Abstract
The identification of a constantly increasing number of genes whose mutations are causally implicated in tumor initiation and progression (cancer genes) requires the development of tools to store and analyze them. The Network of Cancer Genes (NCG 3.0) collects information on 1494 cancer genes that have been found mutated in 16 different cancer types. These genes were collected from the Cancer Gene Census as well as from 18 whole exome and 11 whole-genome screenings of cancer samples. For each cancer gene, NCG 3.0 provides a summary of the gene features and the cross-reference to other databases. In addition, it describes duplicability, evolutionary origin, orthology, network properties, interaction partners, microRNA regulation and functional roles of cancer genes and of all genes that are related to them. This integrated network of information can be used to better characterize cancer genes in the context of the system in which they act. The data can also be used to identify novel candidates that share the same properties of known cancer genes and may therefore play a similar role in cancer. NCG 3.0 is freely available at http://bio.ifom-ieo-campus.it/ncg.
INTRODUCTION
The pivotal role of genomic instability in causing human cancer is an old concept that dates back to the first cytogenetic studies on cancer cells (1,2). Until very recently, however, the number of genes whose somatic mutations are causally implicated in cancer initiation and progression was very low. This was mainly due to the difficult and lengthy process of identifying genetic modifications through traditional sequencing methods. The recent development of next-generation sequencing techniques has boosted the discovery of novel cancer genes. In the last 5 years, high-throughput mutational screenings were performed on several cancer types and led to the identification of mutations in both coding and non-coding regions of the cancer genome. So far, around 30 high-throughput and whole genome sequencing experiments on cancer samples have been published, which identified around 1500 mutated genes that are potentially actively involved in cancer development (3–30). This constant delivery of new sequencing data is radically changing our understanding of cancer genetics, showing that the genomic landscape of cancer is complex and varies among and within cancer types (31). One way to reduce the complexity implies the identification of properties that are shared among cancer genes and distinguish them from the rest of human genes (32). For example, cancer genes tend to be singletons, i.e. they preserve one gene copy in the genome, and to encode hubs, i.e. proteins that engage numerous physical connections with other proteins (33). Moreover, recessive cancer genes appeared early in evolution and are involved in basic cellular processes, while oncogenes originated later and mainly act as regulators and signal transducers (34). Thus, the identification of specific properties of cancer genes may help in better rationalizing their role in cancer. While several databases collect mutational and functional information on cancer genes, no description of their systems-level properties is currently available. The Network of Cancer Genes (NCG) was originally created to collect information on duplicability and network properties of cancer genes (35).
Information on duplicability and network properties of cancer genes (35). In the current version of the database (NCG 3.0), we included 1494 cancer genes that have been identified so far, and provide an update of the human gene set and the protein interactome. We also added novel features, such as the functional description of cancer genes as well as information about their interaction with microRNAs (miRNAs). Finally, we developed a new web interface to allow more intuitive and flexible queries of the database. Since properties of cancer genes can be also exploited to identify new candidates that may be involved in cancer (36), we present a case study that involves two paralogs, the DNA methyltransferase alpha (DNMT3A) and beta (DNMT3B). On the basis of the information that can be retrieved in NCG 3.0, we hypothesize that, upon the acquisition of somatic mutations, these genes may play an active role in cancer development.
UPDATES OF DATA SOURCES
Cancer genes
NCG 3.0 includes information on 1494 cancer genes that were extracted from 30 distinct studies (Table 1). The genes were divided into three groups, according to the experimental evidence that supported their involvement in cancer. The first group includes 498 genes from the latest release of the Cancer Gene Census (CGC, 444 genes, 31 March 2011) (37) and from the census of amplified cancer genes (77 genes) (38). Both these resources are literature-based collections of genes whose mutations and/or amplifications have been proven to be causally implicated in tumorigenesis. Among the 444 genes from CGC, 346 are defined as dominant (i.e. heterozygous mutations are sufficient to cause tumorigenesis) and 96 as recessive (i.e. homozygous mutations are required to initiate tumorigenesis) (37). The second group is formed of 698 candidate cancer genes from 18 high-throughput mutational screenings (HTMS) on 16 distinct cancer types, such as breast (15,30), colorectal (30), pancreatic (13–15), lung (10,15), renal (8), prostatic (15), bladder (12), and ovarian (15) cancers, glioblastoma (20,21), leukemia (11), lymphoma (23), sarcoma (4), myeloma (6), medulloblastoma (22), head and neck squamous cell carcinoma (3,27), and melanoma (29). These genes represent the subsets of mutated genes that were identified as bearing driver mutations, i.e. mutations that confer growth advantage and are actively involved in tumor development (31). The third group of cancer genes contains 457 genes that have been identified in eleven whole genome sequencing (WGS) screenings of cancer genomes, such as breast (26), lung (16,25), prostatic (5), liver cancer (28), glioblastoma (7), leukemia (17,19), myeloma (6) and melanoma (24).
