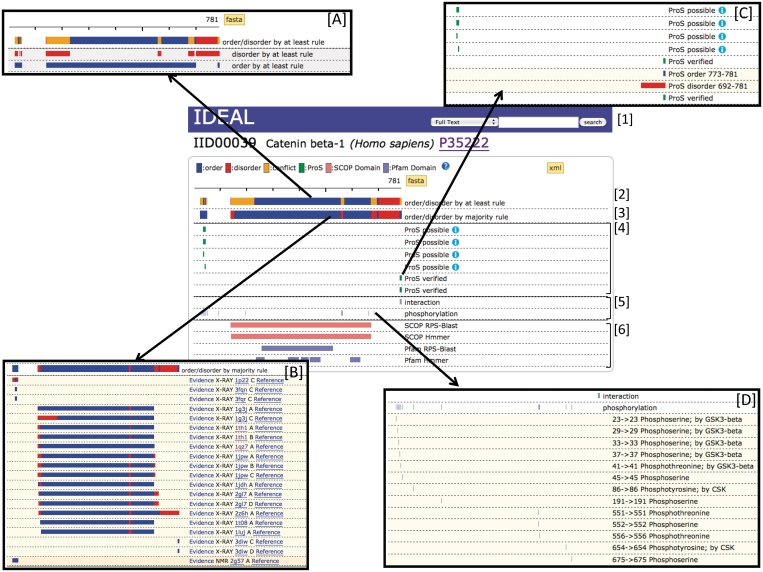
IDEAL annotation for catenin β-1. The identifier, IID, protein name, source organism and the link to UniProt are shown below the blue bar which contains the search tool ([1]). Bars [2] and [3] show a summary of the annotated regions using the ‘at least rule’ and ‘majority rule’ criteria. Ordered, disordered and conflict regions are colored in blue, red and orange, respectively. The inner boxes (A) and (B) show a detailed breakdown of the regions in [2] and [3], respectively. These diagrams appear by clicking on the bars. The inner box (B) provides the link to the experimental evidence and the technique supporting the order/disorder regions. ProSs are represented in section [4] as the green bars, which can be expanded by clicking to reveal the inner box (C). Interaction sites and post-translational modification sites are shown in section [5]. These bars expand to show the detailed information in the inner box (D). The bars in section [6] show the domain assignments.
Representation of each entry
IDEAL provides a user-friendly web interface for each entry. An example, a page for catenin β-1, is shown in Figure 1. The annotated regions are presented in a bar diagram to help make annotations intuitively understandable. Two color bars at the top ([2] and [3] in Figure 1) summarize ordered/disordered information in the distinctive ways shown. A protein may have multiple PDB entries and other information without accompanying PDB entries from different experimental techniques such as CD, H/D exchange, etc. Because IDEAL contains all PDB entries together with the other structural information associated with a query protein, all the associated information are not necessarily consistent due to different experimental conditions and other reasons. To summarize these diverse situations, IDEAL uses two representations:
The bar [2] in Figure 1 shows the summary of ordered/disordered regions by the ‘at least rule’. Here an ordered (blue bar) or disordered (red bar) site is shown if the site has at least one ordered or one disordered annotation. When a single site has both an ordered and a disordered annotation, the site is in ‘conflict’ (orange bar). The inner box [A] in Figure 1, opened by clicking the bar, shows the detailed breakdown of the annotations. The first and the second bars correspond to the at least ordered regions, and the at least disordered regions, respectively. All of the data sources supporting each of order/disorder regions can be presented by cricking the ‘majority rule’ bar explained below.
The bar at [3] shows the summary of ordered/disordered regions by the ‘majority rule’, in which majority decision is employed to show the annotation. The inner-box (B), opened by clicking the bar, shows all the evidence of annotations used to in the majority vote. They include ordered and disordered regions derived from the literature and PDB structures. The experimental methods supporting the order/disorder regions (‘X-ray’, ‘NMR’, etc) are also shown together with the links to the PubMed Abstracts (‘Reference’).
A unique feature of IDEAL is the explicit description of IDRs with the ability to undergo structural transformation, the ProSs, which are shown by the green bars ([4] in Figure 1). Each of the bars expands by a click to show the ordered and disordered regions that account for the ‘verified ProS’ status [inner box (C)]. For ‘possible ProS’, only ordered regions are presented. Note that a verified ProS should match one of the conflict regions in the bar at [2].
