Abstract
This work presents the Apo–Holo DataBase (AH-DB, http://ahdb.ee.ncku.edu.tw/ and http://ahdb.csbb.ntu.edu.tw/), which provides corresponding pairs of protein structures before and after binding. Conformational transitions are commonly observed in various protein interactions that are involved in important biological functions. For example, copper–zinc superoxide dismutase (SOD1), which destroys free superoxide radicals in the body, undergoes a large conformational transition from an ‘open’ state (apo structure) to a ‘closed’ state (holo structure). Many studies have utilized collections of apo–holo structure pairs to investigate the conformational transitions and critical residues. However, the collection process is usually complicated, varies from study to study and produces a small-scale data set. AH-DB is designed to provide an easy and unified way to prepare such data, which is generated by identifying/mapping molecules in different Protein Data Bank (PDB) entries. Conformational transitions are identified based on a refined alignment scheme to overcome the challenge that many structures in the PDB database are only protein fragments and not complete proteins. There are 746 314 apo–holo pairs in AH-DB, which is about 30 times those in the second largest collection of similar data. AH-DB provides sophisticated interfaces for searching apo–holo structure pairs and exploring conformational transitions from apo structures to the corresponding holo structures.
INTRODUCTION
Interactions between proteins and other molecules play an important role in living cells. Many studies have shown that proteins usually undergo conformational transitions upon binding to other molecules (1–5). Such transitions can be broadly categorized into three types: (i) secondary structure transitions in which residues change their secondary structures, such as from α-helix to β-strand, upon binding; (ii) disorder/order transitions in which disordered regions acquire stable structures upon binding and (iii) other spatial motions in which protein segments move around flexible linkers. These conformational transitions have been identified in various proteins [such as hormones (6), proteinase inhibitors (7), prion peptides (8) and ribosomal subunits (9)] and are involved in many important biological processes [such as catalytic processes (10), DNA replication (11), ligand binding (5), protein–protein recognition (12), signal transduction (13) and transcriptional regulation (14)]. The studies of conformational transitions rely greatly on comparing the structure of a protein before binding (apo structure) with that after binding (holo structure). A comprehensive library of apo–holo structure pairs is useful in studying conformational transitions and those biological processes in which they are involved.
Currently available resources of apo–holo structure pairs, despite of their wide usage, are rather short. Most studies describe a preparation procedure but do not provide important intermediate data, such as alignments of paired structures. Hence, subsequent researchers have to compile their own apo–holo structure pairs from scratch and may not be able to reproduce the same collection owing to revisions of the Protein Data Bank (PDB) database (15). Another issue is one of scale. Many studies adopt only tens or hundreds of pairs, which do not suffice for a large-scale analysis. In these small-scale collections, a protein is typically associated with a single apo–holo structure pair, ignoring the fact that the same protein may interact with different molecules via different binding sites (16). A more serious problem in existing apo–holo collections is that preparation procedures vary from study to study owing to various requirements of different analyses, such as those for protein–protein (5,17), protein–DNA (18) and protein–peptide interactions (19). Furthermore, the selection of a representative apo–holo structure pair of a protein may involve manual intervention, making the preparation complicated and more difficult to repeat. Therefore, an easy-to-use platform to solve these problems that is applicable to a wide range of large-scale analyses is greatly desirable.
This work presents the Apo–Holo DataBase (AH-DB), which provides sophisticated interfaces for searching apo–holo structure pairs and exploring conformational transitions from apo structures to the corresponding holo structures. AH-DB contains 746 314 apo–holo structure pairs of 3638 proteins from 702 organisms. It categorizes molecules into proteins, nucleic acids, ligands and ions and identifies/maps molecules in different PDB entries to generate pairs of apo and holo structures. Conformational transitions of secondary structure and disorder/order are identified based on a refined alignment scheme to overcome the challenge that many structures in the PDB database are only protein fragments and not complete proteins. The interface of AH-DB provides much flexibility for searching apo–holo structure pairs and a highly interactive means of exploring the paired structures. Users can search AH-DB by proteins, binding partners and miscellaneous constraints, such as formation of homo-dimers upon binding. Additionally, AH-DB is the first platform that allows users to find bindings with molecules of multiple types: for example, users can easily extract apo–holo structure pairs upon binding with both DNAs and ions. AH-DB also provides an exploratory tool using which researchers can quickly see the apo and holo structures superimposed on each other in space. The highly integrated interface enables users to choose conformational transitions of interest and instantly see them in sequence and in structure. All of the identified conformational transitions and important intermediate data, such as sequence/structure alignments, can be downloaded for further analysis.