Table 1.
Primary data correspond to the list of cancer genes as they were extracted from the original papers. Five genes are not present in NCG 3.0 because they could not be mapped to current version of Entrez IDs. Dominant and recessive genes refer to the definition reported in the cancer gene census (37). Ancient and recent genes refer to genes that originated between the last universal common ancestor and opisthokonts and with metazoans or later, respectively. HTMS, high-throughput mutation screening; WGS, whole genome sequencing
| Cancer genes | Dominant | Recessive | Amplified | HTMS | WGS | Total |
|---|---|---|---|---|---|---|
| Primary data | 348 | 98 | 77 | 699 | 457 | 1,499 |
| Present in NCG 3 | 346 | 96 | 77 | 698 | 454 | 1,494 |
| Duplicated (%) | 58 (17.7) | 10 (10.5) | 23 (29.9) | 102 (14.8) | 64 (14.3) | 230 (15.7) |
| Ancient (%) | 184 (55.6) | 63 (66.3) | 53 (69.9) | 377 (54.6) | 273 (61.2) | 837 (57.1) |
| Recent (%) | 147 (44.4) | 32 (33.7) | 24 (31.1) | 313 (45.4) | 173 (38.8) | 629 (42.9) |
| Hubs (%) | 185 (58.6) | 71 (75.5) | 50 (70.4) | 231 (42.6) | 116 (34.6) | 532 (44.7) |
| miRNA targets (%) | 53 (15.3) | 14 (14.6) | 17 (22.1) | 53 (7.6) | 28 (6.2) | 118 (7.9) |
| miRNA hosts (%) | 12 (3.5) | 6 (6.3) | 2 (2.6) | 28 (4.0) | 17 (3.7) | 55 (3.7) |
Human gene set and orthology information
We gathered information on the human gene set from the combination of Gencode v.7.0 (39) and RefSeq v. 46 (40). Gencode is an initiative that aims at identifying and mapping all human protein coding genes (41). It has been used as the reference gene set for the Encode project, for the 1000 Genomes Project and for the design of the capture baits for whole exome sequencing (39). This gene set is therefore likely to include all genes present in current and future mutational screenings of whole cancer exomes. Starting from 20 700 genes in Gencode v.7.0 (corresponding to 84 408 protein sequences), we removed multiple isoforms that map to the same gene locus by aligning all protein sequences to the reference human genome (hg18) and retaining only the longest isoform (33). At the end of this pipeline, we were able to retrieve 19 560 unique genes. The 1140 missing genes corresponded to transcripts that spanned more than one gene. To recover them, we repeated the same pipeline with 20 750 genes in RefSeq (corresponding to 34 571 protein sequences). The union of these two gene lists led to the final dataset of 20 531 unique human genes.
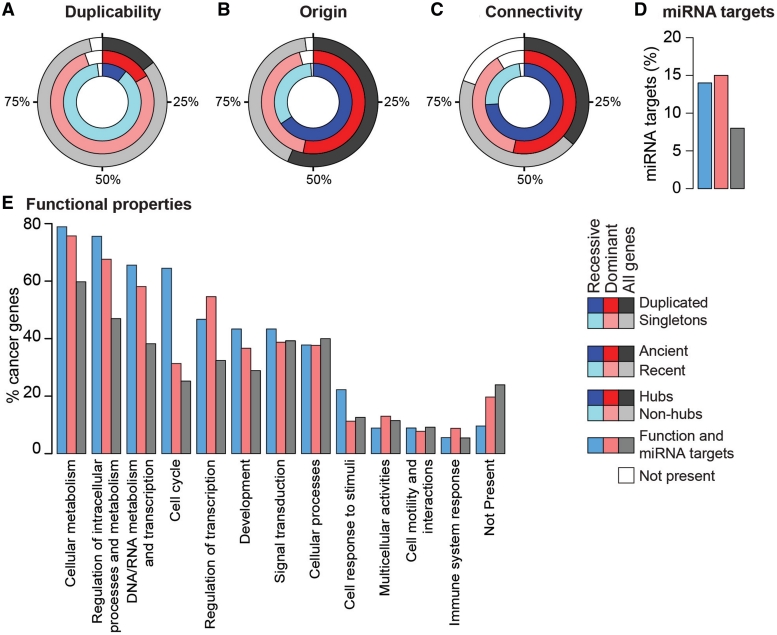
To measure the duplicability of each gene, we identified all additional hits on the genome that span at least 60% of the gene length (33), and found that 4311 human genes (21% of the total) have at least one additional copy. Consistently with previous reports (33,34), we found that cancer genes duplicate significantly less than the rest of human genes (15.7% of the total, p-value= 3.0 10−7, Pearson’s Chi-squared Test), and dominant genes are more duplicated than recessive genes (Figure 1A and Table 1).
