Abstract
The Central Aspergillus Data REpository (CADRE; http://www.cadre-genomes.org.uk) is a public resource for genomic data extracted from species of Aspergillus. It provides an array of online tools for searching and visualising features of this significant fungal genus. CADRE arose from a need within the medical community to understand the human pathogen Aspergillus fumigatus. Due to the paucity of Aspergillus genomic resources 10 years ago, the long-term goal of this project was to collate and maintain Aspergillus genomes as they became available. Since our first release in 2004, the resource has expanded to encompass annotated sequence for eight other Aspergilli and provides much needed support to the international Aspergillus research community. Recent developments, however, in sequencing technology are creating a vast amount of genomic data and, as a result, we shortly expect a tidal wave of Aspergillus data. In preparation for this, we have upgraded the database and software suite. This not only enables better management of more complex data sets, but also improves annotation by providing access to genome comparison data and the integration of high-throughput data.
INTRODUCTION
Aspergillus is a genus of fungus, which has significant impact on human and animal welfare worldwide. Among this group are potent pathogens (1–3) causing invasive diseases, allergens, mycotoxin producers causing crop spoilage (4), as well as species used to our benefit in the food and drinks industries (5–7). Given the medical and industrial importance of Aspergillus, The Central Aspergillus Data REpository (CADRE) project was initiated in 2001 to support the international Aspergillus research community by gathering all genomic information for this genus into one public resource.
Our first report in 2004 (8), presented data from a pilot study of clinical isolate Aspergillus fumigatus Af293 (9). A. fumigatus is the most common mould pathogen of humans, causing both life-threatening invasive diseases of immunocompromised patients and allergic disease in patients with atopic immune systems (3). With few effective antifungal drugs available (10,11), there was a need within the medical community to better understand this organism. CADRE was used to bring together the work of an international collaboration to sequence and annotate a selected region of this clinical isolate. Building on the success of this initial study, the full genome was sequenced (12) and managed within CADRE.
Over the last decade, 11 other genomes have been sequenced, 8 of which are currently available within CADRE. CADRE facilitates visualization and analyses of data using the Ensembl (13) software suite. Much of our data have been extracted from GenBank (14) and augmented using both automated and manual efforts, with support from AspGD (15), specific annotation projects and the general Aspergillus community. The expansion of data within CADRE has enabled us to support other projects directly, such as Ensembl Genomes (16) and the European-led Aspergillus nidulans reannotation project (17).
As the interest in comparing strains and species increases, we will manage genomes as they arise and aim to become a broad resource, providing data for all sequenced Aspergilli strains. CADRE is not the only Aspergillus resource currently available. Other resources, however, such as AspGD that provides in-depth annotation using manual efforts and the Aspergillus Comparative database (http://www.broadinstitute.org/annotation/genome/aspergillus_group/MultiHome.html) that provides tools for viewing comparative data are narrower in their use of selected strains. In order to offer a wider range of data, we have taken the first steps to improving our initial website: we have focused on the user interface and relationships within the data content. We have made a significant jump in the Ensembl software version to provide users with a better page layout and to provide access to comparative data. In our collaboration with the Ensembl Genomes team, we have also established evolutionary relationships and performed genome-wide comparisons across the current genome content. With new data and an improved interface, users now have access to much richer Aspergillus information within CADRE.
SOURCE DATA AND CURRENT RELEASE
CADRE 3 has been implemented using Ensembl 59 and Ensembl Genomes 6 software suites. Ensembl is an annotation system and software suite devised for handling annotated eukaryotic genomes, its focus being mainly on higher eukaryotes. Ensembl Genomes complement this system by providing software to handle smaller eukaryotes and prokaryotes. We have used both systems and extended them by implementing specific plugins for CADRE.