DATA COLLECTION
The construction of AH-DB comprises two stages: molecular mapping and residue mapping. The molecular mapping refers to procedures for pairing protein structures, called ‘chains’ in PDB, among different PDB entries; residue mapping refers to the alignment of two paired protein structures and identification of conformational transitions.
Molecular mapping
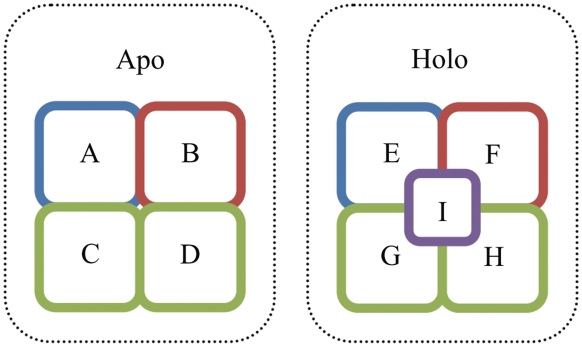
AH-DB first collects protein chains from all PDB entries, each of which represents a complex structure, that are determined by X-ray diffraction and by nuclear magnetic resonance (NMR). If two protein chains in two complexes are equivalent, they can be selected as ‘target protein’ and the other molecules of the complex pair can be split as ‘core molecules’ that in both complexes and ‘added molecules’ that in only one complex (Figure 1). Here two protein chains are equivalent if their sequence alignment by BLAST (20) satisfies that: (i) start and end at sequence ends (full-length alignment), (ii) has no gap and (iii) e-value < 0.001. A candidate of an apo–holo structure pair of the target protein is formed if ‘apo complex’ has no added molecule and ‘holo complex’ has at least an added molecule. The added molecules are potential binding partners of the target protein. For accommodating compounds that require assistant molecules to facilitate the structure determination, AH-DB also provides apo–holo structure pairs in which the mapping of ligands and/or ions is ignored. Ligands/ions are extracted from the HETATM records in PDB files of which the equivalence are identified by their identifiers in PDB. Pseudo ligands, such as water molecules and selenomethionines, are excluded. The list of pseudo ligands used in AH-DB can be found in the help page (http://ahdb.ee.ncku.edu.tw/help.html#pseudo_ligand). Because nucleic acids lack a unique identifier, any nucleic acid is regarded equivalent in AH-DB. According to the aforementioned definitions, two complexes in a group may produce multiple candidates of apo–holo structure pairs as shown in Figure 1. These candidates are further screened in the residue mapping.
Figure 1.
Schema of molecular elements in AH-DB. This figure shows an apo complex of four apo structures of molecules A, B, C and D; a holo complex of four holo structures of molecules E, F, G, H upon binding with I. Molecules with identical color indicate that they are equivalent. For example, A–E is a candidate apo–holo structure pair. If the molecule A/E is the ‘target protein’, the remaining molecules in both complexes (B, C, D, F, G and H) are called ‘core molecules’ and the molecules in only holo complex (I) is called ‘added molecules’. If C/D/G/H is the target protein, all of C–G, C–H, D–G and D–H pairs are formed with different core molecules. For example, the corresponding core molecules of C–G pair are A, B, D, E, F and H.
