Abstract
The PlantMetabolomics (PM) database (http://www.plantmetabolomics.org) contains comprehensive targeted and untargeted mass spectrum metabolomics data for Arabidopsis mutants across a variety of metabolomics platforms. The database allows users to generate hypotheses about the changes in metabolism for mutants with genes of unknown function. Version 2.0 of PlantMetabolomics.org currently contains data for 140 mutant lines along with the morphological data. A web-based data analysis wizard allows researchers to select preprocessing and data-mining procedures to discover differences between mutants. This community resource enables researchers to formulate models of the metabolic network of Arabidopsis and enhances the research community's ability to formulate testable hypotheses concerning gene functions. PM features new web-based tools for data-mining analysis, visualization tools and enhanced cross links to other databases. The database is publicly available. PM aims to provide a hypothesis building platform for the researchers interested in any of the mutant lines or metabolites.
INTRODUCTION
PlantMetabolomics.org stores the data from an NSF-funded multi-institutional consortium that is developing metabolomics as a functional genomics tool for elucidating the functions of Arabidopsis genes without visible phenotype. The consortium has established mass spectrometry-based metabolomics platforms that detect ∼2000 metabolites, of which ∼1000 are chemically defined (1). The consortium generates the Arabidopsis biological material at a single location followed by distribution to the analytical laboratories for targeted and untargeted analyses. Phase 1 focused on investigating the robustness of the Arabidopsis metabolome and defining the conditions that minimize the environmental and developmental effects. Subsequently, the consortium profiled the metabolome of specific T-DNA knockout alleles for these targeted genes (2). These MSI-compliant metabolomics data (3,4) are integrated with phenotypic data and data concerning protein function, transcription and other studies to help users generate hypotheses concerning the functions of the targeted genes. The datasets complement the Arabidopsis developmental (5) and ecotype (6) LC-MS datasets at AtMetExpress.
The updated PlantMetabolomics.org database features new datasets and morphological information for the plant community along with new web-based analysis tools. These tools include clustering and classification tools to distinguish between different mutants as well as determining which metabolites best differentiate the mutant. New visualization tools include ratio plots of metabolites and CytoscapeWeb (7) pathway visualization of metabolites on the AraCyc pathways (8). PlantMetabolomics (PM) also offers web services for the concentration data and annotation sharing.
DATABASE CONTENTS
PlantMetabolomics.org contains mass spectrometry-based metabolomics concentration data for 140 novel single-knockout gene mutant lines in Arabidopsis. Fifty-three lines are novel since the last release and 35 were repeated to increase the number of replications. Approximately 998 known metabolites and 2020 unknown metabolites were detected using seven different MS-based platforms for each of these mutant lines. The number of replicates for each line was also increased from three replicates to six replicates.
The database has also added morphological image data including features of the mutants’ leaves, cotyledons and roots at 16 days after imbibitions (DAI) and mature seeds using an Olympus stereomicroscope with reflected and transmitted light sources and a high-resolution digital color image and scanning electron microscope. Digital camera images of the roots of all the Arabidopsis thaliana tissue were collected at 6, 9, 13 and 16 days after imbibitions (DAI) in pixels, and these were converted from pixels to root length measurements using Image J software (9). A user can select a gene and compare its morphological images with the images from the wild-type samples using a side-by-side image analysis tool in the database, which is accessible when the user searches for a gene of interest from the home page or uses the search functionality to search for a gene.
