Abstract
We have recently developed the Inferred Biomolecular Interaction Server (IBIS) and database, which reports, predicts and integrates different types of interaction partners and locations of binding sites in proteins based on the analysis of homologous structural complexes. Here, we highlight several new IBIS features and options. The server's webpage is now redesigned to allow users easier access to data for different interaction types. An entry page is added to give a quick summary of available results and to now accept protein sequence accessions. To elucidate the formation of protein complexes, not just binary interactions, IBIS currently presents an expandable interaction network. Previously, IBIS provided annotations for four different types of binding partners: proteins, small molecules, nucleic acids and peptides; in the current version a new protein–ion interaction type has been added. Several options provide easy downloads of IBIS data for all Protein Data Bank (PDB) protein chains and the results for each query. In this study, we show that about one-third of all RefSeq sequences can be annotated with IBIS interaction partners and binding sites. The IBIS server is available at http://www.ncbi.nlm.nih.gov/Structure/ibis/ibis.cgi and updated biweekly.
INTRODUCTION
Analysis of protein interactions is crucial for understanding the mechanisms of cell function. There are many different computational approaches to predict protein interactions but comprehensive interactome mapping for many organisms is still far from complete (1). Given that the number of structures of protein complexes increases by a few hundred every month, low-throughput and high-resolution X-ray/NMR methods can be utilized to complement and verify interactions obtained from high-throughput screens and to infer interactions for unknown proteins. A number of servers have been developed for predicting protein binding sites from structures by locating the binding pockets, by identifying sequence and structural features of homologous proteins, which are important for binding (1–8). A powerful homology inference approach to infer protein interactions has been introduced previously (9–12) and implemented in several of the most recent servers (13–15). However, annotations transferred from one homologous protein to another may result in incorrect assignment for remote homologs and even for close homologs if they have different binding specificities. To verify and guide predictions based on inference, one needs to ensure similarity between the unknown query protein and on the observed binding sites detected in homologs. Here, we offer an updated version of the Inferred Biomolecular Interactions Server (IBIS, http://www.ncbi.nlm.nih.gov/Structure/ibis/ibis.cgi) database and server (16), which imposes a number of rigorous criteria in its underlying computational methods in order to increase the reliability of homology-based inference of interactions. IBIS provides annotations of binding partners and locations of binding sites for protein–protein, protein–small molecule, protein–nucleic acid, protein–peptide and recently added protein–ion interactions. To ensure biological relevance of binding sites to the query, IBIS clusters similar binding sites found in homologous proteins based on their sequence and structure conservation, further validates them using various approaches, and finally ranks binding sites to assess how well they match the query. The IBIS user interface is designed to allow quick access to detailed information about binding sites while at the same time providing a comprehensible overview of the oftentimes complex interaction data.
There are important new features in the latest version of IBIS, which we will describe in more detail in what follows. To show interaction networks and imply protein complexes rather than only binary interactions as in the previous version, there is a ‘network graphic’ that summarizes all interactions for a given query. The different interaction categories for a query are presented in a tabular form, so a user may easily find binding sites of interest. A new search facility has been implemented so that a user can submit not only a Protein Data Bank (PDB) code (17) but also GenBank identifiers or protein accessions for sequences without known structures, which are used to search the structure database for homologous complexes. We have added a few options for easy downloads of IBIS data including an FTP site to download IBIS data for all PDB protein chains. The IBIS server is now linked to the Macromolecular Modeling Database (MMDB) structure pages that allow users to see inferred interactions for each structure entry. In this article we also describe new results concerning coverage of the set of RefSeq protein sequences by IBIS binding site annotations.