Residue mapping
This step aims to map residues of paired target protein chains and identify conformational transitions upon binding the corresponding added molecules. The greatest challenge in this step is that the paired structures may contain only fragments—with, for example, fewer than 30 residues—of the target protein, which may comprise hundreds of residues. Directly aligning these two sequence fragments may lead to incorrect local alignments. Thus, the alignment scheme is refined by introducing the complete protein sequence that is obtained from the UniProt database (21) as a bridge between the two sequences that are obtained from PDB entries. The two PDB sequences are aligned with the UniProt sequence using BLAST (20). A candidate of an apo–holo structure pair is removed if: (i) either sequence falls outside the UniProt sequence boundaries or (ii) the mapped regions of the PDB sequences to the UniProt sequence do no overlap. Finally, AH-DB contains 746 314 apo–holo pairs of 3638 proteins from the PDB release of 2 March 2011. Table 1 shows the detailed statistics of AH-DB.
Table 1.
Number of apo–holo structure pairs in AH-DB
| #Pairsa | #Non-redundant pairsb | #Proteinsc | |
|---|---|---|---|
| Consider ligand and ion mappingd | 296 208 | 18 395 | 2836 |
| Ignore ligand mappinge | 426 464 | 11 315 | 2528 |
| Ignore ion mappingf | 362 042 | 18 227 | 2987 |
| Ignore ligand and ion mappingg | 292 345 | 10 966 | 2032 |
| Unionh | 746 314 | 26 517 | 3638 |
aNumber of apo–holo structure pairs.
bNumber of apo–holo structure pairs with distinct target protein and added molecules, namely redundant pairs that have identical target protein and added molecules are removed.
cNumber of proteins involving in the apo–holo structure pairs.
dBoth ligand and ion mapping are considered while pairing complexes.
eLigand mapping is ignored while pairing complexes.
fIon mapping is ignored while pairing complexes.
gBoth ligand and ion mapping are ignored while pairing complexes.
hUnion of the above four conditions of ligand/ion mapping.
Next, the secondary structures and disorder/order states of residues are assigned to identify the conformational transitions that occur upon binding. The definition of transitions between a structure pair is identical to (4). Secondary structures are assigned using the DSSP algorithm (22), according to hydrogen bond patterns that are derived from the atomic coordinates. Each residue is labeled as one of the three types of secondary structure elements (SSEs)—alpha-helix (H), beta-strand (E) and coil (C). A conformational transition of secondary structure is identified if the two residues in the apo and holo structures that are mapped to the same residue in the UniProt sequence have different secondary structures. According to the secondary structure, this transition can be further split into sub-categories, such as helix-to-sheet, sheet-to-coil and others. Another conformational transition is the disorder transition. Residues in the ATOM records (even with zero occupancy) of a PDB entry are regarded as ordered; residues in the SEQRES but not in the ATOM records are regarded as disordered. A disorder-to-order transition is identified if a residue in the apo structure is disordered and the corresponding residue in the holo structure is ordered.
Finally, AH-DB adopts two algorithms to generate the superimposition of the apo and holo structures of a target protein according to the residue mapping obtained in the previous steps. The set of residue pairs in an apo–holo structure pair are regarded as a set of paired vectors in the 3D vector space, where a residue is represented by its alpha carbon. For a NMR structure, the coordinates of the first model is used. The first algorithm (23) is a conventional least-square method that minimizes root-mean-square deviation (RMSD); while the second algorithm, THESEUS (24), is a maximum likelihood method that down-weights variable structural regions for a better superimposition.
DATABASE INTERFACE
The home page provides various search functions for extracting AH-DB data. Users can search for apo–holo structure pairs by the target protein, added molecules and miscellaneous constraints, such as the requirement that the added molecules must contain a protein that is identical to the target protein, namely the target protein and the added protein form a homo-dimer. Multiple keywords and logical operators (AND, OR and NOT) of them are allowed. In AH-DB, users can easily extract apo–holo structure pairs upon binding multiple types of molecules, such as binding with both DNAs and ions, and those undergo conformational transitions upon binding. The apo–holo structure pairs that satisfy the user-specified conditions will be listed on the next page with basic information, including the target protein, composition of the added molecules, conformational transitions and resolution of structure determination. Users can sort by any combination of these fields to select, for example, the pair that undergoes the most conformational transitions or that has the best resolution of structure determination. This page also allows users to download all intermediate data, including sequences in the FASTA format, DSSP results, PDB entries and sequence alignment into an archive for further analysis.