Below the ProS annotation, the miscellaneous information from UniProt, is summarized. These bars can be clicked on to open up the detailed information shown in box [D]. The results of the domain assignment ([6] in Figure 1) show the SCOP and Pfam domains identified by the reverse PSI-Blast and HMMer. The bars show a summary of the results and expand to show the details.
The XML files
The XML files are provided and can be downloaded by clicking on the xml link button at the top right of the page [2]. A definition of the XML schema is available at http://idp1.force.cs.is.nagoya-u.ac.jp/IDEAL/help.html.
FUTURE WORK
It took about 1 year to annotate more than 120 proteins. We now plan to accelerate the annotation rate. We also expect to collect more ProSs, and investigate the interaction mechanism of the ProS. To do this, we aim to develop an interface showing the binding partner proteins associated with ProSs and to illustrate their interaction networks. As in any databases, updating the contents is a key issue. We will address this by developing an update system to keep information in IDEAL as current as possible.
FUNDING
Grant-in-Aid for Scientific Research on Innovative Areas, ‘Target recognition and expression mechanism of intrinsically disordered proteins’ from the Ministry of Education, Culture, Sports, Science, and Technology (MEXT) of Japan. Funding for open access charge: MEXT.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Masahito Umezaki and Tomoko Sato for their contributions at the beginning of the project. The authors also thank Keiichi Homma for his valuable suggestions.
REFERENCES
- 1.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 2.Uversky VN, Dunker AK. Understanding protein non-folding. Biochim. Biophys. Acta. 2010;1804:1231–1264. doi: 10.1016/j.bbapap.2010.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J. Mol. Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 4.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat. Rev. Mol. Cell. Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 5.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 6.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, et al. DisProt: the database of disordered proteins. Nucleic Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, Reid A, et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bell S, Klein C, Muller L, Hansen S, Buchner J. p53 contains large unstructured regions in its native state. J. Mol. Biol. 2002;322:917–927. doi: 10.1016/s0022-2836(02)00848-3. [DOI] [PubMed] [Google Scholar]
- 11.Dawson R, Muller L, Dehner A, Klein C, Kessler H, Buchner J. The N-terminal domain of p53 is natively unfolded. J. Mol. Biol. 2003;332:1131–1141. doi: 10.1016/j.jmb.2003.08.008. [DOI] [PubMed] [Google Scholar]
- 12.Kumar R, Betney R, Li J, Thompson EB, McEwan IJ. Induced alpha-helix structure in AF1 of the androgen receptor upon binding transcription factor TFIIF. Biochemistry. 2004;43:3008–3013. doi: 10.1021/bi035934p. [DOI] [PubMed] [Google Scholar]
- 13.Lee H, Mok KH, Muhandiram R, Park KH, Suk JE, Kim DH, Chang J, Sung YC, Choi KY, Han KH. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J. Biol. Chem. 2000;275:29426–29432. doi: 10.1074/jbc.M003107200. [DOI] [PubMed] [Google Scholar]
- 14.Nagadoi A, Nakazawa K, Uda H, Okuno K, Maekawa T, Ishii S, Nishimura Y. Solution structure of the transactivation domain of ATF-2 comprising a zinc finger-like subdomain and a flexible subdomain. J. Mol. Biol. 1999;287:593–607. doi: 10.1006/jmbi.1999.2620. [DOI] [PubMed] [Google Scholar]
- 15.Receveur-Brechot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins. 2006;62:24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- 16.Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B(betabeta) Nat. Struct. Biol. 2000;7:570–574. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- 17.UniProt_Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukuchi S, Hosoda K, Homma K, Gojobori T, Nishikawa K. Binary classification of protein molecules into intrinsically disordered and ordered segments. BMC Struct. Biol. 2011;11:29. doi: 10.1186/1472-6807-11-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs) J. Mol. Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 21.Gould CM, Diella F, Via A, Puntervoll P, Gemund C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, Chica C, et al. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 2010;38:D167–D180. doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tompa P, Fuxreiter M, Oldfield CJ, Simon I, Dunker AK, Uversky VN. Close encounters of the third kind: disordered domains and the interactions of proteins. Bioessays. 2009;31:328–335. doi: 10.1002/bies.200800151. [DOI] [PubMed] [Google Scholar]
- 23.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gough J, Karplus K, Hughey R, Chothia C. Assignment of homology to genome sequences using a library of hidden Markov models that represent all proteins of known structure. J. Mol. Biol. 2001;313:903–919. doi: 10.1006/jmbi.2001.5080. [DOI] [PubMed] [Google Scholar]