Abstract
T-DNA insertion mutants are very valuable for reverse genetics in Arabidopsis thaliana. Several projects have generated large sequence-indexed collections of T-DNA insertion lines, of which GABI-Kat is the second largest resource worldwide. User access to the collection and its Flanking Sequence Tags (FSTs) is provided by the front end SimpleSearch (http://www.GABI-Kat.de). Several significant improvements have been implemented recently. The database now relies on the TAIRv10 genome sequence and annotation dataset. All FSTs have been newly mapped using an optimized procedure that leads to improved accuracy of insertion site predictions. A fraction of the collection with weak FST yield was re-analysed by generating new FSTs. Along with newly found predictions for older sequences about 20 000 new FSTs were included in the database. Information about groups of FSTs pointing to the same insertion site that is found in several lines but is real only in a single line are included, and many problematic FST-to-line links have been corrected using new wet-lab data. SimpleSearch currently contains data from ∼71 000 lines with predicted insertions covering 62.5% of the 27 206 nuclear protein coding genes, and offers insertion allele-specific data from 9545 confirmed lines that are available from the Nottingham Arabidopsis Stock Centre.
INTRODUCTION
Since the genome sequence of the model plant Arabidopsis thaliana was completed in the year 2000 (1), the determination of gene function is a key task in the Arabidopsis research community. Insertional mutagenesis approaches have been proven to be a valuable tool for reverse genetics (2). Several large collections of A. thaliana lines containing independent insertions of Agrobacterium T-DNA in the plant genome have been established. T-DNA integration in the plant genome results in stable mutations, which may perturb gene functions. An important goal is to saturate the A. thaliana genome with T-DNA insertion lines for each (or at least most) of the 27 206 nuclear protein-coding genes that are currently annotated (3). A popular strategy to determine the position of a T-DNA insertion in the genome of a given insertion line is to generate Flanking Sequence Tags (FSTs). Using PCR-based methods, genomic DNA fragments flanking the T-DNA are amplified (4), sequenced and subsequently mapped to the genome. When this is applied to a large collection of lines, the population can easily be searched for mutants of interest.
GABI-Kat is the second largest FST-indexed T-DNA insertion line collection of A. thaliana, which is publicly available since 2002 (5). User access to the collection and extensive metadata for the included mutants and alleles is provided by GABI-Kat SimpleSearch, the web interface of the corresponding database (6). The interface can either be used to search the collection for mutant alleles of interest, or more importantly to access different kinds of information about specific GABI-Kat lines. Lines containing insertions of interest can be ordered via a web order form, which is also provided in SimpleSearch. In contrast to other FST databases, GABI-Kat SimpleSearch offers information on confirmation success of lines along with sequences derived from the T-DNA/genome junction of an offspring generation, segregation data and primer information for the respective insertions (7).
Since its availability in 2002, the database as well as the interface has been continuously improved. However, during the last 18 months several significant improvements and extensions have been implemented that are beneficial for users that rely on SimpleSearch when working with GABI-Kat insertion alleles. These enhancements include an extended data set, allow an easier and more comfortable user access, and provide more detailed and more reliable meta-information about the insertion alleles in the GABI-Kat collection. In detail, the recent improvements are (i) an update to the most recent Ath genome annotation dataset TAIRv10, (ii) an improved insertion site prediction and gene hit definition, (iii) enhanced information about confirmation success and line availability. Together with the correction of problematic FST-to-line links using new experimental data, these enhancements result in an increased overall quality and reliability of the collection and its database.
