Abstract
The Gene Wiki is an open-access and openly editable collection of Wikipedia articles about human genes. Initiated in 2008, it has grown to include articles about more than 10 000 genes that, collectively, contain more than 1.4 million words of gene-centric text with extensive citations back to the primary scientific literature. This growing body of useful, gene-centric content is the result of the work of thousands of individuals throughout the scientific community. Here, we describe recent improvements to the automated system that keeps the structured data presented on Gene Wiki articles in sync with the data from trusted primary databases. We also describe the expanding contents, editors and users of the Gene Wiki. Finally, we introduce a new automated system, called WikiTrust, which can effectively compute the quality of Wikipedia articles, including Gene Wiki articles, at the word level. All articles in the Gene Wiki can be freely accessed and edited at Wikipedia, and additional links and information can be found at the project's Wikipedia portal page: http://en.wikipedia.org/wiki/Portal:Gene_Wiki.
INTRODUCTION
The goal of the Gene Wiki project is to create a continuously updated, community-reviewed and collaboratively-written review article for every human gene. In an era of genome-scale profiling, scientists are increasingly faced with the challenge of quickly learning about the core functions of unfamiliar genes. Writing gene-specific review articles is a daunting task when faced with more than 20 000 human genes, more than 21 million citations already in PubMed, and nearly a million new biomedical articles published every year. Since the writing of review articles is an inherently manual process, building and maintaining these review articles is a formidable challenge requiring intense and sustained human effort. In 2008, we initiated the Gene Wiki project to harness the intelligence of the broad scientific community to achieve this goal (1).
The Gene Wiki currently exists in the form of more than 10 000 articles in the online encyclopedia Wikipedia. Each Gene Wiki article can be unambiguously linked to an entry in the NCBI Gene database (2). Like the rest of Wikipedia, anyone can edit a Gene Wiki article through an open and collaborative hypertext authoring system. Individual contributions can be attributed to either an anonymous computer identifier or a specific Wikipedia user account, and a full revision history is stored for every article.
The Gene Wiki was created by automatically seeding ‘stub’ articles into Wikipedia with data primarily gathered from NCBI Gene (1). By systematically creating these stubs with a uniform layout and baseline level of content, we hypothesized that the community at large would maintain and expand these gene articles. Building upon the early success of this initial seeding process, the Gene Wiki articles were subsequently automatically enhanced with links to interacting genes and image galleries for relevant protein structures (3). In addition, a tool was created that allows users to generate properly formatted Gene Wiki stub articles for genes not yet in the Gene Wiki (http://biogps.org/plugin/493/genewiki-code-creator/).
Here, we describe the most recent automated updates to the Gene Wiki articles, provide an analysis of the Gene Wiki's continued community-driven growth and describe a new tool that effectively computes and displays information about the quality of Gene Wiki article content called WikiTrust.
UPDATE TO Gene Wiki AUTOMATION
While human editing is the core strength and motivation for the Gene Wiki, automated processes that insert and format structured data from the primary databases play an important role in the development and maintenance of each article (Figure 1). Recently we completed an extensive update to the program (known as the Protein Box Bot) that keeps the structured data on Gene Wiki articles current (http://code.google.com/p/genewiki/). Executing this updated bot resulted in 115 034 distinct changes to the information boxes on 10 204 Gene Wiki articles.