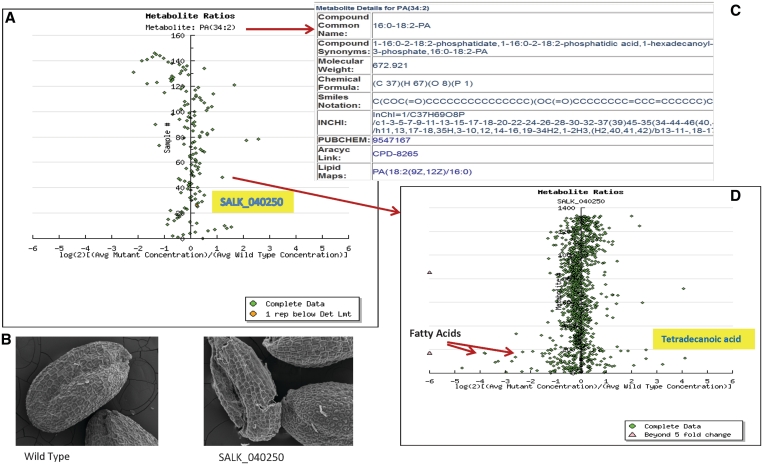
New annotation links to LipidMaps (10) have been added for metabolites. Structurally known metabolites have been annotated with metabolic pathway information from the AraCyc database (version 8.0) (8). This annotation helps users understand how changes in a metabolite might affect the metabolism of the entire organism. Figure 1 shows an example of the new annotation and the images.
Figure 1.
(A) Log-ratio plot of a metabolite (PA 34:2), where each point shows the ratio of the concentration of the given metabolite in mutant samples versus the wild-type samples. The highlighted mutant line (SALK_040250) looks interesting as it is away from the central vertical axis and thus depicts difference between mutant samples and the wild-type samples. (B) The user can instantly access the stereomicroscopic images for this mutant and compare them with wild-type samples. Seed images at ×250 zoom of mutant's seeds look a little distorted as compared to the wild-type seeds (Seed image courtesy of Jennifer Robinson). (C) The user can also access the details of the metabolites including cross links to other databases. (D) Clicking on any of the points in the log-ratio plot in (A) shows the log-ratio plot of all the metabolites for that mutant. For example, some fatty acids including tetradecanoic acid look interesting for this mutant as they are away from the central vertical axis and show large fold change between the wild type and mutant samples.
Analysis tools for metabolomics
PlantMetabolomics.org includes new web-based data analysis tools to aid a researcher in generating hypothesis about the metabolomics signature of a mutation. The data analysis wizard provides various options to normalize and preprocess data along with many choices of multivariate data analysis methods along with step-by-step guidance on the analysis pipeline. Default choices are provided at each step, and the downstream analyses are made available only after the necessary preprocessing steps have been successfully performed. All the analysis results and figures are made available for download at the end of the analysis. The data analysis tool is developed with PHP and the R programming environment (11).
Data preprocessing
The data preprocessing steps involve missing value imputation and normalization. For missing value imputation, the user selects a threshold to eliminate metabolites that have a higher percentage of missing values than the threshold (e.g. for a threshold of 50%, a metabolite with four or more missing values out of six will be removed from further computation). For cases where there are fewer missing values, the missing values will be imputed by means of the concentration for that metabolite over the remaining values. The next step is data normalization. Data normalization weights the metabolites to emphasize different attributes of the data. Common choices described in (12), Range Scaling, Pareto Scaling and Auto Scaling, help weight metabolites equally regardless of overall abundance. Log transformation is used to correct for heteroscedascity and make multiplicative effects additive. The equations and a discussion of each method are accessible from the ‘?’ icon in the data analysis wizard. After the preprocessing and normalization steps, a user can choose one or more of the analysis tools to analyze the data. Examples have been provided at each data mining step to help users interpret their results.
Clustering analysis
Biologists can generate hierarchical clustering plots to see which mutants are statistically close to each other and have similar metabolic profiles. Multiple choices for distance measure (Euclidean and Manhattan) and for the linkage method (Ward, complete, single, average, median and centroid) are available. The goal is to group or segment a collection of samples (mutants) into subsets or ‘clusters’, such that those within each cluster are more closely related to one another than objects assigned to different clusters. The result of clustering is presented as a dendrogram that a user can download from the PM Web site. Figure 2A shows an example of a dendrogram using hierarchical clustering analysis tool with average linkage and Euclidean distance parameters.