RESULTS AND DISCUSSION
General database content
The SimpleSearch database contains data about GABI-Kat insertion mutants, the derived FSTs and their mapping to the A. thaliana genome sequence. In addition and in contrast to other FST databases like SIGnAL (8) or FLAGdb++ (9), SimpleSearch focuses on metadata concerning GABI-Kat lines and insertion alleles. The database contains data about confirmation attempts for predicted insertion alleles, genetic segregation data of the resistance phenotype provided by the T-DNA allowing an estimation of the number of insertion loci per line, and sequences of successfully confirmed alleles from the offspring generation that allow to determine the real site of insertion much more accurately than the crude FST sequences. Moreover, the user is provided with information about line availability (alleles in dead lines are lost although the respective FST exists in databases), and about insertions that could not be confirmed (failed insertions, which are predicted from FSTs but are not existing in following generations). The updated version of SimpleSearch offers significantly improved data quality due to intensive and ongoing quality management and manual curation (see below). GABI-Kat FSTs are produced by an adaptor-ligation PCR method (4) (see also the Methods and FAQ pages on http://www.gabi-kat.de/) and are mapped to the A. thaliana genome using BLAST (10). Users can access the FST data, select alleles of interest and place insertion requests. Upon request the GABI-Kat lines are segregated on selective medium, genomic DNA is prepared and the predicted insertion sites are confirmed by PCR and sequencing, using an insertion site specific primer and a T-DNA border primer. The obtained ‘confirmation sequences’ are again mapped to the genome. If the insertion site deduced from confirmation sequencing matches the position predicted from the respective FST, the insertion is regarded as confirmed.
Update of the database to TAIRv10
Until recently, the SimpleSearch database (7) was based upon the TIGR version 5 annotation dataset of the A. thaliana genome. The TIGRv5 annotation consisted of individual BAC sequences and contains an outdated set of gene annotation data (11). SimpleSearch has now been updated to the current genome annotation data, namely TAIR version 10, which is based on pseudochromosomes (ftp://ftp.arabidopsis.org/home/tair/home/tair/Sequences/whole_chromosomes/). In this context all FSTs from the GABI-Kat collection were newly mapped to the genome sequence, and putative insertion positions were deduced. The increase in the number of successfully mapped FSTs results from using optimized mapping parameters, reduced gaps in the genome sequence and also from the generation of new FSTs for fractions of the GABI-Kat population with formerly low FST yield.
About 20 000 new FSTs have been submitted to EMBL/GenBank. These contribute to a total of now 130 000 FSTs at EMBL/GenBank, which are also accessible via SimpleSearch. The reliability of the insertion site prediction was improved by deducing the insertion position from the annealing site of the T-DNA border-specific primer and the nucleotide positions from the original trace-file of the FST (Figure 1) (12,13). Particularly in cases where no T-DNA sequence is detectable in the FST, the prediction of the insertion positions is more accurate now. This advantage is evident when compared to insertion positions deduced from the called and trimmed sequence only (8). However, deletions at the T-DNA border to genome junction still cause errors in the prediction, which can only be resolved by detailed analysis of the confirmation sequences. Nevertheless, we now explicitly predict a defined pseudochromosome position as insertion position, and not only a locus that is described by a gene code or BAC name.
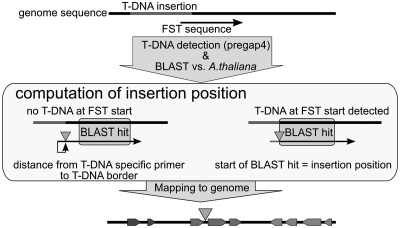
Figure 1.
Workflow of the improved insertion site prediction. The insertion position is determined using the best BLAST hit of the FST sequence vs. the A. thaliana genome sequence, and the location of the T-DNA within the FST sequence determined by pregap4. If no T-DNA is detected at the start of the FST sequence, the insertion site is located x bases upstream of the BLAST hit, where x is the number of bases before the start of the BLAST hit minus the distance of the T-DNA specific primer to the T-DNA border. Otherwise, the start of the BLAST hit is considered as insertion position.
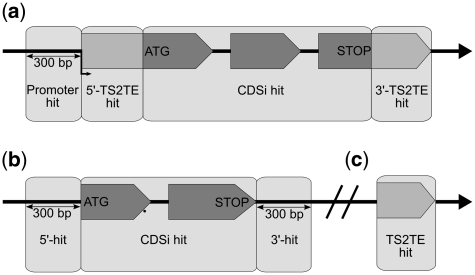
For a better assessment of the relevance of a predicted insertion allele, we used data from TAIRv10 to further qualify hits with respect to annotated genes. TAIRv10 contains information about the untranslated region of the mRNA (UTR) for the majority of the protein coding genes, as well as information about RNA-coding genes. Our former definition defined a ‘gene hit’ as an insertion site prediction between 300-bp upstream of the ATG and 300-bp downstream of the stop codon of a gene, whereas a ‘CDSi hit’ had a predicted insertion between ATG and stop codon (CDSi for coding sequence plus introns). We extended this definition by including hits in the transcribed region of a gene as well as the promoter region (Figure 2). ‘TS2TE hits’ have an insertion between transcription start and transcript end, and ‘promoter hits’ have the predicted insertion position within 300-bp upstream of the transcription start. This definition applies for protein- as well as for RNA-coding genes. If no UTR is annotated for a given gene, we still use the old gene hit definition. All hits that are not linked to a gene keep the insertion type ‘genome hit’ (Figure 2). Due to the fact that the old ‘gene hit’ definition covered a larger genome area than the TS2TE area of the genome, the total number of insertions that qualify as ‘gene hits’ went slightly down, although the number of predicted insertions in the database did increase. The updated insertion types allow a more reliable selection of insertion alleles that may be NULL alleles.