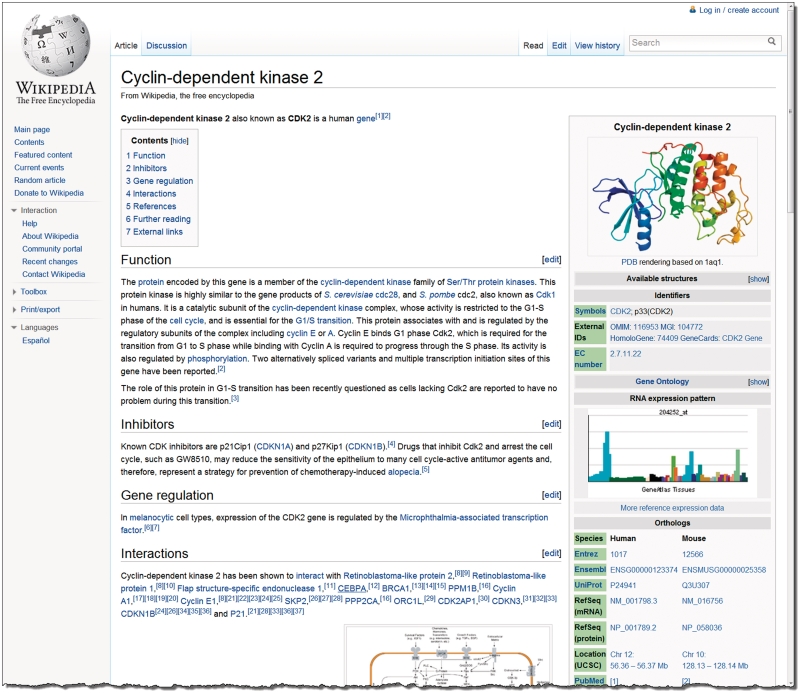
Figure 1.
The Gene Wiki article about Cyclin-dependent kinase 2. Programmatically gathered and updated data such as protein structure diagrams, Gene Ontology annotations and links to related database entries are displayed in the information box on the right. The manually authored text that forms the main body of the article is organized into subsections as indicated by the table of contents on the top left. Note that the article had to be truncated for space considerations and thus the bottom portion, including the references section, is not displayed.
The most common changes were updates to the chromosome location data (now corresponding to build hg19) for the gene in both human and mouse genomes, which occurred on 90% of the articles in the Gene Wiki. These updates largely reflected the change in genome assemblies since the Gene Wiki effort was initiated (4). In addition, an effort was made to supply protein structure illustrations to all genes with sufficient structural information, resulting in the generation of nearly 500 new images using the PyMOL program (The PyMOL Molecular Graphics System, Version 1.3, Schrödinger, LLC) and the insertion of 2853 images already in the public domain into Gene Wiki articles. The Gene Ontology annotations were also updated in >70% of the articles, reflecting the ongoing expansion of gene annotation databases since the last update.
Automated updates such as these remove the need for human editors to spend time synchronizing structured content with authoritative databases. Instead, authors can focus on providing more valuable contributions to the body of the article on the functions and disease relevance of the gene and corresponding protein. In addition, the automated updates keep every article factually accurate, even the ones that receive little attention. By keeping these articles up-to-date, we sustain the relevancy and accuracy of each article, potentially drawing more viewers (and thus more editors). It also prevents articles that have not yet been expanded from becoming too far obsolete, maintaining their use as ‘seed’ articles for further expansion. To this end, the bot is now running on a monthly schedule, ensuring the structured data is never significantly out-of-date.
CONTENT
As of 1 September 2011, the Gene Wiki contained 10 369 articles, representing approximately one half of the known human protein-coding genes in NCBI Gene. The number of articles has increased by more than 2000 since the automated article creation process concluded in February 2008. Members of the community created all of these new articles manually. Taken together, these articles now contain approximately 78 megabytes of data and nearly 1.42 million words (not including references). The largest article, for the protein insulin, contains about 6450 words, roughly equaling 26 pages of text.
Importantly, the text of the Gene Wiki articles is complemented by extensive citations of the primary literature. Overall, the Gene Wiki contains in-line references to 37 578 PubMed citations with about 200 new citations added each month. These citations allow interested readers to easily explore the literature cited in support of statements made in a Gene Wiki article.
In addition to literature citations, Gene Wiki articles contain many hyperlinks to other Wikipedia articles on related topics. In total, the hypertext of Gene Wiki articles contains more than 109 000 links to other articles in Wikipedia. The average number of links per article is 11 and the maximum is 420 (for insulin). As detailed in previous work, the number of links per article follows a power law distribution in which a few hub genes are very well connected while the majority are only loosely connected (3).
We see similar patterns for many aspects of the Gene Wiki as we see for the hyperlinks. Power law-like distributions are found when examining the number of words, revisions and references to PubMed citations per article, as well as the number of contributions per editor (see Supplementary Data). In general, there are a large number of fairly small articles and a few very large articles, and there are a large number of editors but only a small number of heavy contributors. These trends are consistent with a variety of studies of other open data systems including the many different language Wikipedias (5), social bookmarking systems (6,7) and a sample of more than 100 wikis operating in many different domains (8).