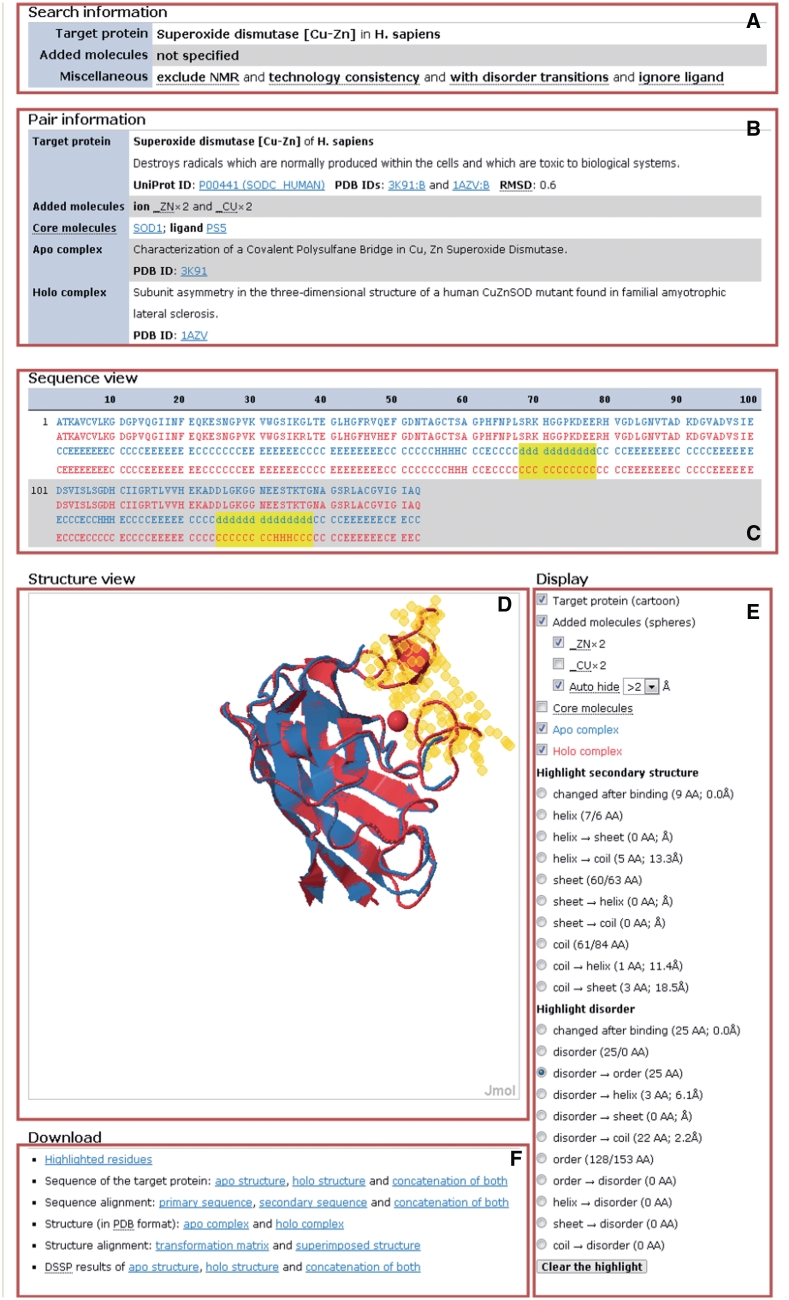
Clicking the ‘view’ links takes users to the ‘pair page’ (Figure 2), which shows details of an apo–holo structure pair. The pair page consists of six areas. The search information (Figure 2a) shows user-specified conditions imposed at the beginning of the search. The pair information (Figure 2b) is basic information about the molecular elements (target protein, added molecules, core molecules, apo complex and holo complex) of the selected apo–holo structure pair, including the organism, names, functional annotations and links to other databases. The sequence view (Figure 2c) shows the alignment of the primary and secondary structures of the target protein in the apo and holo structures. The structure view (Figure 2d) uses a Jmol plug-in (available at http://www.jmol.org/) to render the superimposed structure of the apo and holo complexes. The most powerful function of the sequence and structure views is the instantly response to users’ input in the ‘display’ area (Figure 2e). The display area provides controls to adjust the presence and absence of each molecular element in the structure view. Moreover, users can highlight residues with a specific secondary structure, disorder/order state, or specific conformational transitions upon binding. The display area provides the quantity of the highlighted residues and their distances, defined by the closest heavy atom pairs, to the added molecules. The highlighted residues are immediately displayed in the sequence and structure views and can be downloaded in the download area (Figure 2f). The distances between the added molecules and the highlighted residues are also updated as the added molecules are changed. This sophisticated interface is necessary to allow users more comprehensively to explore the complicated relations among molecules and conformational transitions. The download area provides various data, such as the superimposed structure, to download.
