Abstract
Recent exponential growth in the throughput of next-generation DNA sequencing platforms has dramatically spurred the use of accessible and scalable targeted resequencing approaches. This includes candidate region diagnostic resequencing and novel variant validation from whole genome or exome sequencing analysis. We have previously demonstrated that selective genomic circularization is a robust in-solution approach for capturing and resequencing thousands of target human genome loci such as exons and regulatory sequences. To facilitate the design and production of customized capture assays for any given region in the human genome, we developed the Human OligoGenome Resource (http://oligogenome.stanford.edu/). This online database contains over 21 million capture oligonucleotide sequences. It enables one to create customized and highly multiplexed resequencing assays of target regions across the human genome and is not restricted to coding regions. In total, this resource provides 92.1% in silico coverage of the human genome. The online server allows researchers to download a complete repository of oligonucleotide probes and design customized capture assays to target multiple regions throughout the human genome. The website has query tools for selecting and evaluating capture oligonucleotides from specified genomic regions.
INTRODUCTION
Using next-generation DNA sequencing (NGS) technologies, there has been a dramatic increase in intermediate-scale, targeted resequencing applications. This is a generally useful approach for discovering polymorphisms and mutations of interest among candidate regions and validating novel variants and mutations from complete genomes and exomes (1,2). NGS-based targeted resequencing also has immediate application as a clinical diagnostic for identifying pathogenic mutations in medical conditions such as inherited diseases and cancer. Therefore, it has become increasingly important to develop accessible, cost effective and flexible methods that can be used to design customized capture assays targeting any region throughout the entire human genome beyond coding sequences. Currently there is very little available with regard to conducting targeted resequencing of non-coding human genome regions. We present a genome-wide solution towards targeted resequencing of loci from the human genome. Relying on a capture technology we developed, our genome-wide design covers the human genome with in-solution capture probes. As a result, it provides both exome coverage as well as facilitating the analysis of non-coding regions such as promoters and regulatory sequences. These non-coding regions are of increasing interest with regard to disease-related polymorphisms and mutations.
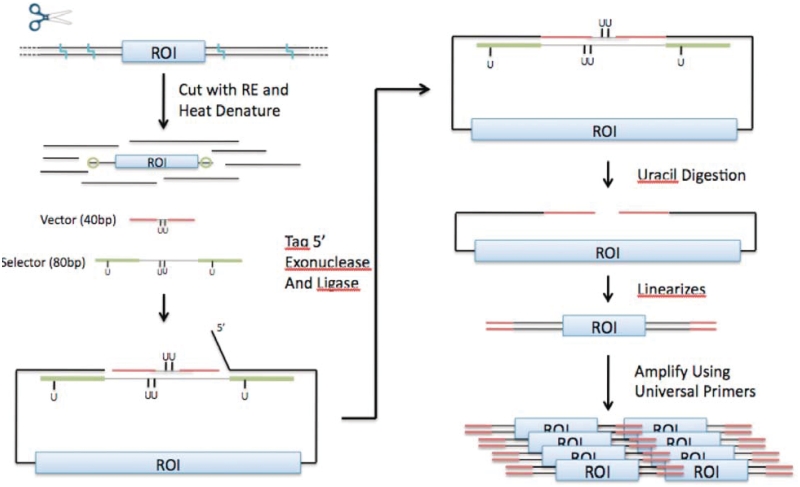
As recently described by Natsoulis et al. (3), this in-solution capture approach enables targeted resequencing of large sets of genomic loci targets up to 1 Mb and potentially higher. Using highly multiplexed pools of single-stranded 80-mer capture oligonucleotides to circularize target genomic regions en masse (Figure 1), this capture assay enables the amplification of target-specific regions with a universal set of PCR primers common to all targets. A capture oligonucleotide contains two single-stranded capture arm sequences that mediate circularization by hybridizing specifically to the complementary flanking sequences of a genomic target. A fixed sequence general motif links the capture arm oligonucleotides to form a complete capture oligonucleotide with 80 bp length. Each circularization reaction also incorporates a 40-bp universal vector oligonucleotide that complements the capture oligonucleotide's general motif. This vector provides the universal primer sequences necessary for downstream amplification. Previously, we designed a set of capture oligonucleotides spanning the human exome (http://oligoexome.stanford.edu) and demonstrated that customized capture assays could be easily developed using this resource (3).