USER INTERFACE REDESIGNED
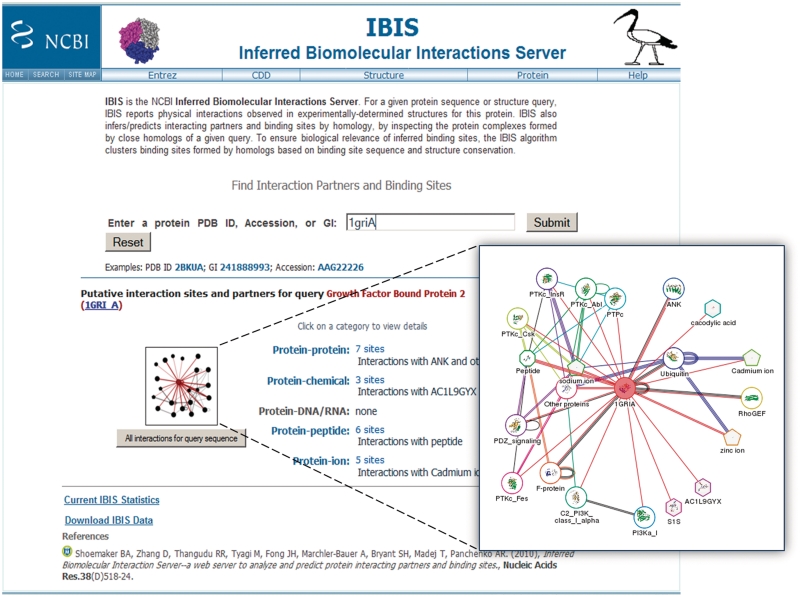
The server's webpage is redesigned to allow users easier access to data for the different interaction types. A new entry page gives a quick summary of available results and accepts protein sequence accessions. To describe the role IBIS can play in annotating and discovering putative interactions, we now give a brief walkthrough of the system as a typical user might encounter it. From the IBIS homepage, there is a single query box that accepts a protein accession, GenBank identifier or protein structure identifier (PDB accession) (Figure 1). If a protein accession is entered, an intermediate page is displayed showing the best result of a cBLAST (18) search of the protein sequence against all protein sequences with known structures. The sequence identity and fraction of this sequence aligned is shown along with a link pointing to the alignment. If this aligned range on the protein query is of interest, one proceeds by following the main link to ‘view interactions’. From here, a summary of interactions is displayed for the ‘template’ structure—homolog with the known structure closest to the query protein. This summary page gives a convenient starting point for quickly winnowing down the types of interactions of interest. For the example from Figure 1, the structure identifier ‘1griA’ has been entered and the summary shows four types of interactions: protein–protein, protein–chemical (small molecule), protein–peptide and protein–ion.
Figure 1.
IBIS interaction summary results page. The IBIS summary results page is shown for a protein accession or protein structure query entered into the search box on the page. After submitting a query, an immediate summary of results is shown below with counts and examples given for each interaction type. In addition, a summary network graphic is displayed, which can be expanded (see inset) as an alternative overview. Interaction details can be explored by clicking on an interaction type.
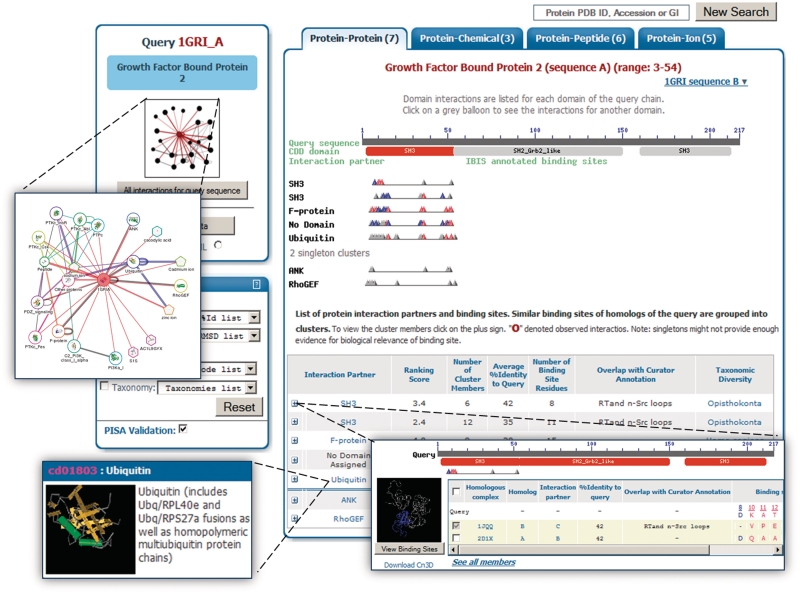
To explore these putative interactions one can follow the link of one of the types of interactions such as protein–protein. A redesigned layout of the interactions is displayed in three main boxes (Figure 2). The main box on the center-right of the page presents lists of the search results in several levels of detail. At the top is a graphic of the query sequence (if the original query does not have a known structure, the best homologous structural template serves as the query) with a line of conserved domains [CDD (19)], if present, shown in red and below that IBIS binding site annotations are displayed. The example in Figure 2 shows a query consisting of three CDD domains. Interaction partners of the first SH3 domain of the query (SH3, Ubiquitin, F-protein, RhoGEF and ANK) are shown in this figure, although one can navigate over the interaction partners for different domains by following the link to the CDD grey boxes representing the domain annotations. At the same time, the network graphic displays the combined set of interaction partners for all domains of the query protein.