Figure 1.
Properties of cancer genes. Circles represent the fraction of cancer, dominant, and recessive genes that are (A) singleton or duplicated, (B) ancient or recent, (C) hubs or non-hubs. Ancient genes originated between the last universal common ancestor and opisthokonts; recent genes originated with metazoans or later. (D) miRNA targets were derived from TarBase (50) and miRecords (51). Approximately 15% of dominant and recessive cancer genes are targets of miRNAs, compared with 8% of all cancer genes. (E) Cancer genes were associated to one of the 12 functional categories based on the corresponding GO terms (34).
We updated the information on orthology relationships between human and other species to eggNOG v. 2.0 (42). Orthologs were used to identify the evolutionary origin of 1,466 cancer genes (98% of the total), defined as the deepest internal node of the tree of life where an ortholog could be found (34). We confirmed that recessive cancer genes are mostly ancient genes that have orthologs in prokaryotes, plants and fungi, while a higher fraction of dominant cancer genes appeared with metazoans or later (Figure 1B and Table 1).
Human interaction network
We rebuilt the human interactome by integrating the latest releases of five sources of protein–protein interactions, such as HPRD (43), BioGRID (44), IntAct (45), MINT (46) and DIP (47). Only primary interactions were used while putative interactions inferred from orthology relationships were discarded. The final dataset comprised 98 492 binary interactions between 13 531 proteins, supported by 25 915 independent publications. The resulting human interactome contains 13% more proteins and 44% more interactions than the previous version of NCG, and reports interaction data for 1189 cancer genes (79% of the total). We confirmed that cancer genes are enriched in hubs, defined as top 25% most connected proteins (at least 15 interactions) (34), and are therefore more connected than the rest of human genes (Figure 1C and Table 1).
NEW FEATURES OF NCG 3.0
Interactions between cancer gene and miRNAs
miRNAs are endogenous short nucleotide sequences that interfere with RNA transcripts in the cytoplasm, thus regulating protein synthesis (48). Alterations in miRNA expression have been reported during cancer initiation, progression and metastasis (49). In addition, miRNAs regulate well-known cancer genes, such as PTEN, NRAS, KRAS, (48). Because of this involvement in cancer, we included information about the interactions between miRNAs and their target cancer genes. Primary data were extracted from TarBase v.5.0 (50) and miRecords v.1.0 (51), which collect experimentally supported interactions between miRNAs and their targets. The non-redundant integration of the two databases returned a list of 54 miRNAs that regulate 118 cancer genes (8% of the total, Table 1). This fraction is significantly higher compared to the rest of human genes (∼4%, p-value = 10−11, Pearson's Chi-squared Test), and becomes even higher when only recessive or dominant cancer genes are considered (∼15%, Figure 1D and Table 1). This enrichment reflects at least partly the fact that cancer genes are heavily studied and therefore more information are available for them compared to other genes. In addition to miRNA targets, we also retrieved information on 55 cancer genes that host miRNAs within their genomic locus (Table 1).
Functional classification of cancer genes
To derive the gene functional classification, we first extracted 1108 cancer genes (74% of all the total) that have at least one term at levels 5 and 6 of the biological process branch of the Gene Ontology (GO) tree (52). We then grouped all these terms into 12 functional categories, in order to provide a more general description of the gene function (34). Dominant and recessive cancer genes have different functional distributions. In particular, while dominant genes are mostly involved in the regulation of transcription, recessive genes are associated with basic cellular functions such as cell cycle, cell response to stimuli, cellular and DNA/RNA metabolism (Figure 1E).