Figure 1.
Schema for target-specific capture and amplification by selective genomic circularization. This schema for the Natsoulis et al. (3) capture protocol describes the major steps for conducting capture and amplification of a target region. The light blue squiggles at the top of the figure indicate restriction enzyme recognition sites that are cut by the addition of a single restriction enzyme. ROI stands for region of interest (i.e. target region), green bars indicate capture arms, green circles indicate capture arm hybridization sites and red bars indicate universal primer sequence. The protocol described by this figure is performed separately for each restriction enzyme.
In brief, the full protocol includes the following key steps (Figure 1): (i) genomic DNA is subject to restriction enzyme digestion by a single enzyme; (ii) the addition of capture oligonucleotides pools that are specific to a given restriction enzyme and the vector sequence circularizes genomic targets; (iii) 5′ exonuclease cleaves unbound 5′ sequence (the 5′ flap); (iv) circularization is completed by ligation; and (v) a uracil glycosylase reaction linearizes circularized molecules to produce capture regions flanked by universal primer sequences. These molecules can then be uniformly amplified by PCR and prepared for sequencing.
As has been described, this assay successfully targets up to 1 Mb of human sequence and can accommodate the highly multiplexed capture of thousands of loci (3). Additionally, the technology achieves both high-sensitivity and high-specificity human genomic capture across target regions up to 800 bp in length. On-target regions of >10-fold coverage make up >85% of the original targets. Off-target capture as we recently demonstrated was <5%. Based on a published cost assessment (3), the overall assay is significantly less costly than common capture methods such as multiplex PCR and in-solution capture. The procedure uses <100 ng of DNA per individual sample, which makes it ideal for clinical applications with limited sample material, and the capture assay can be completed in several days. Finally, this capture assay can be adapted for multiple sequencing platforms. The most recent application as described by Natsoulis et al. (3) uses next-generation Illumina technology for downstream sequencing, but it may be adapted for use with other next-generation sequencers, as we have previously demonstrated with Roche's 454 sequencer (4).
This selective capture protocol introduces several molecular constraints that must be considered in identifying capture arm sequences (Figure 1). To complete the ligation in Step 4, the termini of a captured DNA sequence must lie flush to the 40-bp vector oligonucleotide. The 3′ capture arm of a successful capture oligonucleotide must therefore hybridize precisely at the 3′ terminus of a restriction fragment containing the genomic region of interest. The 5′ exonuclease in Step 3 enables flexible placement of the 5′ capture arm by removing the 5′ flap produced by genomic DNA that extends beyond the capture arm. These molecular mechanisms complicate capture arm design and render the procedure intractable by standard primer design software. Designing capture arms that cover any given human genome target represents a major challenge to disseminating this technology to interested users.
To facilitate designing customized targeted resequencing assays for any human genome region, we have created the Stanford Human OligoGenome Resource, a database of oligonucleotide capture sequences that span the human genome. Using our previous experience in designing and implementing assays, we improved the design method to avoid issues which decrease capture efficiency (3). This unique resource has extensive utility given that it provides coverage for capture targets beyond the 3% of the coding region portion (e.g. exome) of the human genome. The OligoGenome website (http://oligogenome.stanford.edu/) provides an interface for browsing, filtering and downloading capture oligonucleotide sequences based upon user queried genomic regions and annotation-based constraints. The capture oligonucleotide designs and the search tools expedite the experimental design of customized captures assays and provides researchers with the ability to query both inside and outside of the coding regions of the human genome. Given its low cost and limited infrastructure requirements (3), this resource greatly improves the accessibility of highly multiplexed genomic target capture and resequencing for researchers.
MATERIALS AND METHODS
Capture oligonucleotide sequence generation
We created the Capture Oligonucleotide Annotation and Creation in Human (COACH) ruby suite to generate capture oligonucleotides for the human genome in silico. The suite has two primary modules: a Capture Oligonucleotide Generator (COG) that finds putative capture arm sites and a Refactoring Engine for INvalid Selection (REINS) that removes sites which fail to pass all specified constrains. As input, the program takes a 2-bit genome file, a set of restriction enzymes and one or more bed-formatted annotation files. The suite processes the restriction enzymes independently and outputs a set of capture oligonucleotides that maximizes genome coverage for each enzyme.