Figure 2.
IBIS interaction results page. The main interaction results page is shown for the growth factor bound protein 2 structure query, 1gri. There are three main boxes on the page and this figure shows three additional boxes (larger blue borders), which can be displayed by following the links indicated by the dashed lines. The main box on the center-right has an upper sequence overview graphic of the binding sites inferred as potential interaction partners with the query. In the lower panel of the box is a table with summary information of each binding site cluster. Expanding a row of the table reveals the alignment of the binding sites. Additional information can be displayed for each interacting entity shown in the lower left. Finally, there are two boxes on the left side, the top giving an expandable network image overview and the bottom box giving several search filters for refining the results on the right.
For more information on these interactions, the table of binding site clusters below shows one expandable line for each interaction. The summary statistics on each line, when expanded, reveal that each ‘interaction’ is actually a clustered group of similar interactions. Expanding the SH3 binding site cluster shows two non-redundant structural evidences of this interaction, for example, an interaction between chains B and C coming from PDB structure 1jqq. In fact there are a total of six instances of this interaction from two structures, which can be viewed with the link ‘See all members’. For further details of an interaction interface, the expanded cluster line in the lower table includes a link to the helper application, Cn3D, for an annotated visualization of the binding surface in 3D computer graphics (http://www.ncbi.nlm.nih.gov/Structure/CN3D/cn3d.shtml). The table also helps the user assess the applicability of the binding site clusters for annotating a given query and lists ranking score, percent identity, overlap with manually curated sites and validation by the PISA (Protein Interfaces, Surfaces, and Assemblies) algorithm (20).
The third box on the interaction results page is the search refinement box in the lower left-hand side. It includes several dynamically updated options for quickly refining the result list. The result list can be refined by focusing on particular PDB structures or on particular organisms or taxonomic groups. The checkmarks are used to draw the user's attention to the appropriate lines of binding site clusters, which contain the requested items. All refinements can easily be removed or switched back and forth. Finally, the default practice of showing only interactions that have been validated by PISA can be turned off to reveal all interactions found. This can be useful to better understand all possible data that are available, but should be used with care as it permits crystal-packing interactions.
NETWORK IMAGES
To elucidate the formation of protein complexes, not just binary interactions, IBIS currently presents a network image of all types of interactions. Interactions of the binding partners of the query protein (partners of partners) are obtained using up to four representative structures taken from the binding site clusters and used as queries in IBIS. The current schematic only displays interactions between biomolecules that interact with the original query protein; to obtain comprehensive interaction data for any of these partners, one should re-query IBIS with that protein. These interaction images are found both on the initial search results summary page (Figure 1) as well as on the main interaction results page in the upper left-hand box (Figure 2).
Nodes of various shapes depict the different biomolecule types. Each protein, chemical and ion node represents interaction partners from binding site clusters and is identified by the name of the cluster representative as shown on the table of interaction partners (for proteins, this is the name of the CDD superfamily). All nucleotide partners are grouped into a single node labeled ‘DNA/RNA’. The query node is highlighted by red color and all other nodes are assigned a unique color. Edges, including self-loops, indicate interactions between protein nodes and other types of nodes. Multiple lines are drawn between two nodes to show that more than one binding site cluster has been found for that interaction, black lines are used for observed interactions and the colored lines for inferred interactions. The thumbnail version of the network has an identical layout and simplified graphics compared to the larger image. Schematics are created using the Graphviz library with Node placement computed using a force-directed layout algorithm (http://www.graphviz.org/).
NEW DATA TYPE: PROTEIN–ION INTERACTIONS
Previously, IBIS provided annotations for four different types of binding partners: proteins, small molecules, nucleic acids and peptides. Now a new interaction type is included—protein–ion interactions. This is one of the most abundant types of interactions and currently more than one hundred thousand protein chains/domains can be annotated with protein–ion interactions. Protein–ion interactions are parsed from the structure data in the same way as protein–small molecule interactions with the changes that the capture radius of the interaction is reduced from 4 Å for all other types to 3 Å for protein–ions. An ion molecule must consist of at most one non-hydrogen atom and there is no minimum on the number of protein binding site residues with an ion as was imposed for other interaction types (16). Protein–ion interactions are typically distinguished from protein–small molecule interactions by the former’s tendency toward much higher levels of conservation across many species.