Network representation
We developed a novel web interface and added several new possibilities of querying the database, thus allowing the user to search for specific cancer genes, for lists of genes and miRNAs related to cancer, and for the presence of cancer genes in genomic regions of interest. In addition, it is now possible to retrieve the interactions between a certain miRNA and the cancer genes that are associated to it, as well as all cancer genes that are miRNA targets or that host miRNAs within their genomic locus. Finally, we adopted CytoscapeWeb (53) for the visualization of protein–protein and miRNA–target interaction networks, and for displaying the evolutionary origin of cancer genes and their orthologs in other species.
IDENTIFICATION OF CANDIDATE CANCER GENES USING NCG
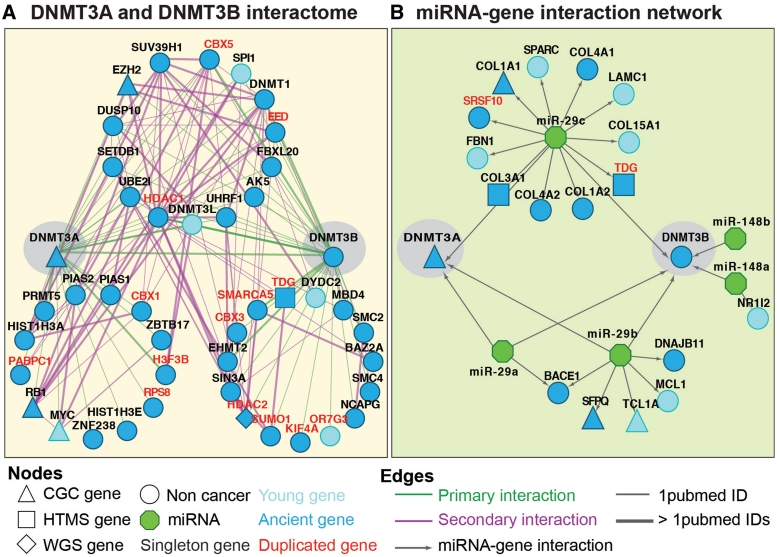
In addition to providing a comprehensive description of the properties of cancer genes, NCG can be used to identify new possible candidates, based on the hypothesis that genes with systems-level properties similar to known cancer genes may also play similar roles in cancer (36). Following this idea, two almost identical paralogs, GNAQ GNA11, have recently been associated to the same tumor type (54). Here, we hypothesize that a similar association can be drawn for DNMT3A and its paralog DNMT3B. Both these methyltransferases cause de novo methylation during the differentiation of hematopoietic cells and are involved in hematopoietic stem-cell renewal (55). DNMT3A is an already known cancer gene, part of the cancer gene census and frequently mutated in leukemia (56). Although its role in tumorigenesis is still unclear, it has been hypothesized that mutations in DNMT3A either induce altered gene expression or affect genome stability (56). To date there is no evidence that also DNMT3B is mutated in cancer, although NCG 3.0 provides several indications that this might be indeed the case. First, both DNMT3A and DNMT3B originated with eukaryotes and are associated with the same GO terms, thus indicating functional redundancy. Second, the encoded proteins DNMT3A and DNMT3B share 14 interactors, which, in turn, are connected through secondary interactions (Figure 2A). The resulting highly interconnected module involves key players in epigenetic changes of chromatin, and consequent regulation of gene expression. Third, both genes are targets of three miRNAs of the miR-29 family (Figure 2B). The downregulation of these miRNAs induces aberrant expression of both DNMT3A and DNMT3B in non-small-cell lung cancer (57) and the upregulation of both paralogs has been observed in several tumor types (58). We hypothesize that the aberrant expression can be related to the acquisition of mutations in the sequence of these genes.
Figure 2.
Functional redundancy of DNMT3A and DNMT3B. (A) Protein-protein interaction network of DNMT3A and DNMT3B. The two proteins share 14 interactors, which are mostly involved in the epigenetic control of chromatin and in the regulation of gene expression. Primary interactions are physical interactions of a protein directly with DNMT3A or DNMT3B, while secondary interactions are physical interactions between their primary interactors. (B) Interaction network between DNMT3A and DNMT3B and target miRNAs. Both genes are regulated by members of the miR-29 family, whose expression is altered in cancer and is inversely correlated with the gene expression (57). CGC, cancer gene census; HTMS, high-throughput mutational screenings; WGS, whole genome sequencing.
FUNDING
Funding for open access charge: Associazione Italiana Ricerca sul Cancro (AIRC) and Fondazione Cariplo (to F.D.C.).
ACKNOWLEDGMENTS
The authors thank all members of the Ciccarelli lab for testing NCG and providing useful suggestion to improve it.