Figure 2.
Pair page in AH-DB. (A) Search information; (B) pair information; (C) sequence view; (D) structure view, in which apo complex is colored blue, holo complex is colored red and added molecules are rendered as spheres; (E) display controls; (F) download links.
CASE STUDY
The metal-binding loop of copper–zinc superoxide dismutase (SOD1), which destroys free superoxide radicals in the body, has been shown have disorder-to-order transition after binding the ions (25). This section uses superoxide dismutase as an example to demonstrate the usage of AH-DB. In this sample, ‘superoxide dismutase’ was used as the keyword of target protein and ‘copper AND zinc’ was used as the keyword of added molecules to ask that the added molecules must contain both copper and zinc ions. Furthermore, the ‘exclude NMR’ constraint was enabled to exclude NMR structure, the ‘with disorder transitions’ constraint was enabled to focus on disorder transitions and the ‘ignore ligand’ constraint was enabled to screened unwanted added molecules such as malonate ions. In the page of search results, users can sort the apo–holo structure pairs by number of disorder/order transitions having greater than or equal to five residues (Td) or the worse resolution of the apo and holo structures (Qr).
In this example, the first apo–holo structure pair was selected (3K91:B-1AZV:B, Figure 2). The structure of 3K91 is an apo structure of a single SOD1 dimer used to characterize the covalent polysulfane bridge between the two subunits (26). The structure of 1AZV is a metal-bound structure used to analyze SOD1's Cu and Zn binding sites (27). Figure 2b shows that the added molecules are two copper ions and two zinc ions, as expected for a SOD1 dimer. Figure 2e shows that this apo–holo structure pair contains 25 residues undergo disorder-to-order transitions upon binding the copper and zinc ions. Clicking the radio button of ‘disorder → order’ highlights the 25 residues in sequence and in structure. The sequence view (Figure 2c) shows the 25 residues locate in two segments (68–78 and 125–138). This result perfectly matches the study that was performed by Galaleldeen et al. (25), in which the authors used another metal-bound structure (2C9V) and had to crystallize two metal-free structures for their analysis. The structure view (Figure 2e) shows that the apo structure (colored blue) lacks stable structures of the highlighted residues for Jmol to display, and with the holo structure (colored red) superimposed together, users can easily figure out that the two disordered segments are close to a zinc ion.
DATABASE STATISTICS AND COMPARISON WITH OTHER STUDIES
Although conformational transitions have been discussed in the literature for decades, the first large-scale analysis of the phenomenon was conducted in a surprisingly recent study of disorder transitions by Fong et al. (28). The used dataset was then refined to construct the ComSin database (29), which is the only collection of structure pairs with >10 000 entries. ComSin has 24 910 entries—about 1/30th of the number in AH-DB and focuses on only disorder transitions in protein–protein interactions. These differences show the advantages of AH-DB in both scale and flexibility to meet various requirements of different analyses. This section provides some statistics about AH-DB that elucidate the difference in scale between AH-DB and other collections of apo–holo structure pairs.