To gain some insight into the underlying content of the Gene Wiki articles, we conducted a semantic analysis of article text using the MetaMap concept recognition system (9). Using MetaMap we identified a wealth of relationships between genes and biomedical concepts in the text of Gene Wiki articles (Figure 2). The analysis and application of relational data such as this mined from Gene Wiki text is an area of ongoing research.
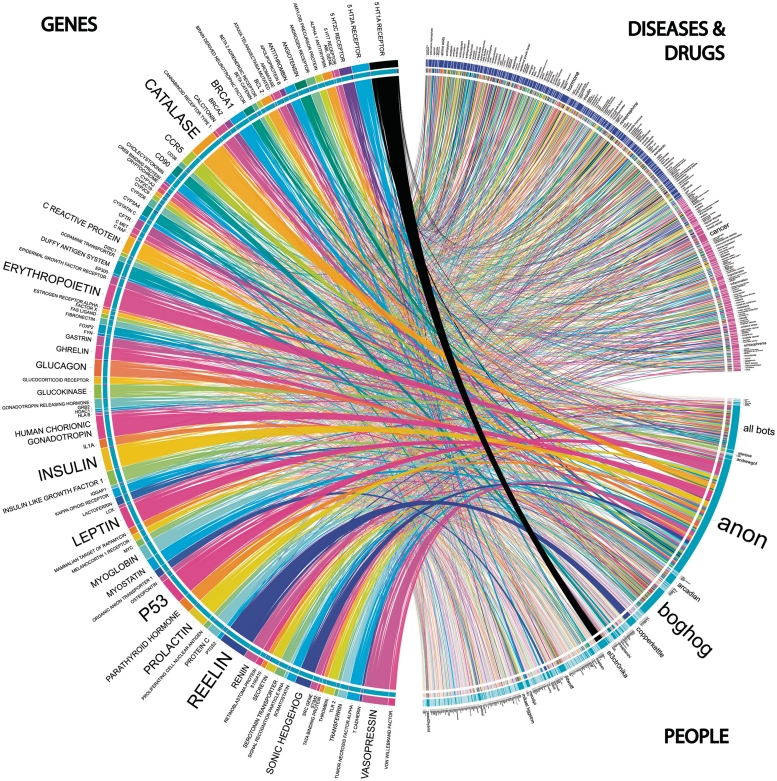
Figure 2.
The top 100 most heavily linked genes, their connections to topics classified as diseases or drugs, and to the people that contributed most heavily to each article. (‘anon’ represents anonymous editors and ‘all bots’ aggregates automated edits to the articles). The thickness of the band connecting a gene to a person indicates the relative number of edits made to the article for that gene by that person.
COMMUNITY
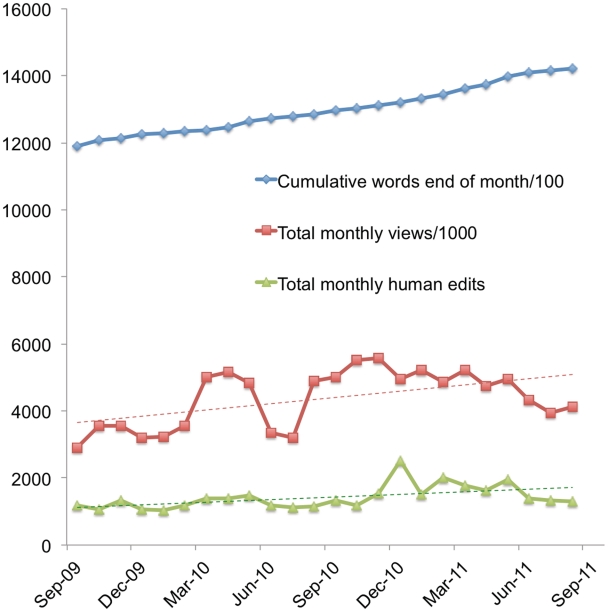
Nearly all of the textual content of the Gene Wiki is the result of manual effort by unpaid volunteers. The reasons why people spend their time contributing to the Gene Wiki and Wikipedia are diverse and not well understood. However, examining the progress of successful web-based volunteer-driven content-creation projects like Wikipedia, it is clear that a positive feedback loop between content value, content use and content production is a necessary aspect of the overall system. Within the context of the Gene Wiki, we roughly quantified these three pillars of a community intelligence system using the amount of text as an indicator of value, article views as an indicator of use, and edits as an indicator of production. Figure 3 illustrates the steady increase in the value of the Gene Wiki as indicated by increasing total word count. In addition it demonstrates less uniform but still evident increases in the amount of editing activity and article views per month between 1 September 2009 and 1 September 2011.