Figure 2.
Definition of gene hits at GABI-Kat. (a) For protein-coding genes with annotated UTR-regions in TAIRv10, we differentiate between CDSi hits (insertion position between ATG and STOP), 5′- and 3′-TS2TE hits (insertion position in the 5′- or 3′-UTR) and promoter hit [insertion position up to 300-bp upstream of transcription start (TS)]. (b) If the UTR is not annotated in TAIRv10 (and for pseudogenes), insertion positions 300-bp up or downstream of ATG and STOP are considered as 5′- and 3′-hits. (c) For RNA genes and transposable elements, TS2TE hits are annotated, if the insertion is located between TS and transcript end.
Overall, the total number of lines with predicted insertions was increased to 71 235. Table 1 gives a summary of the current data content of the SimpleSearch database.
Table 1.
Summary of data in the GABI-Kat SimpleSearch databasea
| Data type | Number of entries |
|---|---|
| FSTsb | ∼133 000 |
| Linesb | 71 235 |
| with segregation data | 15 289 |
| available at NASC | 9644 |
| Insertions with predicted insertion positionc | 88 580 |
| analysed with final resultd | 16 081 |
| delivered to individual users | 6816 |
| confirmed and available at NASCe | 9653 |
| Distinct genes covered | 21 005 |
| protein coding genes | 19 120 |
| ncRNA coding genes | 182 |
| pseudogenes | 420 |
| transposable element genes | 1283 |
| Gene hits available only from GABI-Katf | 2114 |
| Confirmed ‘GABI-Kat only’ hits at NASC | 1201 |
| ‘GABI-Kat only’ hits to be adressedg | 765 |
| Distinct CDSi covered | 13 037 |
aNumbers as of 15 September 2011.
bDatabase release version 24 (affects FSTs and lines that are in the database, the data values for the items in the database are updated every 24 h).
cInsertions are different from lines, because a line can contain several insertions. Example: 011F01, which is confirmed for a genome hit at F26P21 (Chr4) and a TS2TE hit in At5g05180.
dA final result can be ‘confirmed’, but also ‘failed to confirm’ or ‘part of a contamination group’ are considered.
eFor each confirmed insertion there are confirmation sequences available which are generated from the amplicon that spans the T-DNA/genome sequence junction.
fOnly hits that may cause a NULL allele (CDSi hits and hits in the 5′-UTR) are counted. Only lines in the accession Columbia-0 are considered, which is the accession used by the main FST-based insertion line collections.
gAbout 150 ‘GABI-Kat only’ alleles are either in the queue already and wait for mature T3 seed, or did fail to confirm.
Increase of quality and accuracy of the database content
Due to the manual handling steps during the generation of the GABI-Kat population, it is an unpleasant fact that some links between FST sequences and insertion lines are wrong. Reasons for these errors can be mix-ups during plant growth in the greenhouse, handling errors during sequence generation, or alike. Incorrectly assigned FST-sequences were detected when the confirmation rate in a set of 96 lines (corresponding to a microtitre plate) was much lower than expected from average values. Based on the knowledge about the FST production workflow, we deduced different types of errors that might have happened and reassigned the FST-sequences to the correct lines after wet-lab validation of the hypothesis. This reassignment of FST-sequences has meanwhile been performed for the majority of problematic microtitre plates and contributed to a significant increase (∼3%) of the confirmation rate of insertion alleles from the collection. The corrected FST-to-line connections have been integrated into the database and are available to users through SimpleSearch. It should be noted that SimpleSearch is currently the only source of these corrections. Until other FST databases have been updated, the now detected and corrected errors in the original FST dataset are still ‘proliferated’ from e.g. the old FST data in sequence databases.