REFERENCES
- 1.von Hansemann DP. On primary cancer of the liver. Berl. klin. Wschr. 1890;27:353–356. [Google Scholar]
- 2.Boveri T. Zur Frage der Entstehung Maligner Tumoren. Jena: Gustav Fischer. 1914;1:1–64. [Google Scholar]
- 3.Agrawal N, Frederick MJ, Pickering CR, Bettegowda C, Chang K, Li RJ, Fakhry C, Xie TX, Zhang J, Wang J, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in NOTCH1. Science. 2011;333:1154–1157. doi: 10.1126/science.1206923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barretina J, Taylor BS, Banerji S, Ramos AH, Lagos-Quintana M, Decarolis PL, Shah K, Socci ND, Weir BA, Ho A, et al. Subtype-specific genomic alterations define new targets for soft-tissue sarcoma therapy. Nat. Genet. 2010;42:715–721. doi: 10.1038/ng.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet JP, Ahmann GJ, Adli M, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clark MJ, Homer N, O'Connor BD, Chen Z, Eskin A, Lee H, Merriman B, Nelson SF. U87MG decoded: the genomic sequence of a cytogenetically aberrant human cancer cell line. PLoS Genet. 2010;6:e1000832. doi: 10.1371/journal.pgen.1000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463:360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greif PA, Eck SH, Konstandin NP, Benet-Pages A, Ksienzyk B, Dufour A, Vetter AT, Popp HD, Lorenz-Depiereux B, Meitinger T, et al. Identification of recurring tumor-specific somatic mutations in acute myeloid leukemia by transcriptome sequencing. Leukemia. 2011;25:821–827. doi: 10.1038/leu.2011.19. [DOI] [PubMed] [Google Scholar]
- 12.Gui Y, Guo G, Huang Y, Hu X, Tang A, Gao S, Wu R, Chen C, Li X, Zhou L, et al. Frequent mutations of chromatin remodeling genes in transitional cell carcinoma of the bladder. Nat. Genet. 2011;43:875–878. doi: 10.1038/ng.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, et al. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jones S, Zhang X, Parsons DW, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Kamiyama H, Jimeno A, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J, et al. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869–873. doi: 10.1038/nature09208. [DOI] [PubMed] [Google Scholar]
- 16.Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, Yue P, Zhang Y, Pant KP, Bhatt D, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 17.Ley TJ, Mardis ER, Ding L, Fulton B, McLellan MD, Chen K, Dooling D, Dunford-Shore BH, McGrath S, Hickenbotham M, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Zhao H, Zhang X, Wood LD, Anders RA, Choti MA, Pawlik TM, Daniel HD, Kannangai R, Offerhaus GJ, et al. Inactivating mutations of the chromatin remodeling gene ARID2 in hepatocellular carcinoma. Nat. Genet. 2011;43:828–829. doi: 10.1038/ng.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McLendon R, Friedman A, Bigner D, Van MEG, Brat D, Mastrogianakis G, Olson J, Mikkelsen T, Lehman N, Aldape K, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, et al. The genetic landscape of the childhood cancer medulloblastoma. Science. 2010;331:435–439. doi: 10.1126/science.1198056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pasqualucci L, Trifonov V, Fabbri G, Ma J, Rossi D, Chiarenza A, Wells VA, Grunn A, Messina M, Elliot O, et al. Analysis of the coding genome of diffuse large B-cell lymphoma. Nat. Genet. 2011 doi: 10.1038/ng.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordonez GR, Bignell GR, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shah SP, Morin RD, Khattra J, Prentice L, Pugh T, Burleigh A, Delaney A, Gelmon K, Guliany R, Senz J, et al. Mutational evolution in a lobular breast tumour profiled at single nucleotide resolution. Nature. 2009;461:809–813. doi: 10.1038/nature08489. [DOI] [PubMed] [Google Scholar]
- 27.Stransky N, Egloff AM, Tward AD, Kostic AD, Cibulskis K, Sivachenko A, Kryukov GV, Lawrence M, Sougnez C, McKenna A, et al. The mutational landscape of head and neck squamous cell carcinoma. Science. 2011;333:1157–1160. doi: 10.1126/science.1208130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Totoki Y, Tatsuno K, Yamamoto S, Arai Y, Hosoda F, Ishikawa S, Tsutsumi S, Sonoda K, Totsuka H, Shirakihara T, et al. High resolution characterization of a hepatocellular carcinoma genome. Nat. Genet. 2011;43:464–469. doi: 10.1038/ng.804. [DOI] [PubMed] [Google Scholar]