The current release (January 2011) contains annotation pertaining to nine genomes initially extracted from GenBank: A. fumigatus Af293, A. fumigatus A1163 (18), A. nidulans FGSC A4 (19), A. oryzae RIB 40 (20), A. niger CIB 513.88 (21), A. clavatus NRRL 1 (18), Neosartorya fischeri NRRL 181 (18), A. flavus NRRL 3357 and A. terreus NIH 2624. The assemblies for A. niger, A. oryzae and A. nidulans are based on data extracted from published and unpublished material. A. nidulans assembly and revised annotation have since been published (17) and submitted to GenBank (BN001301-BN001308). At the time of writing, eight new records, representing each A. oryzae chromosome, have been submitted by RefSeq (BA000049-BA000056). These have arisen from assembly description files (AGP) files recently made available by the Japanese consortium (20). As these new data affect the assembly only, we will seek to update our current A. oryzae assembly to reflect the RefSeq records.
SEARCH AND DISPLAY SOFTWARE
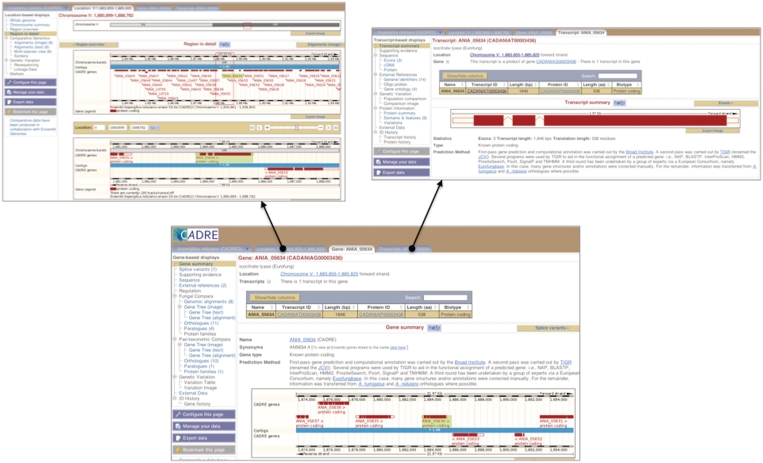
The CADRE home page provides links to all the genomes as well as a search box for searching each genome given general text or specific identifiers. For each genome, CADRE provides access to graphical views of assembled sequence and mapped features such as genes and transcripts. As the resource has grown, the predominant view has become the ‘GeneView’ (Figure 1) that imparts detailed information about a particular gene. The initial page presents a summary with information pertaining to public locus, genome location, related predicted transcripts and proteins, gene description and prediction methods. New features to this page are tabs along the top and a context-dependent menu down the left hand side. The tabs link to background information, further information on related transcripts (TranscriptView) and a view of the gene within the assembly (LocationView). The menu down the left hand side provides a set of links to familiar annotations, such as sequence and database cross-references (e.g. AspGD), as well as to new ones that present information regarding comparative analyses. It also includes a link to a pop-up window for data export, which allows the user to extract a region or feature of interest. As this new menu is common to all implementations of Ensembl software, where data or tools are currently unavailable (e.g. this may arise where Ensembl Genomes software is not inline with Ensembl), the appropriate links are disabled.
Figure 1.
Screenshots of GeneView, TranscriptView and LocationView. The delivery of gene annotation (bottom) has changed within CADRE due to the increasing volume of information and cross-linking. To ease data assimilation, it has been split and managed (i) using tabs along the top to link to pages providing further information on transcripts (top right) and the location within the assembly (top left), and (ii) introducing a new menu on the left hand side, providing links to database cross-references and secondary data resulting from analyses.
TranscriptView (Figure 1) displays information about a particular transcript. The centre of the page provides a summary of the transcript structure and a link to protein information where applicable. Protein-related information consists of predicted peptide statistics and cross-references to family- or domain-based signatures [e.g. PRINTS (22) and Pfam (23)] detected by InterProScan (24). Links to protein information are also included in the left hand side menu. Other links provided in this menu provide highlighted exon, cDNA and protein sequences as well as database cross-references. The database cross-references section includes mapped gene ontology (GO) terms (25) and related entries in external protein databases such as UniProt (26).