To generate the capture probes for the Stanford Human OligoGenome Resource, we used the UCSC hg19 genome build for chromosomes 1–22; X and Y (5). The coordinates for these regions exactly match the coordinates of NCBI genome Build 37. We chose the four restriction enzymes MseI, BfaI, Sau3AI and CViQI to define our in silico-cut sites based upon empirical results from Natsoulis et al. (3). Finally, we used UCSC dbSNP131 annotations to define common variants (6) and a 24-mer mapability track from ENCODE to provide an application-specific repeat mask (7,8). For a given 24-mer in the human genome, the mapability track indicates how many other 24-mers in the genome have a sequence that differs by two or fewer bases. Determination of exon coverage relied on the Consensus Coding Sequence (CCDS) Project (9) for hg19.
COG uses a greedy algorithm that guarantees selection of capture arms that maximize genomic coverage given the constraints in REINS. COG significantly improves upon the TargetedOligoDesign program described in Natsoulis et al. (3), which evaluated a fixed set of oligonucleotide capture arms for each target region. For each chromosome, COG defines a set of genomic target regions such that each region is bounded by adjacent restriction sites. Within each target region, COG finds the pair of plus strand capture arms that would achieve greatest coverage of the region and submits them to REINS for validation. It continues to generate capture arm sites in decreasing order of coverage until REINS either validates a pair of sites or until no further sites are available. The same procedure is repeated for minus strand capture arms. It also tests for a combination of minimally overlapping plus and minus strand capture arms. COG compares the three capture sequence sets returned by this process and outputs the set that achieves the greatest coverage of the queried region. In the case of a tie for coverage, it selects the set that covers the fewest bases redundantly. If no valid set of capture arms is available, COG does not produce any output for that target region.
In order to ensure highly sensitive and specific capture, REINS applied the following, stringent constraints to the capture oligonucleotide sequences generated for the Stanford Human OligoGenome Resource. These rules correspond to the empirical best practices empirically determined by Natsoulis et al. (3):
Capture arms are 20 bp in length;
The sequences in a pair of capture arms must have the same polarity with respect to the reference genome;
3′ capture arms must be flush to a restriction site; and
The maximum size of a DNA molecule targeted by a capture oligonucleotide is 800 bp and the minimum size is 100 bp.
Also, REINS applies rules based on genome-specific annotations to improve capture performance in human genomic target sequences. REINS rejects capture arm sequences that would hybridize to regions containing known variants from dbSNP131. Additionally, because certain common variants disrupt restriction sites of interest or introduce new restriction sites, it ensures that capture arms mediate circularization both in the presence and in the absence of these variable cut sites. REINS uses the 24-mer mapability track described above to detect capture arms with non-specific hybridization, which leads to inefficient reactions or non-specific, off-target capture. To prioritize highly specific capture arms, we ran COACH three times, using different mapability constraints based on the 24-mer mapability track to create three tiers of oligos: (i) Tier 1: oligos must fall within uniquely mapable regions; (ii) Tier 2: oligos must fall within regions mapping to fewer than 10 other regions; and (iii) Tier 3: no mapability restriction. We used capture arms from Run 2 to fill in gaps in coverage left after Run 1, and similarly filled remaining gaps with oligos from Run 3. The combination of these genome-specific rules and parameters constitutes a stringent constraint engine that enforces capture oligonucleotide quality.
Quality control annotation for capture oligonucleotides
We generated annotations for each capture oligonucleotide produced by COACH to serve as a proxy for capture efficiency and capture specificity. Among them are parameters which we previously had demonstrated are important for mediating on-target and efficient capture. The following annotations apply to each capture arm for any given oligonucleotide, and all repeat annotations are specific to the human genome: (i) number of exact sequence matches present in the human genome; (ii) number of matches differing by one base, (iii) number of matches differing by two bases; and (iv) GC content. Parameters 1–3 influence the on-target capture efficiency. As was previously demonstrated, one can reduce off-target capture by avoiding repetitive regions of the genome in either one or both of the capture arm sequences. We used bowtie (10) to determine in silico the number of off-target regions per oligonucleotide capture arm sequences. We considered an off-target capture to occur if the capture arms aligned between 100 and 1000 bp from each other with zero mismatches and had the correct relative orientation. We defined these positions as paralogs (P) of the intended capture site.