Figure 3.
Monthly growth of words in Gene Wiki articles, page views per month and edits per month between 1 September 2009 and 1 September 2011.
While word counts provide a useful quantitative indicator, the real value of the Gene Wiki to the scientific community is its use as a novel supplement to institutionally curated gene annotation databases. When a Gene Wiki article is first created, it already provides a useful integration of information from several of these databases. As the articles grow, they provide an increasingly rich, textual integration of knowledge pertinent to each gene that could only be achieved by manual effort. Since Wikipedia mandates that ‘all material added to articles must be attributable to a reliable, published source’ (http://en.wikipedia.org/wiki/Wikipedia:Verifiability), this growing corpus of text is also a growing corpus of references to the primary scientific literature. Scientists can thus use the Gene Wiki to come up to speed quickly about individual genes, to obtain links to other gene annotation resources, and as a dynamic, manually curated, gene-centric index of the literature. Given these less tangible attributes, the amount of use is likely the best indicator of actual value.
In the 2 years examined here, the Gene Wiki article set in aggregate was viewed approximately 104.9 million times, equal to an average of 422 views per article per month. This represents an increase from approximately 300 views per article per month reported in the first half of 2009 (3). The most views accumulated for a particular article in one month was 188 247 for the article on human chorionic gonadotropin (HCG) in June of 2011. As with article size and links, the distribution of views per article remained highly skewed with the most popular genes being viewed tens of thousands of times per month while some genes were viewed only a handful of times.
As both an explanation for the large and increasing number of article views and as another indicator of relative value, we performed an analysis of the status of Gene Wiki articles as viewed through the Google search engine. On 17 August 2011, 92% of Gene Wiki articles appeared on the first page of results when searching Google with the official gene symbol for the corresponding gene. This was an increase from the 85% reported in 2009 and the 66% reported in 2008 providing clear evidence of the increasingly important place that Gene Wiki articles have earned in the context of the Web.
In what we hypothesize to be a direct consequence of the increasing value and visibility of the Gene Wiki articles, we also observed a trend toward increasing editing activity (Figure 3). In the 2 years examined here, we identified 34 069 edits by 6830 distinct editors [based on account name or Internet Protocol (IP) address if anonymous]. In addition to these manual edits, 113 different bots made another 35 380 automated edits (not including the Protein Box Bot edits discussed above). In total, these revisions resulted in a net expansion of about 9 MB of content corresponding to more than 230 000 words of article text.
QUALITY (WikiTrust)
One of the most common criticisms of Wikipedia, especially for scientific topics, is the uncertain reliability of community-contributed content. Wikipedia does not explicitly take into account the academic credentials of editors, and in most cases each editor's background is unknown.
To better assess the trustworthiness of authors and the text that they contribute, we used a reputation system for Wikipedia called WikiTrust (10). In WikiTrust, authors gain reputation when subsequent editors preserve their contributions, and text gains reputation when it is preserved by edits by high-reputation authors. Experimental data showed a strong correlation between the reputation of authors and the quality of their future contributions, as well as between the reputation of text [called ‘trust’ in (10)] and its future persistence in Wikipedia. Author and text trust, together with other signals, can be used to identify Wikipedia vandalism, with a recall of 90% and a precision of 43% (11). Thus, 90% of the vandalism present in Wikipedia is correctly labeled as such by WikiTrust; furthermore, the percentage of edits labeled as vandalism by WikiTrust that are truly vandalism is 43%. These performance figures were obtained by comparing the WikiTrust classifier output with the opinions of human subjects, gathered via Amazon Mechanical Turk; more details on the evaluation setup can be found in (11). The low precision is due in part to the relative rarity of vandalism, which comprises only ∼5% of Wikipedia revisions.