LocationView (Figure 1) presents a high-level assembly view of the contigs, supercontigs or scaffolds, which constitute a genome as well as the features that have been mapped onto them. Three levels of detail are displayed within the view: the first presents an overview of the best assembled feature (e.g. chromosome), the second focuses on assembly components (e.g. contig) and the third presents features mapped at sequence level (e.g. 55 000 bp of a contig). Predicted genes are displayed parallel to the assembly in accordance with their position and orientation. The position of other features, such as tRNAs, repeats and start/stop codons, can also be shown alongside the genes. As with GeneView and TranscriptView, a new menu is provided to the left. Within this menu, links to summaries of the genome and selected chromosome are provided. Two new links (Alignments and Multi-species view) are also included for viewing the results of whole genome alignments and will be discussed further in the following section.
EXPLORING NEW FEATURES IN CADRE
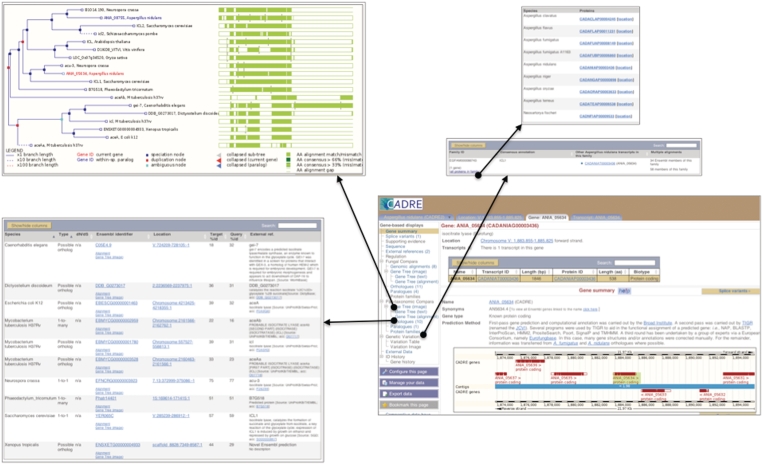
While GeneView, TranscriptView and LocationView remain central to viewing genomic data within CADRE, several new features have been added to enable the incorporation of comparative data analyses. GeneView now includes fungal and pan-taxonomic comparative data (which can be accessed via the new context-dependant menu at the left hand side), and if no correlating data has been successfully detected, the hyperlinks are disabled. As we have collaborated with the Ensembl Genomes project, the ‘Pan-taxonomic compara’ section (Figure 2) draws together data resulting from the analyses of representative genomes from all clades managed within the Ensembl and Ensembl Genomes databases (http://www.cadre-genomes.org.uk/info/docs/compara/homology_method.html#species). It is worth noting that these species are selected by the Ensembl Genomes management team to provide an overview of relationships across the broad taxonomic range, hence, not all Aspergilli are included. Data for orthologues, paralogues and gene trees have been calculated using the EnsemblCompara GeneTrees pipeline (27) that handles clustering, multiple sequence alignment and phylogenetic tree generation for translated genes. Protein family information is also available and has been generated by the Putative ORthologues Clusters (PROC) algorithm (http://www.ebi.ac.uk/integr8). This algorithm was developed as part of the Integr8 project (28) in order to group genes by the similarity of their longest protein product.
Figure 2.
Screenshot of ‘Pan-taxonomic Compara’ data. CADRE now includes the means of viewing data resulting from comparative analyses for Aspergillus and several other representative genomes from the taxonomic range. This information can be accessed from the new context-dependent menu within GeneView and includes images of gene trees (top left) as well as lists of orthologues (bottom left), paralogues and protein families (top right).