Database construction
The Stanford Human OligoGenome Resource (http://oligogenome.stanford.edu) runs on a 2 × 2.27 GHz Quad Core Intel Xeon E5520 server, with 24 GB memory and Ubuntu 9.10 operating system. The web application is implemented in Ruby on Rails 2.3.8, running under Passenger 2.2.15. The underlying database is MySQL 5.0.42 community edition, which is hosted on a separate database server. Query and data download is via any current web browser. Recommended browsers and versions are: Internet Explorer 7.0+; Firefox 3.0+; Safari 5.0+; or Chrome (any version).
RESULTS
Coverage of the human genome
The Stanford Human OligoGenome Resources achieves 92.1% in silico coverage of the entire human genome using the four restriction enzymes MseI, BfaI, Sau3AI and CViQI. In total, ∼21.8 million probes capture 2.85 billion nucleotide positions at least once. Of these probes, 20.2 million that cover 88.4% of the genome are predicted to have a unique capture site due to the absence of paralogous regions (Table 1). Nearly 720 000 probes cover the CCDS-coding regions (99.65% coverage) for the 22 April 2011 release of CCDS (Table 2). Approximately 70 000 of these capture oligonucleotides have only one predicted target site, providing 97.12% coverage of the CCDS-annotated coding regions at high specificity. At least 77.2% of the genome is covered by capture oligonucleotides from two or more different restriction enzymes (91.5% of CCDS regions), which allows for experimental redundancy. As ∼50% of the human genome is highly repetitive, these total coverage numbers indicate that the capture design successfully bridges short repetitive sequences such as Alu elements by placing capture arms in uniquely mapping region on either side of these regions. Average capture lengths of a given genomic target region are also listed in Table 1.
Table 1.
Summary statistics for all capture oligonucleotides designed to target human genome Build 37/hg19
| Statistics for whole genome capture | BfaI | CviQI | MseI | Sau3AI | Total |
|---|---|---|---|---|---|
| Tier 1 only | |||||
| Total number of oligos | 4 049 706 | 2 999 049 | 4 825 988 | 3 246 400 | 15 121 143 |
| Average capture length (bases) | 401 | 483 | 269 | 430 | 381 |
| Total bases covered (megabases) | 1614 | 1441 | 1295 | 1388 | 2 311 |
| Percent of genome covered | 52.14 | 46.54 | 41.83 | 44.83 | 74.64 |
| Percent of oligos with U0 > 1 | 4.71 | 5.13 | 5.08 | 5.06 | 4.99 |
| Percent of oligos with paralogs > 0 | 0.07 | 0.07 | 0.06 | 0.07 | 0.07 |
| Percent of genome covered with paralogs removed | 52.10 | 46.50 | 41.80 | 44.80 | 74.60 |
| Tiers 1, 2 and 3 combined | |||||
| Total number of oligos | 5 787 809 | 4 362 946 | 6 757 372 | 4 938 767 | 21 846 894 |
| Average capture length (bases) | 410 | 496 | 280 | 426 | 391 |
| Total bases covered (megabases) | 2160 | 1978 | 1760 | 1938 | 2852 |
| Percent of genome covered | 69.79 | 63.89 | 56.85 | 62.61 | 92.14 |
| Percent of oligos with U0 > 1 | 23.99 | 24.60 | 23.23 | 28.60 | 24.92 |
| Percent of oligos with paralogs > 0 | 6.96 | 6.48 | 7.25 | 8.90 | 7.39 |
| Percent of genome covered with paralogs removed | 64.91 | 59.41 | 52.32 | 57.67 | 88.43 |
Tier 1 oligonucleotides are the subset of targeting molecules generated with the strictest repeat masking parameters based upon k-mer mapability. Tiers 1, 2 and 3 represent all oligonucleotides in the database. This table illustrates that the looser mapability masking parameters used in Tiers 2 and 3 allowed for increased coverage but with a higher probability of having off-target binding and amplification.
Table 2.