Using the WikiTrust system, we conducted a comparative analysis of the quality of Gene Wiki articles as compared to general Wikipedia articles where ‘general’ indicates any Wikipedia article other than a Gene Wiki article. The analysis compared the 10 028 Gene Wiki articles with at least seven revisions with a random sample of 50 130 Wikipedia articles also with at least seven revisions. Of the seven most recent revisions of every analyzed article, we considered the middle five (thus discarding the oldest, and most recent), as on those the WikiTrust quality metrics are more precise.
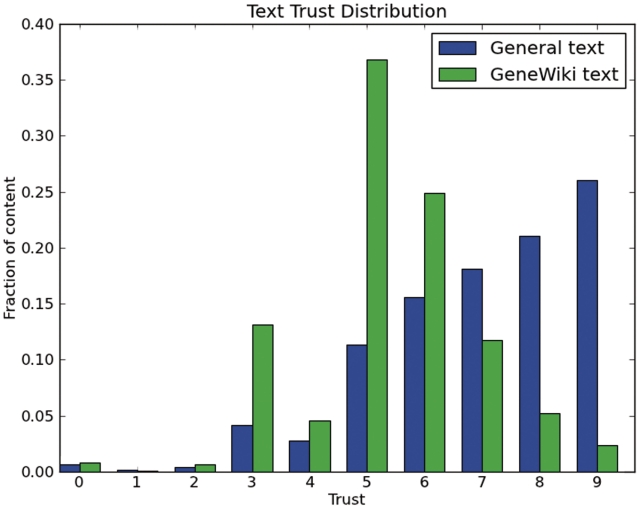
In the WikiTrust system, newly added text has a trust value in the range of 0–3, depending on the author's reputation. Fully trusted text has a trust value of 9 (the maximum value we assign in WikiTrust). We calculated the trust distributions of Gene Wiki articles versus a random sampling of Wikipedia articles (Figure 4). More precisely, the distribution of text trust scores were computed for each article revision and weighted according to the length of time that specific revision was ‘live’.
Figure 4.
Trust distributions of Gene Wiki revisions versus general (non-Gene Wiki) Wikipedia revisions.
These results illustrated that the most common trust values in Gene Wiki articles were intermediate ones, while general Wikipedia articles most commonly had the highest trust values. Trust values increase with each edit, and WikiTrust prevents any individual author from raising the trust multiple times in a row (12). The difference in trust distribution could be thus explained by the Gene Wiki articles having a smaller number of edits, by a smaller number of editors, compared with general Wikipedia articles.
WikiTrust scores can be accessed directly from the WikiTrust API (http://www.wikitrust.net/) and can be visualized using a plugin to the Firefox web browser (Figure 5). In the visual interface, text with a trust value of nine is displayed on white background, while text with lower trust values are highlighted with an orange background. The shade of orange is proportional to trust scores where more intense color corresponds to lower text trust (10).
Figure 5.
Screenshot of Firefox plugin displaying WikiTrust information for the Wikipedia article on the gene CDK2.
In addition to the distributions of trust per article, we used the WikiTrust system to compute the total likelihood that a visitor to the Gene Wiki would see an article displaying vandalized content. Again, to measure this probability we considered articles with at least seven revisions, and we considered the middle five of these last seven revisions. We calculate the likelihood of seeing vandalism as the fractional time that the Gene Wiki was in a vandalized state (as detected by WikiTrust):
where  is the fractional time in a vandalized state,
is the fractional time in a vandalized state, 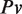 is the probability that any given edit is vandalism,
is the probability that any given edit is vandalism, 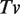 is the average lifetime of a vandalized revision, and Tnv is the average lifetime of a non-vandalized revision. For Gene Wiki articles, Pv was 0.011, Tv was 3.43 days and Tnv was 115.4 days, leading to a total fractional time of vandalism of 0.00032. This figure indicated that only about 3 article views out of 10 000 would display vandalized content. In contrast, general Wikipedia articles have Pv = 0.042, Tv = 10.75 days and Tnv = 74.2 days, leading to FTv = 0.0063, or approximately 63 article views out of 10 000. Viewed from a different perspective, we can see that the cumulative production of ‘good’ revisions has far outstripped the insertion of vandalism in the Gene Wiki over the past 2 years (Figure 6). These results were based on the WikiTrust vandalism detection algorithm, which has a precision of ∼43% (i.e., it over-predicts vandalism by 57%), so the real frequencies of vandalism were actually lower. Nevertheless, assuming that the precision of vandalism detection is roughly the same for Gene Wiki and for general Wikipedia articles, these results indicated that a user was 19 times more likely to visit a vandalized article on the general Wikipedia than on the subset of Gene Wiki articles.