- 29.Wei X, Walia V, Lin JC, Teer JK, Prickett TD, Gartner J, Davis S, Stemke-Hale K, Davies MA, Gershenwald JE, et al. Exome sequencing identifies GRIN2A as frequently mutated in melanoma. Nat. Genet. 2011;43:442–446. doi: 10.1038/ng.810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wood LD, Parsons DW, Jones S, Lin J, Sjoblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 31.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ciccarelli FD. The (r)evolution of cancer genetics. BMC Biol. 2010;8:74. doi: 10.1186/1741-7007-8-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rambaldi D, Giorgi FM, Capuani F, Ciliberto A, Ciccarelli FD. Low duplicability and network fragility of cancer genes. Trends Genet. 2008;24:427–430. doi: 10.1016/j.tig.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 34.D'Antonio M, Ciccarelli FD. Modification of gene duplicability during the evolution of protein interaction network. PLoS Comput. Biol. 2011;7:e1002029. doi: 10.1371/journal.pcbi.1002029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Syed AS, D'Antonio M, Ciccarelli FD. Network of cancer genes: a web resource to analyze duplicability, orthology and network properties of cancer genes. Nucleic Acids Res. 2010;38:D670–D675. doi: 10.1093/nar/gkp957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anelli V, Santoriello C, Distel M, Koster RW, Ciccarelli FD, Mione M. Global repression of cancer gene expression in a zebrafish model of melanoma is linked to epigenetic regulation. Zebrafish. 2009;6:417–424. doi: 10.1089/zeb.2009.0612. [DOI] [PubMed] [Google Scholar]
- 37.Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nat. Rev. Cancer. 2010;10:59–64. doi: 10.1038/nrc2771. [DOI] [PubMed] [Google Scholar]
- 39.Coffey AJ, Kokocinski F, Calafato MS, Scott CE, Palta P, Drury E, Joyce CJ, Leproust EM, Harrow J, Hunt S, et al. The GENCODE exome: sequencing the complete human exome. Eur. J. Hum. Genet. 2011;19:827–831. doi: 10.1038/ejhg.2011.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl. 1):S41–S49. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Muller J, Szklarczyk D, Julien P, Letunic I, Roth A, Kuhn M, Powell S, von Mering C, Doerks T, Jensen LJ, et al. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res. 2010;38:D190–D195. doi: 10.1093/nar/gkp951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human protein reference database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. The BioGRID interaction database: 2011 update. Nucleic Acids Res. 2010;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2009;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2009;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Inui M, Martello G, Piccolo S. MicroRNA control of signal transduction. Nat. Rev. Mol. Cell. Biol. 2010;11:252–263. doi: 10.1038/nrm2868. [DOI] [PubMed] [Google Scholar]
- 49.Croce CM. Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 2009;10:704–714. doi: 10.1038/nrg2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Papadopoulos GL, Reczko M, Simossis VA, Sethupathy P, Hatzigeorgiou AG. The database of experimentally supported targets: a functional update of TarBase. Nucleic Acids Res. 2009;37:D155–D158. doi: 10.1093/nar/gkn809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape Web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, Obenauf AC, Wackernagel W, Green G, Bouvier N, et al. Mutations in GNA11 in uveal melanoma. N. Engl. J. Med. 2010;363:2191–2199. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tadokoro Y, Ema H, Okano M, Li E, Nakauchi H. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J. Exp. Med. 2007;204:715–722. doi: 10.1084/jem.20060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Payton JE, Baty J, Welch J, et al. DNMT3A mutations in acute myeloid leukemia. N. Engl. J. Med. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fabbri M, Garzon R, Cimmino A, Liu Z, Zanesi N, Callegari E, Liu S, Alder H, Costinean S, Fernandez-Cymering C, et al. MicroRNA-29 family reverts aberrant methylation in lung cancer by targeting DNA methyltransferases 3A and 3B. Proc. Natl Acad. Sci. USA. 2007;104:15805–15810. doi: 10.1073/pnas.0707628104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Li B, Wang B, Niu LJ, Jiang L, Qiu CC. Hypermethylation of multiple tumor-related genes associated with DNMT3b up-regulation served as a biomarker for early diagnosis of esophageal squamous cell carcinoma. Epigenetics. 2011;6:307–316. doi: 10.4161/epi.6.3.14182. [DOI] [PMC free article] [PubMed] [Google Scholar]