Summary statistics describing the in silico percent capture of CCDS regions by the combined set of oligonucleotide probes
| Statistics for CCDS capture for all tiers | BfaI | CviQI | MseI | Sau3AI | Total |
|---|---|---|---|---|---|
| Total number of oligos covering CCDS target area | 182 483 | 178 338 | 158 445 | 200 019 | 719 285 |
| Average capture length (bases) | 521 | 550 | 419 | 489 | 497 |
| Total bases covered (megabases) | 25.286 | 23.270 | 22.162 | 24.04 | 31.70 |
| Percent of CCDS covered | 79.49 | 73.15 | 69.67 | 75.58 | 99.65 |
| Percent of oligos with paralogs > 0 | 2.89 | 2.85 | 3.00 | 3.03 | 2.94 |
| Percent of CCDS covered with paralogs removed | 76.96 | 70.85 | 67.36 | 73.02 | 97.12 |
Exonic regions prove possible to capture with high sensitivity and specificity due to their high k-mer complexity.
Capture oligonucleotide human genome mapping
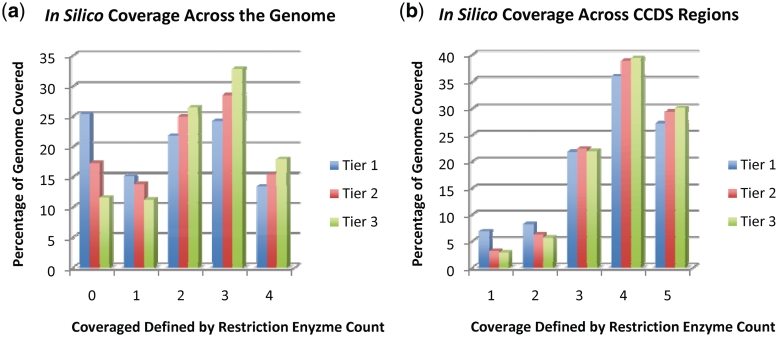
As described in the ‘Materials and Methods’ section, we established three tiers of mapability to assess off-target capture. Tier 1 oligonucleotides are the subset of targeting molecules generated with the strictest repeat masking parameters based upon k-mer mapability (Table 1). Tiers 2 and 3 have fewer constraints on their presence in the genome and are more susceptible to off-target capture. Combined Tiers 1, 2 and 3 represent all oligonucleotides in the database. Figure 2 additionally illustrates the advantage of the multitiered approach to repeat masking the oligonucleotide capture sites. Tier 1 provides highly specific capture oligonucleotides with reduced coverage, while the addition of subsequent tiers with reduced repeat masking achieve higher coverage at the cost of less efficient reactions through off-target capture.
Figure 2.
In silico coverage by the set of capture oligonucleotides from the Human OligoGenome Resource. Coverage is across (a) the whole genome and (b) the regions defined by CCDS in each successive tier of 24-mer repeat masking. Tier 1 oligonucleotides are the subset of targeting molecules generated with the strictest repeat masking parameters based upon k-mer mapability. Tiers 1, 2 and 3 represent all oligonucleotides in the database. The restriction enzyme count on the x-axis is the number of restriction enzymes for which the OligoGenome database contains an oligonucleotide that can capture a given base. Zero depth indicates the set of positions for which no capture oligonucleotides exist. As expected, fewer repeat mask restrictions lead to a greater number of positions covered by multiple restriction enyzmes' oligonucleotides.