is the average lifetime of a vandalized revision, and Tnv is the average lifetime of a non-vandalized revision. For Gene Wiki articles, Pv was 0.011, Tv was 3.43 days and Tnv was 115.4 days, leading to a total fractional time of vandalism of 0.00032. This figure indicated that only about 3 article views out of 10 000 would display vandalized content. In contrast, general Wikipedia articles have Pv = 0.042, Tv = 10.75 days and Tnv = 74.2 days, leading to FTv = 0.0063, or approximately 63 article views out of 10 000. Viewed from a different perspective, we can see that the cumulative production of ‘good’ revisions has far outstripped the insertion of vandalism in the Gene Wiki over the past 2 years (Figure 6). These results were based on the WikiTrust vandalism detection algorithm, which has a precision of ∼43% (i.e., it over-predicts vandalism by 57%), so the real frequencies of vandalism were actually lower. Nevertheless, assuming that the precision of vandalism detection is roughly the same for Gene Wiki and for general Wikipedia articles, these results indicated that a user was 19 times more likely to visit a vandalized article on the general Wikipedia than on the subset of Gene Wiki articles.
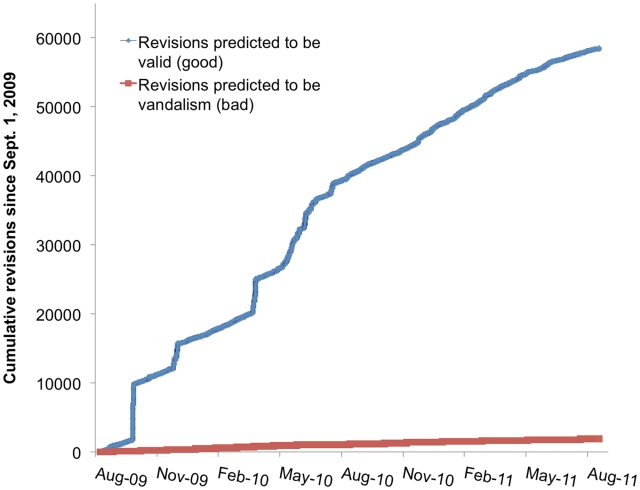
Figure 6.
Cumulative revisions to Gene Wiki articles with trust assessments between 1 September 2009 and 1 September 2011. Note that the figure includes both manual and bot edits. The sharp edit spikes that occur near the beginning of the chart are the result of bots. Overall about half of the edits are manual and half are automated.
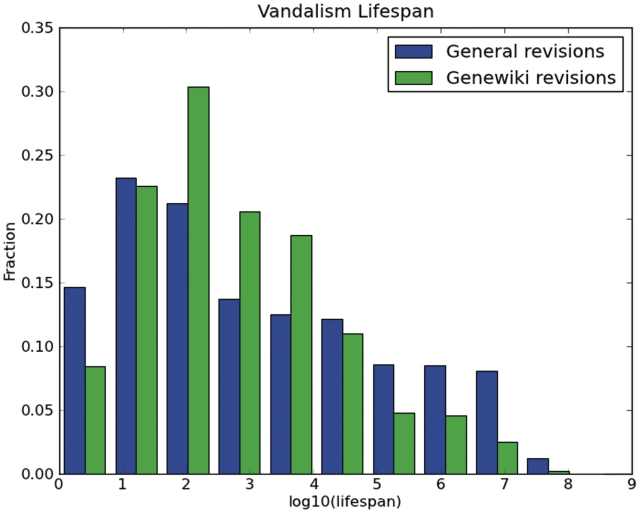
We also compared the longevity distribution of vandalism on the general Wikipedia, and on the Gene Wiki (Figure 7). These results illustrated that fewer acts of vandalism survive in the Gene Wiki for longer than 105seconds, or approximately 1 day. Similar results hold for revisions that were ultimately reverted, a measure that is not affected by the limited precision and recall of vandalism detection. Such revisions were five times more likely to be encountered by a visitor at a random time on the general Wikipedia, compared to the Gene Wiki.