NEW DATA DOWNLOAD AND ACCESS OPTIONS
The web interface of IBIS has provided a convenient way to study the interactions of a protein of interest in an interactive fashion. For more systematic studies of multiple protein queries, however, we now offer a couple of options to streamline this endeavor. We have added a few options for easy downloads of IBIS data: an FTP site to download IBIS data for all PDB protein chains; and per-query results from the website in the form of Excel and XML data files. For data download, a single archive file is created with each biweekly update and placed in an FTP directory: ftp://ftp.ncbi.nlm.nih.gov/pub/mmdb/ibis/. The directory contains a README.txt file with specific instructions, the tar file that includes individual files for each protein structure query with interactions, and one example file to better illustrate what is contained within the tar file. Statistics of all interactions are monitored and a statistics table is provided for each update on IBIS home page.
INTEGRATION WITH NCBI PROTEIN STRUCTURE SERVICES
The IBIS server is linked to the MMDB (18) structure page that allows users to see inferred interactions for each structure entry. Using NCBI's integrated Entrez search service (21), one can potentially begin from a wide variety of databases and end up looking at a protein structure record. That record gives useful information on the biological unit of the structure, and its observed interactions, but for a complete picture of all relevant, inferred interactions from structurally related proteins, there is a link in the upper-right corner of the page to IBIS. Conversely, in IBIS the full structure record can be viewed for any protein structure by following the link in its accession. This linking gives a convenient way to zoom in to a structure record for more contextual detail and to zoom out to the IBIS record for a broader picture of all relevant interactions inferred from many related structures.
IBIS APPLICATION: ANNOTATION OF ALL REFSEQ PROTEIN SEQUENCES
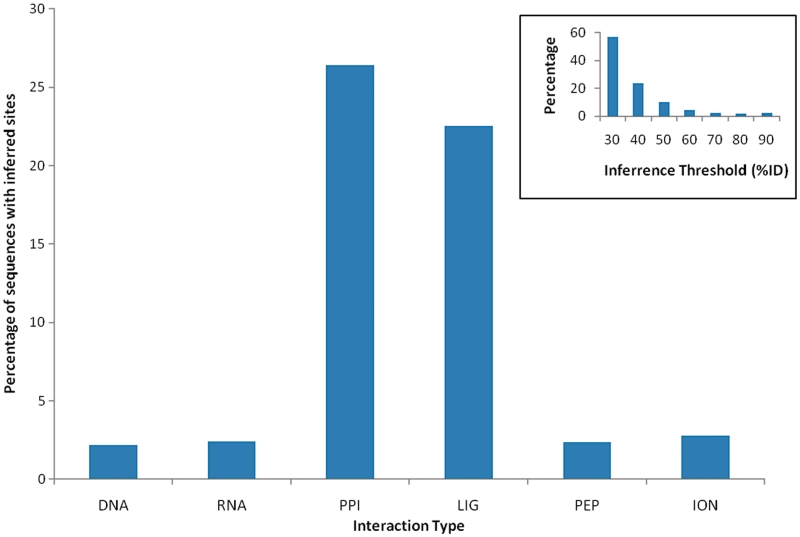
IBIS has been used to make high-quality binding site annotations on all RefSeq protein sequences (22). We took all protein sequences (12 903 605) from RefSeq release 47 and tried to annotate each sequence with IBIS binding sites. A protein sequence is annotated by IBIS if and only if the average percent identity between the query and binding site cluster members is >30%. We found that about one-third of the RefSeq proteins (3 876 072) can be annotated with at least one IBIS binding site and in total ∼49 million binding site annotations are assigned. As can be seen in Figure 3 about 20–30% of RefSeq sequences are annotated with protein–protein and protein–chemical interactions and ∼5% can be characterized by IBIS protein–DNA/RNA binding sites. Most of these annotations come from the structural complexes of 30–50% sequence identity to RefSeq sequences (Figure 3, inset). IBIS binding site annotations add ∼11% of RefSeq sequences (1 493 256) to the list of 4 418 746 currently available RefSeq sequences annotated with CDD, Swissprot (23) and other types of binding sites. Therefore, all these binding site annotation resources may provide up to 40% RefSeq annotation coverage.