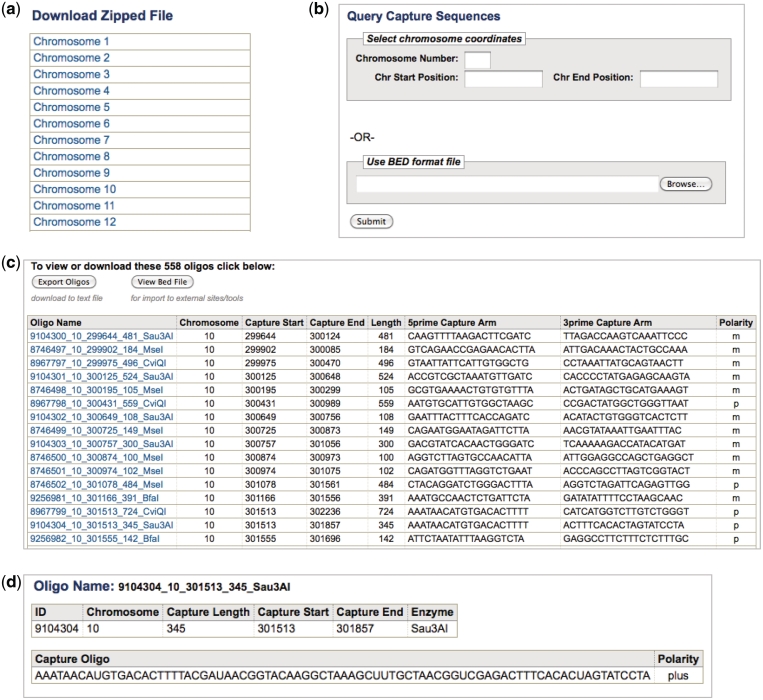
Interface for the Human OligoGenome
The Human OligoGenome Resource website presents an intuitive interface for selecting and downloading capture oligonucleotides for customized assays to mediate targeted resequencing. Users can download all probe sets by selecting gzipped flat files organized by the chromosome (Figure 3a). Users can also select all capture oligonucleotides from specified genomic regions using the Query Capture Seqs tool, which either takes chromosome, start position and end position as input or allows the user to upload a bed file of capture regions as input (Figure 3b). Before submitting the query, the user may also choose to filter results by the repeat annotations discussed below and by tier number. Each row of output from this tool presents information for a single capture oligonucleotide. The first set of fields contains information about the oligonucleotide sequence and genomic target, including chromosome (Chromosome), 1-based capture region start position (Capture Start), and 1-base capture region end position (Capture End). The Length column calculates total capture region length, and the Polarity column identifies the strand with which the capture arms hybridize relative to the reference sequence. The 5prime Capture Arm and the 3prime Capture Arm columns contain the 20 bp sequences for the 5′ capture arm and the 3′ capture arm, respectively (Figure 3c). The website also generates a table describing the in silico coverage of returned capture oligonucleotides across the queried regions, both per region and in total across all regions.
Figure 3.
A brief overview of the OligoGenome website and its query tools. You may (a) download all capture oligonucleotides directly or (b) search for capture oligos that target a specific interval entered on the page or a set of intervals uploaded in bed format. (c) After the submission of queried regions, you may view the returned capture oligonucleotides on the website, download the table in bed format, or export the results to the UCSC Genome Browser to view as a track. (d) Additionally, clicking an oligo name will bring you to a page with additional information, including the full 80-bp capture oligonucleotide.
The output also includes the annotations generated by COACH. These include GC content, the number of exact sequence matches present in the human genome (U0), the number of matches differing by one base (U1), the number of matches differing by two bases (U2) and the number of in silico off target capture regions (Paralogs). Additionally, each Oligo Name field provides a hyperlink to a page that displays restriction enzyme identity (Enzyme) and full capture oligonucleotide sequence (Capture Oligo) for the specified oligo (Figure 3d). The user can download the oligonucleotide entries returned by the Query Capture Seqs tool by clicking on the Export Oligos button at the top of the page, which produces a tab-delimited text file containing all 10 fields described above, as well as the genome build and download date. The user may also choose to export the data to UCSC as a custom track (11). All data on the Human OligoGenome Resource website are freely accessible.
To design capture assays, one selects the regions-of-interest and then downloads the overlapping capture oligonucleotide sequences. We recommend using Tier 1 capture oligonucleotides and then individually selecting lower tier oligonucleotides to fill specific gaps when needed. Also, choosing oligonucleotides with a GC content <75% will improve general capture efficiency. After oligonucleotides are synthesized, they should be pooled in equimolar ratio to each other based on their affiliated restriction enzyme.