The ‘Fungal compara’ section (Figure 3) focuses on relationships between more closely related species and draws together the analyses of fungal genomes managed within the Ensembl Genomes database (Table 1). The results for this section differ from pan-taxonomic analyses as the full fungal component is utilized as opposed to a small number of representatives. As with the Pan-taxonomic section, it presents information regarding orthologues, paralogues and gene trees. Although, it does not provide information on protein families, it does include a link to pairwise genomic alignments between Aspergilli. Such alignments have been generated using Translated Blat Net, an approach used by Ensembl that combines techniques from Translated Blat (29) and BlastZ-net (30,31) pairwise alignment analyses. Within GeneView, this information is displayed in text format but it is also linked to images within LocationView.
Figure 3.
Screenshot of ‘Fungal Compara’ data. ‘Fungal compara’ is a new section within GeneView and provides links to data resulting from comparative analyses for all fungal species within Ensembl Genomes v6. This information can be accessed from the context-dependent menu at the left and includes images of genomic alignments, gene trees (top left) as well as list of orthologues (bottom left) and paralogues.
Table 1.
List of species used for fungal comparative analyses
| Aspergillus clavatus NRRL 1 |
| Aspergillus flavus NRRL 3357 |
| Aspergillus fumigatus Af293 |
| Aspergillus fumigatus A1163 |
| Aspergillus nidulans FGSC A4 |
| Aspergillus niger CBS 513.88 |
| Aspergillus oryzae RIB40 |
| Aspergillus terreus NIH 2624 |
| Neosartorya fischeri NRRL 181 |
| Neurospora crassa ATCC 24698/74-OR23-1A/CBS 708.71/DSM 1257/FGSC 987 |
| Saccharomyces cerevisiae S288C |
| Schizosaccharomyces pombe ATCC 38366/972 |
LocationView has been expanded to accommodate the viewing of whole genome alignments in two main forms. The first is the alignment of two genomes: one is defined as the reference sequence to which matching segments of the other are extracted and aligned. This can be viewed within text or image format in the menu to the left (Figure 4). The second view is the alignment of two or more genomes: again one is defined as the reference to which others are aligned. In this case, they are aligned by assembly component (e.g. scaffold or contig) and the significantly similar regions are highlighted (Figure 4).
Figure 4.
Screenshots of ‘AlignmentsView’ and ‘MultispeciesView’. Genomic alignments are available for Aspergillus species maintained within CADRE and can be accessed from either ‘LocationView’ or ‘GeneView’. ‘AlignmentsView’ (left) presents pairwise alignments as aligned segments of a query sequence to a reference sequence. In this example, A. nidulans is the reference sequence and A. niger is the query. ‘MultispeciesView’ (right) presents pairwise alignments as aligned regions and displays features as they are found in both assemblies. Significantly similar regions are highlighted.
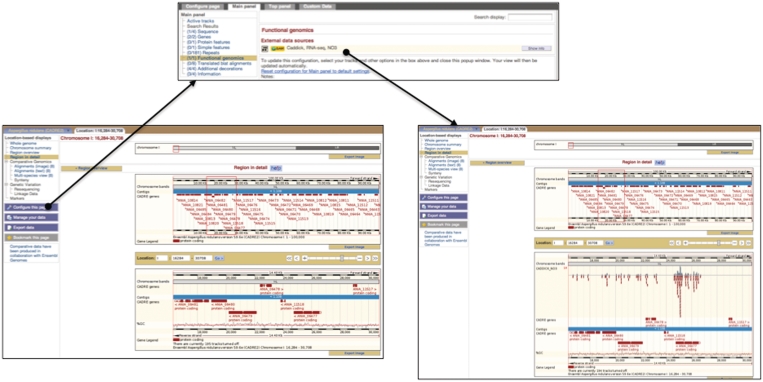
Another significant development within LocationView is the incorporation of functional genomic data from external sources. New low-cost high-throughput sequencing technology has led to the rise of whole transcriptome shotgun sequencing or RNA-seq, which yields information about the entire mRNA content of an organism under specific experimental conditions. In silico, this data can be used to improve gene prediction methods and to improve current annotation. As such information becomes available for Aspergillus, we wish to be able to incorporate it into the graphical views within CADRE, overlaying an annotated genome, and to improve annotation for current and future genomes. As a first step towards these goals, we have incorporated a link to an RNA-seq data set for A. nidulans (Figure 5), which has been made available by the University of Liverpool (courtesy of Mark Caddick).