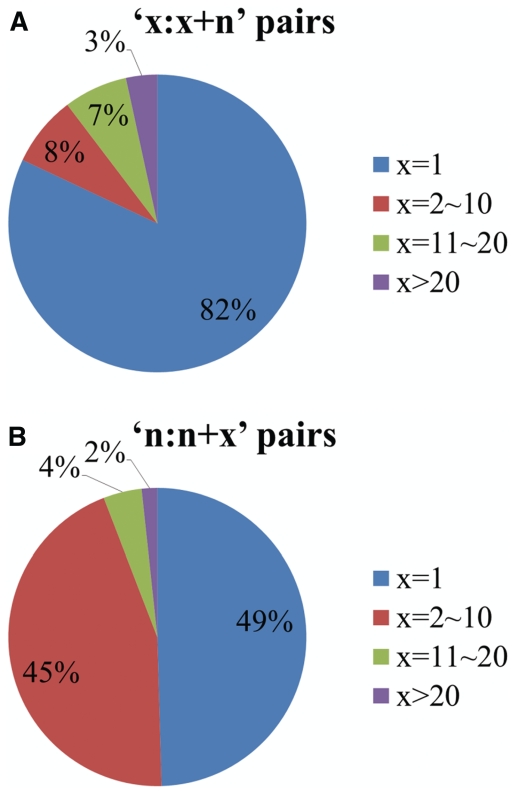
AH-DB has 236 732, 9129, 197 279 and 194 204 apo–holo pairs whose added molecules contain only proteins, nucleic acids, ligands and ions, respectively. The added molecules of the remaining 108 970 apo–holo pairs contain at least two types of molecules. The quantity of pairs with only added nucleic acids, representing protein-nucleic acid interactions, is relatively small, whereas those of the other interactions are much larger and similar to each other. This finding is reasonable owing to the difficulty of determination of the structures of protein–nucleic acid complexes, and does not indicate that proteins interact less with nucleic acids. The 236 732 pairs of protein–protein interactions are far more in number than those in ComSin, suggesting that factors other than considering more molecule types contribute to the scale of AH-DB. Conventional studies have favored controlling the size of the apo complex and/or the quantity of the added molecules to simplify the analysis (4,5,18,28,29). For example, the most frequent constraint is to allow the apo complex to be only a protein monomer, such that identification of the added molecules (binding partners) becomes trivial. Such apo–holo structure pairs are denoted herein as ‘1:1 + n’, where 1 refers to the size of the apo complex and n > 0 represents the number of added molecules. Another frequently imposed constraint is to allow only one molecule to be added, denoted herein as ‘n:n + 1’. The two constraints can also be imposed together to generate ‘1:1 + 1’ apo–holo structure pairs. Here we focused on the pairs whose added molecules contain only proteins. Figure 3 shows the ratio of pairs of different sizes. The ‘1:1 + n’ and ‘n:n + 1’ pairs account for the majority of structure pairs. The 92 381 ‘1:1’ pairs is about three times that in the ComSin database. The rest of the quantity gap results from the fact that the minimum number of contact residues is limited in ComSin.
Figure 3.
Ratio of apo–holo structure pairs of different sizes. (A) Ratio of pairs versus size of apo complex. (B) Ratio of pairs versus number of added molecules.
The aforementioned constraints, such as ‘1:1’, are frequently applied because many studies focus on accuracy, rather than scale, in their analyses. In this regard, AH-DB contains apo–holo structure pairs whose added molecules are questionable. Thus, AH-DB provides many constraints for users to control the extracted apo–holo structure pairs. This design enables AH-DB to meet various analyses of different studies. It also explains the preference of AH-DB for the provision of redundant data rather than for simply filtering them out at risk of losing valuable data. However, including all potential constraints is almost impossible. In this regard, AH-DB is more suitable for use as a first step of data preparation, as it reduces the time taken to extract data from PDB and the basic pairing operations from days (a very conservative estimate) to seconds. AH-DB also enables users to download required data, such as sequences, to perform their own filters such as redundancy elimination. Furthermore, the collection process can be changed to further enlarge AH-DB. For example, the ComSin database utilizes domains to identify targets, which can pair proteins that merely share a common domain. The criteria that govern the molecular mapping cannot be easily changed by applying constraints and doing so requires an extensive refinement of the database architecture. In this regard, AH-DB and ComSin complement each other. Finally, the interface for exploring apo–holo structure pairs in AH-DB is clearly advanced. Even if an apo–holo structure pair is identified outside the AH-DB, viewing it in AH-DB is always worthwhile.