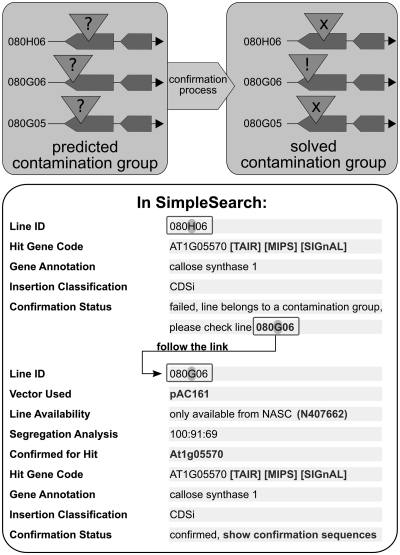
When predictions for very similar insertion sites are found in multiple lines, we combine these lines into ‘contamination groups’. The assumption is that the prediction is only true for one of those lines and the others are caused by contaminations that happened during high-throughput DNA-preparation, PCR, or sequencing. If an insertion that is part of a contamination group is ordered by a user, the whole contamination group is examined experimentally. Finally, the correct insertion is delivered to the user and donated to the Nottingham Arabidopsis Stock Centre (NASC). In some cases, several alleles are confirmed with closely linked insertion positions, and in these cases all alleles except one confirmed allele are removed from the ‘contamination group’. SimpleSearch contains information about failed insertion confirmations and leads the user to the confirmed allele within a (resolved) contamination group (Figure 3). Until September 2011 about 1250 possible contamination groups could be solved and the information is available in SimpleSearch.
Figure 3.
Resolution of contamination groups. A contamination group contains predicted insertions in different lines that share very similar insertion positions (within 50 bp at most). After the confirmation process, only one line is confirmed, the others failed and are considered as contaminations. When searching for insertion alleles in SimpleSearch, the user is guided to confirmed allele if the contamination group is solved.
As a result of various actions addressing data quality, including those mentioned above, the quality of the collection has been improved significantly. This is best quantified with the increase of the confirmation rate from 78% to 84%. Without information from the unique in-house confirmation process that is carried out at GABI-Kat, this big step towards improved reliabilty would have been impossible.
With a total of 21 005 genes that are covered with insertions [counted are ‘gene hits’ if no UTR is annotated, ‘promoter hits’ and ‘TS2TE hits’ (which include ‘CDSi hits’)], the current database release v24 covers about 4000 more genes with (predicted) insertion alleles than described earlier (7). This also includes genes for non-coding RNAs (ncRNAs), which were not annotated in TIGRv5. Due to the small size of many ncRNA-coding genes, the coverage is fairly low (182 of 1290 for ncRNA genes), which also affects the statistics of total gene hit coverage. The total coverage of the nuclear protein coding genes with (predicted) insertion alleles has increased from 64% to 70%.
Changes in the web interface
The static part of the GABI-Kat website is realised with an Open Source CMS system. SimpleSearch is embedded into this website, and the dynamic pages were developed in PHP. Information about individual items in SimpleSearch, e.g. a given line or a list of hits in a given gene, can be accessed by a defined URL format (see the help pages of the website). Both the static and the dynamic part look identical to the user. Besides visual improvement, the information content has been extended. When searching for insertions, SimpleSearch now offers an overview of insertion alleles and their availability. Lines already donated to NASC (blue triangle), insertions that failed in the confirmation process and are therefore unavailable (red triangle), and lines available for entering the confirmation process at GABI-Kat (green triangle) are distinguished. In case that a line with a failed confirmation attempt belongs into a solved contamination group, the correctly confirmed line for the insertion is linked in the SimpleSearch interface, which enables the user to access the confirmed line easily.
In addition to the existing options to search for single lines, FSTs, hits in a given gene or by BLAST, we added a search option that lists all (predicted) insertion positions in a position range on the pseudochromosomes.