Figure 5.
Screenshot of RNA-seq data. Using external data sources, features can be mapped onto existing annotation within ‘LocationView’ (left). This facility can be accessed via ‘Configure this page’, a blue button to the left of the page. On clicking this, a pop-up window (top right) appears with a list of configurable tracks. Selecting ‘Functional genomics’ provides a list of links to data that are provided by external researchers on their own servers and that can be switched on or off (bottom right). Different levels of information can be selected (normal, unlimited or coverage only), but we suggest the use of ‘normal’ or ‘coverage only’ due to the amount of data that has to be transferred from an external server.
FUTURE DIRECTIONS
As always, our aim is to keep our current genome content as up-to-date as possible as well as adding new genomes as they become available. AspGD has released updated annotation for A. nidulans that merges annotation from the current CADRE version (GenBank BN001301-BN001308) with a version made available by the Broad Institute in 2009 (http://www.broadinstitute.org/annotation/genome/aspergillus_group/MultiDownloads.html). We will endeavour to update this annotation as well as the previously mentioned A. oryzae assembly. As the participants of two current collaborative projects, SYBARIS (http://www.sybaris-fp7.eu) and Community Resources for A. fumigatus (gsc.jcvi.org/projects/gsc/a_fumigatus/index.shtml), we also expect to add annotation and RNA-seq data for three new A. fumigatus clinical isolates (Af210, Af10 and CEA10) in the coming months. To improve the incorporation of RNA-seq data, we are currently updating the Ensembl/Ensembl Genomes software as later versions compress the data for better viewing.
There are several areas that we aim to improve, one being the inclusion of user data. At present, Ensembl users are able to upload their own data for viewing within LocationView. This facility is provided by a button (Manage your data) under the new menu to the left of this page; however, it is currently disabled, as it requires further underlying database management. Enabling this facility in future versions will allow users to map their own data sets, such as gff or BAM files, to further enrich their view of existing annotated genomes within CADRE. Another service that will be addressed is the BLAST facility. This has been heavily used in previous versions but we are currently directing users to the BLAST service on our mirror site, Ensembl Genomes. There have been substantial changes to the installation of this service between versions and we are currently bringing this into line for future release.
Finally, in response to the need for metabolic pathway data, we created AsperCyc (http://www.aspercyc.org.uk) (32), a publicly available sister resource containing gene annotation and predicted metabolic pathways. AsperCyc is a stand-alone resource but closely linked with CADRE: i.e. we have used annotated genomes extracted from CADRE and Pathway-tools (v14.5) software (33,34), to predict metabolic pathways for Aspergilli. AsperCyc currently contains eight reference Aspergillus genomes that we have cross-linked with CADRE, and we aim to bring this full circle by linking from CADRE to AsperCyc. As with CADRE, we will add new genomes to AsperCyc as they become available. Some aspects of our work on metabolic pathways are supported by the Aspergillus/Aspergillosis website (http://www.aspergillus.org.uk) and our joint efforts to maintain a medically biased genomics website, Aspergillus Genomes (http://www.aspergillus-genomes.org.uk) (35). It is hoped that, by coupling all these resources, we can greatly enrich and expand the range of data available to the Aspergillus research community.