Figure 3.
Annotation of RefSeq sequences using IBIS binding sites. Percentage of RefSeq sequences with annotated IBIS binding sites for each type of interactions. Percentage of annotated RefSeq sequences at each inference threshold [average percent identity between query and binding site cluster members (inset)].
CONCLUSION
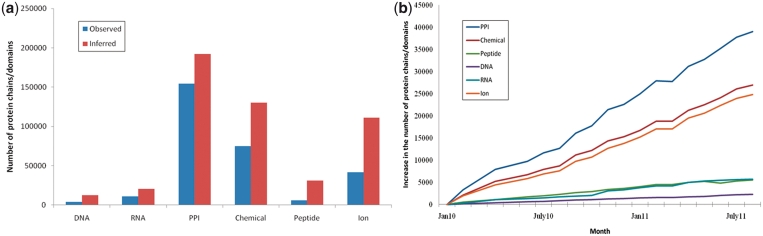
Although recent advances in experimental methods for identification of protein–protein interactions have provided extensive data on protein interaction networks, current ‘interactome’ data sets suffer from a high rate of false positives and low coverage. Complete structural coverage of all protein complexes is desired but still remains a daunting task. Nevertheless, as can be seen in Figure 4, the number of observed and inferred protein interactions based on structure data is increasing rather rapidly with the highest rate of about 2000 domain–domain interactions per month (in the case of protein–protein interactions domain is defined as a unit of interaction in IBIS) and the lowest rate of about 50 protein–DNA interactions per month. Interestingly, the rate with which new interactions are deposited seems to remain fairly constant and points to the need for more extensive sampling of new interactions through the means of structural genomics efforts that have focused so far on structural fold coverage (24).
Figure 4.
IBIS interaction growth. (a) The number of protein chains/domains in IBIS is shown for each interaction type (protein–DNA, protein–RNA, protein–protein, protein–chemical, protein–peptide and protein–ion) and for observed (blue) and inferred (red) interactions. (b) IBIS growth of new interactions (including observed and inferred) starting from January 2010. Growth chart lines are offset from zero and distinguished by interaction type.
The interaction coverage should be complemented with the high reliability of interactions. As was shown in our previous studies, IBIS performance compares very well with other computational methods and can reach 70–80% sensitivity and specificity for protein–small molecule site annotations (25). We also showed that there exists a trade off between specificity and sensitivity between two implementations of our method when only evolutionarily conserved binding site clusters or clusters supported by only one observation (singletons) are used (M. Tyagi et al., manuscript under revision). However, the sensitivity of the conserved binding site approach does not drop as dramatically as the specificity of the singleton approach, so clustering of binding sites remains a valuable and desirable tactic for prediction. Moreover, what is important is that IBIS's accuracy depends critically on its present feature to use all available data on structural complexes and not to be confined by the non-redundant set of complexes as implemented in many other approaches. The method's performance drops significantly if a non-redundant set of structures is employed pointing to the fact that the aggregation of all structural data represents an invaluable source of information and even small characteristic features of binding interfaces should be accounted for by inference. Finally, we show that inferring binding sites from homologous complexes can be very useful to expand functional and interaction annotations in the protein sequence database, with IBIS interaction partner and binding sites currently covering one-third of all RefSeq sequences.
FUNDING
National Institutes of Health/Department of Health and Human Service (DHHS) (Intramural Research program of the National Library of Medicine). Funding for open access charge: National Institutes of Health Intramural Program.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Renata Geer for her help.