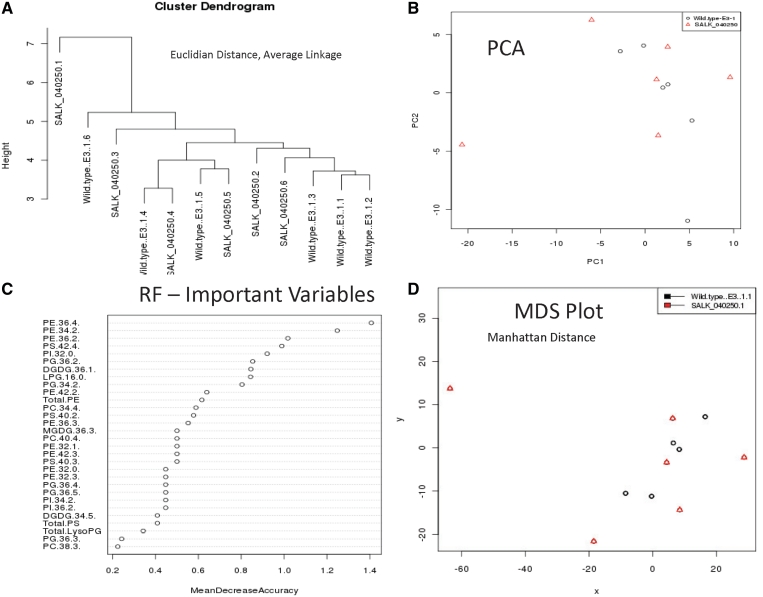
Figure 2.
(A) Hierarchical clustering of lipidomics data from the Welti Lab compares SALK_040250 (At1g61720) mutant line with wild-type samples using Euclidean distance and average linkage method. (B) PCA loadings plot of the first 2 PCs shows that the wild type and mutant are not linearly separable. (C) Important metabolites for the classification between wild type and the mutant line using the Random Forest tool shows that the most important variables are glycerophospholipids with chain lengths of 34 and 36. (D) MDS plot of the mutant and wild-type samples using the Manhattan distance measure that shows that the mutant and wild type are not separable and that there is an outlier in the data.
Multidimensional scaling
A multidimensional scaling (MDS) plot is a commonly used multivariate exploratory data analysis tool. MDS is an exploratory multivariate data analysis method that is used in visualizing the structure of relations between entities by providing a geometrical representation of these relations in a lower dimensional space (13). An MDS plot shows the similarities or dissimilarities in data in two dimensions. In this case, the MDS plot shows statistical distances among samples based on their metabolome signatures (Figure 2D). Commonly used distance measures (Euclidean and Manhattan) are provided for this tool as well.
Principal component analysis
Principal component analysis (PCA) is one of the most commonly used methods used in high-dimensional data analysis (14). PCA provides a low-dimensional view of the multidimensional data by mathematically transforming a number of correlated variables into a smaller set of uncorrelated variables which are called principal components (PCs). A user can generate PCA plot against the first two principal components and also the scree plot that show the percentage of variability explained by subsequent principal components. The PCs are orthogonal and are ordered according to the variance explained. Therefore, the first PC explains the maximum variance. If the variance in the data reflects the true biological difference, then plotting first PC against the second can be used to visualize the separation in the different classes. The original variables that contribute the most to the first few PCs are considered to be the most important. The PCs can be downloaded for further analysis. Figure 2B shows an example of PCA loadings plot for the first 2 PCs.
Random Forest classifier
Random Forests are used in metabolomics for classifying mutants into different classes (15). A Random Forest Classifier is an ensemble of classification trees (16). Random Forests work well for classification when the number of features is much greater than the number of observations, and they have good predictive performance even when most input variables are noisy (17). Of importance to biologists is that the output is easy to understand, because it does not transform the metabolite data and the output ranks variables that are responsible for classification.