Perspective
The GABI-Kat resource has served the A. thaliana community as a valuable tool for reverse genetics since it was made available to the public in 2002. The demand for GABI-Kat insertions was constantly high since then, which is documented by the number of about 72 000 stock requests at NASC until February 2011. Until September 2011 9644 different confirmed GABI-Kat lines have been donated to the stock centre. In addition to continue to confirm and deliver insertion alleles to users, we are currently addressing about 760 lines with new insertion predictions in genes for which no allele is available in the other main FST-based A. thaliana Col-0 insertion line collections.
SimpleSearch offers an easy and well-featured access to data about GABI-Kat lines and confirmed or unconfirmed insertion alleles. To maintain the quality, it is important to constantly curate the database and adopt it to the most recent knowledge about the A. thaliana genome, e.g. an upcoming v11 A. thaliana genome release. In parallel, we are also working on additional topics, which could be improved. One problem arises from (short) FSTs that are assigned to genes of which paralogous copies exist in the genome. In such cases the insertion position prediction and the assignment to a single locus is error-prone. It is a challenge to represent the ‘paralog problem’ in the database, and to address it experimentally quite some wet-lab work would be required. This example shows that there is still room for further improvement. However, the current status and the up-to-date data content of the SimpleSearch database have reached a very comprehensive level through the measures described in this article.
ACCESSION NUMBERS
About 20 000 FSTs have been submitted to EMBL/GenBank: FR799760–FR819654.
FUNDING
German Federal Ministry of Education and Research (BMBF) in the context of the German plant genomics program GABI (Förderkennzeichen 0313855). Funding for open access charge: Bielefeld University.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Yong Li, Mario Rosso, the MPI for Plant Breeding Research and all former co-workers for their contribution to GABI-Kat, and Renate Harder, Helene Schellenberg, Nina Schmidt, Andrea Voigt, Marja-Leena Wilke for technical assistance.
REFERENCES
- 1.Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 2.O’Malley RC, Ecker JR. Linking genotype to phenotype using the Arabidopsis unimutant collection. Plant J. 2010;61:928–940. doi: 10.1111/j.1365-313X.2010.04119.x. [DOI] [PubMed] [Google Scholar]
- 3.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36:D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Strizhov N, Li Y, Rosso MG, Viehoever P, Dekker KA, Weisshaar B. High-throughput generation of sequence indexes from T-DNA mutagenized Arabidopsis thaliana lines. BioTechniques. 2003;35:1164–1168. doi: 10.2144/03356st01. [DOI] [PubMed] [Google Scholar]
- 5.Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenised population (GABI-Kat) for flanking sequence tag based reverse genetics. Plant Mol. Biol. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- 6.Li Y, Rosso MG, Strizhov N, Viehoever P, Weisshaar B. GABI-Kat SimpleSearch: a flanking sequence tag (FST) database for the identification of T-DNA insertion mutants in Arabidopsis thaliana. Bioinformatics. 2003;19:1441–1442. doi: 10.1093/bioinformatics/btg170. [DOI] [PubMed] [Google Scholar]
- 7.Li Y, Rosso MG, Viehoever P, Weisshaar B. GABI-Kat SimpleSearch: an Arabidopsis thaliana T-DNA mutant database with detailed information for confirmed insertions. Nucleic Acids Res. 2007;35:D874–D878. doi: 10.1093/nar/gkl753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 9.Samson F, Brunaud V, Duchene S, De Oliveira Y, Caboche M, Lecharny A, Aubourg S. FLAGdb++: a database for the functional analysis of the Arabidopsis genome. Nucleic Acids Res. 2004;32:D347–D350. doi: 10.1093/nar/gkh134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wortman JR, Haas BJ, Hannick LI, Smith RK, Jr, Maiti R, Ronning CM, Chan AP, Yu C, Ayele M, Whitelaw CA, et al. Annotation of the Arabidopsis genome. Plant Physiol. 2003;132:461–468. doi: 10.1104/pp.103.022251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bonfield JK, Smith KF, Staden R. A new DNA sequence assembly program. Nucleic Acids Res. 1995;23:4992–4999. doi: 10.1093/nar/23.24.4992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Staden R, Beal KF, Bonfield JK. The Staden package, 1998. Methods Mol. Biol. 2000;132:115–130. doi: 10.1385/1-59259-192-2:115. [DOI] [PubMed] [Google Scholar]