REFERENCES
- 1.Stein A, Panjkovich A, Aloy P. 3did Update: domain-domain and peptide-mediated interactions of known 3D structure. Nucleic Acids Res. 2009;37:D300–D304. doi: 10.1093/nar/gkn690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capra JA, Laskowski RA, Thornton JM, Singh M, Funkhouser TA. Predicting protein ligand binding sites by combining evolutionary sequence conservation and 3D structure. PLoS Comput. Biol. 2009;5:e1000585. doi: 10.1371/journal.pcbi.1000585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen YC, Lo YS, Hsu WC, Yang JM. 3D-partner: a web server to infer interacting partners and binding models. Nucleic Acids Res. 2007;35:W561–W567. doi: 10.1093/nar/gkm346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Huang B, Schroeder M. LIGSITEcsc: predicting ligand binding sites using the Connolly surface and degree of conservation. BMC Struct. Biol. 2006;6:19. doi: 10.1186/1472-6807-6-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laurie AT, Jackson RM. Q-SiteFinder: an energy-based method for the prediction of protein-ligand binding sites. Bioinformatics. 2005;21:1908–1916. doi: 10.1093/bioinformatics/bti315. [DOI] [PubMed] [Google Scholar]
- 6.Qin S, Zhou HX. meta-PPISP: a meta web server for protein-protein interaction site prediction. Bioinformatics. 2007;23:3386–3387. doi: 10.1093/bioinformatics/btm434. [DOI] [PubMed] [Google Scholar]
- 7.Talavera D, Laskowski RA, Thornton JM. WSsas: a web service for the annotation of functional residues through structural homologues. Bioinformatics. 2009;25:1192–1194. doi: 10.1093/bioinformatics/btp116. [DOI] [PubMed] [Google Scholar]
- 8.Marino Buslje C, Teppa E, Di Doménico T, Delfino JM, Nielsen M. Networks of high mutual information define the structural proximity of catalytic sites: implications for catalytic residue identification. PLoS Comput. Biol. 2010;6:e1000978. doi: 10.1371/journal.pcbi.1000978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matthews LR, Vaglio P, Reboul J, Ge H, Davis BP, Garrels J, Vincent S, Vidal M. Identification of potential interaction networks using sequence-based searches for conserved protein-protein interactions or "interologs". Genome Res. 2001;11:2120–2126. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kemmer D, Huang Y, Shah SP, Lim J, Brumm J, Yuen MM, Ling J, Xu T, Wasserman WW, Ouellette BF. Ulysses - an application for the projection of molecular interactions across species. Genome Biol. 2005;6:R106. doi: 10.1186/gb-2005-6-12-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Persico M, Ceol A, Gavrila C, Hoffmann R, Florio A, Cesareni G. HomoMINT: an inferred human network based on orthology mapping of protein interactions discovered in model organisms. BMC Bioinformatics. 2005;6(Suppl. 4):S21. doi: 10.1186/1471-2105-6-S4-S21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walhout AJ, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, Thierry-Mieg N, Vidal M. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- 13.Xu Q, Dunbrack RL., Jr The protein common interface database (ProtCID)–a comprehensive database of interactions of homologous proteins in multiple crystal forms. Nucleic Acids Res. 2011;39:D761–D770. doi: 10.1093/nar/gkq1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue LC, Dobbs D, Honavar V. HomPPI: a class of sequence homology based protein-protein interface prediction methods. BMC Bioinformatics. 2011;12:244. doi: 10.1186/1471-2105-12-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang QC, Deng L, Fisher M, Guan J, Honig B, Petrey D. PredUs: a web server for predicting protein interfaces using structural neighbors. Nucleic Acids Res. 2011;39:W283–W287. doi: 10.1093/nar/gkr311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoemaker BA, Zhang D, Thangudu RR, Tyagi M, Fong JH, Marchler-Bauer A, Bryant SH, Madej T, Panchenko AR. Inferred Biomolecular Interaction Server–a web server to analyze and predict protein interacting partners and binding sites. Nucleic Acids Res. 2010;38:D518–D524. doi: 10.1093/nar/gkp842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Addess KJ, Chen J, Geer LY, He J, He S, Lu S, Madej T, Marchler-Bauer A, Thiessen PA, et al. MMDB: annotating protein sequences with Entrez's 3D-structure database. Nucleic Acids Res. 2007;35:D298–D300. doi: 10.1093/nar/gkl952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 21.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lee D, de Beer T, Laskowski R, Thornton J, Orengo C. 1,000 structures and more from the MCSG. BMC Struct. Biol. 2011;11:2. doi: 10.1186/1472-6807-11-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thangudu RR, Tyagi M, Shoemaker BA, Bryant SH, Panchenko AR, Madej T. Knowledge-based annotation of small molecule binding sites in proteins. BMC Bioinformatics. 2010;11:365. doi: 10.1186/1471-2105-11-365. [DOI] [PMC free article] [PubMed] [Google Scholar]