FUNDING
SYBARIS project; EU Framework Programme 7 Collaborative Project (grant number 242220). Funding for open access charge: SYBARIS, FP7 project (grant number 242220).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to thank Prof. David Denning (University of Manchester) for his continuing advice and support; the Ensembl Genomes team for their assistance and collaborative work; visiting students, Joose Rautemaa and Neil Stewart, for their contribution to our work; and Paul Schofield (University IT Services, University Hospital of South Manchester) for his help in maintaining the machines serving CADRE and it's sister resource AsperCyc.
REFERENCES
- 1.Denning DW. Invasive Aspergillosis. Clin. Infect. Dis. 1998;26:781–803. doi: 10.1086/513943. [DOI] [PubMed] [Google Scholar]
- 2.Latgé J-P. Aspergillus fumigatus and Aspergillosis. Clin. Microbiol. Rev. 1999;12:310–350. doi: 10.1128/cmr.12.2.310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marr KA, Patterson T, Denning D. Aspergillosis. Pathogenesis, clinical manifestations, and therapy. Infect. Dis. Clin. North Am. 2002;16:875–894. doi: 10.1016/s0891-5520(02)00035-1. [DOI] [PubMed] [Google Scholar]
- 4.Pitt JI. Toxigenic fungi: which are important? Med. Mycol. 2000;38(Suppl. 1):17–22. [PubMed] [Google Scholar]
- 5.Bennett JW, Klich MA. In: Encyclopedia of Bioprocess Technology: Fermentation, Biocatalysis, and Bioseparation. Flickinger MC, Drew SW, editors. New York: John Wiley and Sons, Inc.; 1999. pp. 213–220. [Google Scholar]
- 6.Nout MJ, Aidoo KE. In: Industrial Applications. Osiewacz HD, editor. Vol. X. Berlin: Springer; 2002. pp. 23–47. [Google Scholar]
- 7.Ruijter GJG, Kubicek CP, Visser J. In: Industrial Applications. Osiewacz HD, editor. Vol. X. Berlin: Springer; 2002. pp. 213–230. [Google Scholar]
- 8.Mabey JE, Anderson MJ, Giles PF, Miller CJ, Attwood TK, Paton NW, Bornberg-Bauer E, Robson GD, Oliver SG, Denning DW. CADRE: the Central Aspergillus Data REpository. Nucleic Acids Res. 2004;32:D401–D405. doi: 10.1093/nar/gkh009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pain A, Woodward J, Quail MA, Anderson MJ, Clark R, Collins M, Fosker N, Fraser A, Harris D, Larke N, et al. Insight into the genome of Aspergillus fumigatus: analysis of a 922 kb region encompassing the nitrate assimilation gene cluster. Fungal Genet. Biol. 2004;41:443–453. doi: 10.1016/j.fgb.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 10.Denning DW, Venkateswarlu K, Oakley KL, Anderson MJ, Manning NJ, Stevens DA, Warnock DW, Kelly SL. Itraconazole resistance in Aspergillus fumigatus. Antimicrob. Agents Chemother. 1997;41:1364–1368. doi: 10.1128/aac.41.6.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moore CB, Walls CM, Denning DW. In vitro activities of terbinafine against Aspergillus species in comparison with those of itraconazole and amphotericin B. Antimicrob. Agents Chemother. 2001;45:1882–1885. doi: 10.1128/AAC.45.6.1882-1885.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, et al. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438:1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 13.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Arnaud MB, Chibucos MC, Costanzo MC, Crabtree J, Inglis DO, Lotia A, Orvis J, Shah P, Skrzypek MS, Binkley G, et al. The Aspergillus Genome Database, a curated comparative genomics resource for gene, protein and sequence information for the Aspergillus research community. Nucleic Acids Res. 2010;38:D420–D427. doi: 10.1093/nar/gkp751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kersey PJ, Lawson D, Birney E, Derwent PS, Haimel M, Herrero J, Keenan S, Kerhornou A, Koscielny G, Kahari A, et al. Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res. 2010;38:D563–D569. doi: 10.1093/nar/gkp871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wortman JR, Gilsenan JM, Joardar V, Deegan J, Clutterbuck J, Andersen MR, Archer D, Bencina M, Braus G, Coutinho P, et al. The 2008 update of the Aspergillus nidulans genome annotation: a community effort. Fungal Genet. Biol. 2009;46(Suppl. 1):S2–S13. doi: 10.1016/j.fgb.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fedorova ND, Khaldi N, Joardar VS, Maiti R, Amedeo P, Anderson MJ, Crabtree J, Silva JC, Badger JH, Albarraq A, et al. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 2008;4:e1000046. doi: 10.1371/journal.pgen.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, Lee SI, Basturkmen M, Spevak CC, Clutterbuck J, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005;438:1105–1115. doi: 10.1038/nature04341. [DOI] [PubMed] [Google Scholar]
- 20.Machida M, Asai K, Sano M, Tanaka T, Kumagai T, Terai G, Kusumoto K, Arima T, Akita O, Kashiwagi Y, et al. Genome sequencing and analysis of Aspergillus oryzae. Nature. 2005;438:1157–1161. doi: 10.1038/nature04300. [DOI] [PubMed] [Google Scholar]
- 21.Pel HJ, de Winde JH, Archer DB, Dyer PS, Hofmann G, Schaap PJ, Turner G, de Vries RP, Albang R, Albermann K, et al. Genome sequencing and analysis of the versatile cell factory Aspergillus niger CBS 513.88. Nat. Biotechnol. 2007;25:221–231. doi: 10.1038/nbt1282. [DOI] [PubMed] [Google Scholar]
- 22.Attwood TK, Bradley P, Flower DR, Gaulton A, Maudling N, Mitchell AL, Moulton G, Nordle A, Paine K, Taylor P, et al. PRINTS and its automatic supplement, prePRINTS. Nucleic Acids Res. 2003;31:400–402. doi: 10.1093/nar/gkg030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2009;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Barrell D, Dimmer E, Huntley RP, Binns D, O'Donovan C, Apweiler R. The GOA database in 2009–an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Magrane M, Consortium U. UniProt Knowledgebase: a hub of integrated protein data. Database. 2011;2011:bar009. doi: 10.1093/database/bar009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kersey P, Bower L, Morris L, Horne A, Petryszak R, Kanz C, Kanapin A, Das U, Michoud K, Phan I, et al. Integr8 and Genome Reviews: integrated views of complete genomes and proteomes. Nucleic Acids Res. 2005;33:D297–D302. doi: 10.1093/nar/gki039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl Acad. Sci. USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tuckwell D, Denning DW, Bowyer P. A public resource for metabolic pathway mapping of Aspergillus fumigatus Af293. Med. Mycol. 2011;49(Suppl. 1):S114–S119. doi: 10.3109/13693786.2010.490243. [DOI] [PubMed] [Google Scholar]
- 33.Karp PD, Paley S, Romero P. The Pathway Tools software. Bioinformatics. 2002;18(Suppl. 1):S225–S232. doi: 10.1093/bioinformatics/18.suppl_1.s225. [DOI] [PubMed] [Google Scholar]
- 34.Karp PD, Paley SM, Krummenacker M, Latendresse M, Dale JM, Lee TJ, Kaipa P, Gilham F, Spaulding A, Popescu L, et al. Pathway Tools version 13.0: integrated software for pathway/genome informatics and systems biology. Brief. Bioinform. 11:40–79. doi: 10.1093/bib/bbp043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mabey Gilsenan JE, Atherton G, Bartholomew J, Giles PF, Attwood TK, Denning DW, Bowyer P. Aspergillus genomes and the Aspergillus cloud. Nucleic Acids Res. 2009;37:D509–D514. doi: 10.1093/nar/gkn876. [DOI] [PMC free article] [PubMed] [Google Scholar]