Figure 7.
Longevity distribution, in seconds, of vandalism on the Gene Wiki and on the general (non-Gene Wiki) Wikipedia.
CONCLUSION
In the past 2 years, the Gene Wiki has expanded in terms of size, editing activity and user community. We have recently made substantial improvements to the machinery that keeps it in sync with external, trusted data sources. We have also identified a reliable method for quantifying the quality of article content. Despite these positive indicators of growth and important technical improvements, the Gene Wiki will always remain a work in progress. Few of the articles in the Gene Wiki could be described as complete with respect to the current state of knowledge, and the total amount of knowledge about all genes is increasing at a rapid rate. This initiative aims to create a continuously updated, community-reviewed and collaboratively written review article for every human gene. If the Gene Wiki is to succeed in this effort, participation by the scientific community will need to be significantly expanded. As with any successful community intelligence initiative, continuing to build this critical mass of editors remains our foremost area of emphasis for the future.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Data 1-3, Supplementary Figures 1-5.
FUNDING
National Institutes of Health (GM089820 to L.A. and A.I.S, and GM083924 to A.I.S). Funding for open access charge: United States National Institutes of Health (grant GM089820).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors acknowledge the entire community of Wikipedia editors and the Molecular and Cellular Biology WikiProject (http://en.wikipedia.org/wiki/WP:MCB) for contributions and feedback. In particular we thank Konrad Koehler for his helpful suggestions and enthusiastic editing. We also thank Martin Krzywinski for assistance creating the Circos diagram in Figure 2.
REFERENCES
- 1.Huss JW, Orozco C, Goodale J, Wu C, Batalov S, Vickers TJ, Valafar F, Su AI. A gene wiki for community annotation of gene function. PLoS Biol. 2008;6:e175. doi: 10.1371/journal.pbio.0060175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2011;39:D52–D57. doi: 10.1093/nar/gkq1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huss JW, Lindenbaum P, Martone M, Roberts D, Pizarro A, Valafar F, Hogenesch JB, Su AI. The Gene Wiki: community intelligence applied to human gene annotation. Nucleic Acids Res. 2010;38:D633–D639. doi: 10.1093/nar/gkp760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ortega F. Thesis. Universidad Rey Juan Carlos, Madrid. 2009. Wikipedia: A Quantitative Analysis. [Google Scholar]
- 6.Golder S, Huberman B. Usage patterns of collaborative tagging systems. J. Inform. Sci. 2006;32:198–208. [Google Scholar]
- 7.Good BM, Tennis JT, Wilkinson MD. Social tagging in the life sciences: characterizing a new metadata resource for bioinformatics. BMC Bioinformatics. 2009;10:313. doi: 10.1186/1471-2105-10-313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stuckman J, Purtilo J. WikiSym '09. Florida: ACM, Orlando; 2009. Measuring the wikisphere. [Google Scholar]
- 9.Aronson AR. American Medical Informatics Annual Symposium. Philadelphia: Hanley & Belfus; 2001. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program; pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 10.Adler BT, Chatterjee K, de Alfaro L, Faella M, Pye I, Raman V. Proceedings of WikiSym '08, International Symposium on Wikis. Porto, Portugal: ACM; 2008. Assigning trust to Wikipedia content. [Google Scholar]
- 11.Adler BT, de Alfaro L, Pye II. PAN lab report, CLEF (Conference on Multilingual and Multimodal Information Access Evaluation) Padua: Italy; 2010. Detecting Wikipedia vandalism using WikiTrust. [Google Scholar]
- 12.Chatterjee K, de Alfaro L, Pye I. AISec '08 Workshop on Security and Artificial Intelligence. Alexandria, Virginia: ACM Press; 2008. Robust content-driven reputation. [Google Scholar]