DISCUSSION
To facilitate targeted resequencing of the human genome, we have developed and released the Human OligoGenome Resource. It covers >92% of the human genome with capture oligonucleotides that can be used in robust in-solution capture assays using the selective genomic circularization method (3). This high level of in silico coverage is partly attributable to our designs capability to straddle over repetitive sequences in the human genome. In particular, the Human OligoGenome Resource provides for the first time a general resource to capture and target resequence non-coding regions such as promoters and regulatory sequences which are of increasing interest in regards to disease-related polymorphisms and mutations. It uses a simple web interface to provide access to capture oligonucleotide sequences for the entire human genome. These sequences facilitate rapid experiment design for using the capture technology as described in Natsoulis et al. (3). The capture oligonucleotides can be ordered and synthesized from any commercial vendor or core oligonucleotide synthesis facility, combined to form highly multiplexed reagent pools and downstream sequencing can be conducted using any NGS platform. These probes also serve as a useful resource for other selective circularization technologies. The recently published paper by Johansson et al. (12) presents a comparable capture method for which the OligoGenome capture oligonucleotides can be easily adapted. The Human OligoGenome Resource site will facilitate previously untenable studies in genetic and clinical resequencing and expedite variant discovery and validation.
FUNDING
National Institutes of Health (RC2 HG005570-01 to G.N., J.M.B., S.G. and H.P.J.; R21CA12848 to G.N. and H.P.J.; 5K08CA96879–6 to H.P.J., DK56339 to H.P.J; 2P01HG000205 to J.M.B., S.G. and H.P.J.; T15-LM007033 to D.E.N.); Doris Duke Clinical Foundation (Clinical Scientist Award to H.J.); Reddere Foundation (to H.J.); Liu Bie Ju Cha and Family Fellowship in Cancer (to H.J.); Wang Family Foundation (to H.J.); Howard Hughes Medical Foundation (to H.J.). Funding for open access charge: National Institutes of Health (RC2 HG005570-01 and 2P01HG000205).
Conflict of interest statement. None declared.
REFERENCES
- 1.Mamanova L, Coffey AJ, Scott CE, Kozarewa I, Turner EH, Kumar A, Howard E, Shendure J, Turner DJ. Target-enrichment strategies for next-generation sequencing. Nat. Methods. 2010;7:111–118. doi: 10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 2.Turner EH, Ng SB, Nickerson DA, Shendure J. Methods for genomic partitioning. Annu. Rev. Genomics Hum. Genet. 2009;10:263–284. doi: 10.1146/annurev-genom-082908-150112. [DOI] [PubMed] [Google Scholar]
- 3.Natsoulis G, Bell JM, Xu H, Buenrostro JD, Ordonez H, Grimes S, Newburger D, Jensen M, Zahn JM, Zhang N, et al. A flexible approach for highly multiplexed candidate gene targeted resequencing. PLOS One. 2011;6:e21088. doi: 10.1371/journal.pone.0021088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dahl F, Stenberg J, Fredriksson S, Welch K, Zhang M, Nilsson M, Bicknell D, Bodmer WF, Davis RW, Ji H. Multigene amplification and massively parallel sequencing for cancer mutation discovery. Proc. Natl Acad. Sci. USA. 2007;104:9387–9392. doi: 10.1073/pnas.0702165104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sequist LV, Gettinger S, Senzer NN, Martins RG, Janne PA, Lilenbaum R, Gray JE, Iafrate AJ, Katayama R, Hafeez N, et al. Activity of IPI-504, a novel heat-shock protein 90 inhibitor, in patients with molecularly defined non-small-cell lung cancer. J. Clin. Oncol. 2010;28:4953–4960. doi: 10.1200/JCO.2010.30.8338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raney BJ, Cline MS, Rosenbloom KR, Dreszer TR, Learned K, Barber GP, Meyer LR, Sloan CA, Malladi VS, Roskin KM, et al. ENCODE whole-genome data in the UCSC genome browser (2011 update) Nucleic Acids Res. 2011;39:D871–D875. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kwak EL, Bang YJ, Camidge DR, Shaw AT, Solomon B, Maki RG, Ou SH, Dezube BJ, Janne PA, Costa DB, et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 2010;363:1693–1703. doi: 10.1056/NEJMoa1006448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johansson H, Isaksson M, Sorqvist EF, Roos F, Stenberg J, Sjoblom T, Botling J, Micke P, Edlund K, Fredriksson S, et al. Targeted resequencing of candidate genes using selector probes. Nucleic Acids Res. 2011;39:e8. doi: 10.1093/nar/gkq1005. [DOI] [PMC free article] [PubMed] [Google Scholar]