CONCLUSION
This work presents the AH-DB database, which provides comprehensive and highly customizable collections of protein structure pairs before and after binding. AH-DB provides more than the collections of apo–holo structure pairs in previous studies, in three respects: (i) scale, (ii) flexibility to meet various requirements and (iii) interface for exploring apo and holo structures. It will be updated monthly. The data in the AH-DB database support analyses of protein disorder, secondary structure transition, catalysis, translational regulation and molecular dynamics. As the structure determination techniques continue to be improved, AH-DB has the potential to greatly expedite and extend analyses in related fields.
FUNDING
The National Science Council, Taiwan (NSC 99-2628-E-006-017). Funding for open access charge: National Science Council of Taiwan (NSC 99-2628-E-006-017).
Conflict of interest statement. None declared.
REFERENCES
- 1.Urbauer JL, Short JH, Dow LK, Wand AJ. Structural analysis of a novel interaction by calmodulin: high-affinity binding of a peptide in the absence of calcium. Biochemistry. 1995;34:8099–8109. doi: 10.1021/bi00025a016. [DOI] [PubMed] [Google Scholar]
- 2.Kim TD, Ryu HJ, Cho HI, Yang CH, Kim J. Thermal behavior of proteins: heat-resistant proteins and their heat-induced secondary structural changes. Biochemistry. 2000;39:14839–14846. doi: 10.1021/bi001441y. [DOI] [PubMed] [Google Scholar]
- 3.Michel EF. Molecular dynamics simulations reveal a disorder-to-order transition on phosphorylation of smooth muscle myosin. Biophys. J. 2007;93:2083–2090. doi: 10.1529/biophysj.106.095802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dan A, Ofran Y, Kliger Y. Large scale analysis of secondary structure changes in proteins suggests a role for disorder to order transitions in nucleotide binding proteins. Proteins. 2010;78:236–248. doi: 10.1002/prot.22531. [DOI] [PubMed] [Google Scholar]
- 5.Goh CS, Milburn D, Gerstein M. Conformational changes associated with protein-protein interactions. Curr. Opin. Struct. Biol. 2004;14:104–109. doi: 10.1016/j.sbi.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Kim Y, Rose CA, Liu Y, Ozaki Y, Datta G, Tu AT. Ft IR and near infared FT Raman studies of the secondary structure of insulinotropin in the solid state: α helix to β sheet conversion induced by phenol and/or by high shear force. J. Pharm. Sci. 1994;83:1175–1180. doi: 10.1002/jps.2600830819. [DOI] [PubMed] [Google Scholar]
- 7.Boden M, Bailey TL. Identifying sequence regions undergoing conformational change via predicted continuum secondary structure. Bioinformatics. 2006;22:1809. doi: 10.1093/bioinformatics/btl198. [DOI] [PubMed] [Google Scholar]
- 8.Levy Y, Hanan E, Solomon B, Becker OM. Helix coil transition of PrP106¡V126: molecular dynamic study. Proteins. 2001;45:382–396. doi: 10.1002/prot.1157. [DOI] [PubMed] [Google Scholar]
- 9.Roll-Mecak A, Cao C, Dever TE, Burley SK. X-ray structures of the universal translation initiation factor IF2/eIF5B: conformational changes on GDP and GTP binding. Cell. 2000;103:781–792. doi: 10.1016/s0092-8674(00)00181-1. [DOI] [PubMed] [Google Scholar]
- 10.Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 11.Russel D, Lasker K, Phillips J, Schneidman-Duhovny D, Velazquez-Muriel JA, Sali A. The structural dynamics of macromolecular processes. Curr. Opin. Cell Biol. 2009;21:97–108. doi: 10.1016/j.ceb.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KFA, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 13.Bai F, Branch RW, Nicolau DV, Pilizota T, Steel BC, Maini PK, Berry RM. Conformational spread as a mechanism for cooperativity in the bacterial flagellar switch. Science. 2010;327:685. doi: 10.1126/science.1182105. [DOI] [PubMed] [Google Scholar]
- 14.Liu J, Perumal NB, Oldfield CJ, Su EW, Uversky VN, Dunker AK. Intrinsic disorder in transcription factors. Biochemistry. 2006;45:6873–6888. doi: 10.1021/bi0602718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prli A, Quesada M, Quinn GB, Westbrook JD. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim R, Guo J-T. PDA: an automatic and comprehensive analysis program for protein-DNA complex structures. BMC Genomics. 2009;10 doi: 10.1186/1471-2164-10-S1-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Meszaros B, Simon I, Dosztanyi Z. The expanding view of protein–protein interactions: complexes involving intrinsically disordered proteins. Phys. Biol. 2011;8:035003. doi: 10.1088/1478-3975/8/3/035003. [DOI] [PubMed] [Google Scholar]
- 18.Gao M, Skolnick J. DBD-Hunter: a knowledge-based method for the prediction of DNA–protein interactions. Nucleic Acids Res. 2008;36:3978. doi: 10.1093/nar/gkn332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.London N, Movshovitz-Attias D, Schueler-Furman O. The structural basis of peptide-protein binding strategies. Structure. 2010;18:188–199. doi: 10.1016/j.str.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 20.Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Apweiler R, Martin M, O'Donovan C, Magrane M, Alam-Faruque Y, Antunes R, Barrell D, Bely B, Bingley M, Binns D. The universal protein resource (UniProt) in 2010. Nucleic Acids Res. 38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z. Iterative point matching for registration of free-form curves and surfaces. Int J Comput Vision. 1994;13:119–152. [Google Scholar]
- 24.Theobald DL, Wuttke DS. THESEUS: maximum likelihood superpositioning and analysis of macromolecular structures. Bioinformatics. 2006;22:2171. doi: 10.1093/bioinformatics/btl332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Galaleldeen A, Strange RW, Whitson LJ, Antonyuk SV, Narayana N, Taylor AB, Schuermann JP, Holloway SP, Hasnain SS, Hart PJ. Structural and biophysical properties of metal-free pathogenic SOD1 mutants A4V and G93A. Arch. Biochem. Biophys. 2009;492:40–47. doi: 10.1016/j.abb.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.You Z, Cao X, Taylor AB, Hart PJ, Levine RL. Characterization of a covalent polysulfane bridge in copper-zinc superoxide dismutase. Biochemistry. 2010;49:1191–1198. doi: 10.1021/bi901844d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Eisenberg D, Hart PJ, Liu H, Pellegrini M, Nersissian AM, Gralla EB, Valentine JS. Subunit asymmetry in the three dimensional structure of a human CuZnSOD mutant found in familial amyotrophic lateral sclerosis. Protein Sci. 1998;7:545–555. doi: 10.1002/pro.5560070302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fong JH, Shoemaker BA, Garbuzynskiy SO, Lobanov MY, Galzitskaya OV, Panchenko AR. Intrinsic disorder in protein interactions: insights from a comprehensive structural analysis. PLoS Comput. Biol. 2009;5:e1000316. doi: 10.1371/journal.pcbi.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lobanov MY, Shoemaker BA, Garbuzynskiy SO, Fong JH, Panchenko AR, Galzitskaya OV. ComSin: database of protein structures in bound (complex) and unbound (single) states in relation to their intrinsic disorder. Nucleic Acids Res. 2010;38:D283. doi: 10.1093/nar/gkp963. [DOI] [PMC free article] [PubMed] [Google Scholar]