The classification trees are built using a bootstrap sample of the data generated by using two-third of the data for sample generation and keeping the remaining one-third of the data for testing. A small subset of the variables is used in building a tree. The random Forest R package provides classification analysis between two or more types of samples (e.g., wild type and a mutant line) (18) and generates the variable importance score plots of the key metabolites (Figure 2C). The list of top 30 key metabolites is also made available along with the annotations for the metabolites. One can click on a metabolite name on this list and see its annotation from various external databases such as KEGG, AraCyc and Lipid Maps. The automatically generated ratio plot shows the metabolite's behavior in the other mutants when compared with wild-type samples. The complete list can be downloaded by clicking at the download file link and used in other applications. The random forest classifier can also be downloaded along with the number of correctly classified and misclassified samples in each class.
Download results
At the end of analysis, the user can download all the results along with comma separated data files and as well as the R code used at each step of the analysis. Examples are also provided at each step to help the users with the interpretation of their results.
Visualization tools for metabolomics
New data visualization plots were added, so that a user can select a metabolite and see its behavior in 140 different mutations in a single plot (as a ratio of mutant and wild-type samples). Similarly, a user can select a gene and see the behavior of all the metabolites (as compared to the wild-type samples). After selecting a gene of interest, a user is taken to gene details page where they are shown the morphological data along with a log-ratio plot of the data. In the log-ratio plot for a gene, each point shows the log-ratio (to base-2) of a metabolite's abundance in the (mutant sample):(wild-type sample). The points are color coded according to the number of missing values for each metabolite and provide an instant data quality check. Clicking on a point in the log-ratio plot takes the user to a page where annotation of that metabolite with the information about its participation in pathways and links to other databases like KEGG (19), LipidMaps (10) and PUBCHEM (20) are shown. The metabolites are annotated with a local copy of the AraCyc database (21) that was updated to the latest release of version 8.0 of AraCyc.
Single metabolic pathways from AraCyc can also be viewed using CytoscapeWeb (7) and PathwayAccess tools (22). From the annotation page, a user can select a pathway that contains their metabolite of interest and view the pathway with their metabolomics data superimposed for any of the experiments in the database.
CONCLUSIONS AND FUTURE DEVELOPMENTS
This updated version of PlantMetabolomics.org provides metabolomics mass spectrometry-based metabolomics data from multiple analytical platforms. A user can analyze this data using our web-based data visualization and mining tools and generate the hypothesis about the functions of gene of their interest. A user can also perform a comparative analysis on a metabolite or metabolic pathway of interest and see their behavior under different mutations. We plan to enhance our coverage mutant lines to 203 novel lines.
The next steps for this database are to create a viewer for extracting the spectra of the measured metabolite from the different platforms and replicates. This will create a valuable resource for mass spectra across many different platforms and gather information on measurement variability. This capability may allow PlantMetabolomics.org to link to the spectral data in the LC-MS Arabidopsis database, AtMetExpress (5) and the GC-MS Golm Metabolomics Database (23). The flexibility of the pathway viewer will also be enhanced to give the user more ways to combine pathways into networks and select data.
AVAILABILITY
The PlantMetabolomics.org database is available online and free to all without restriction at: http://www.plantmetabolomics.org/.
FUNDING
Funding for open access charge: National Science Foundation (grant number MCB 08200823).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The following labs generated the metabolomics data in PlantMetabolomics.org: Oliver Fiehn (UC Davis), B. M. Lange (Washington State University), Lloyd Sumner (Noble Foundation) and Ruth Welti (Kansas State University) as part of the Arabidopsis Metabolomics Consortium. The stereomicroscopic images were generated by Hilal Ilarslan and Jennifer Robinson of Iowa State. The annotations and links to AraCyc were provided by Kate Dreher and Sue Rhee of the Plant Metabolic Network Project and The Arabidopsis Information Resource (TAIR). NSF Research Experience for Undergraduate students, William Van Walbeek and William Petersen developed the Cytoscape Web pathway viewer tool.
REFERENCES
- 1.Bais P, Moon SM, He K, Leitao R, Dreher K, Walk T, Sucaet Y, Barkan L, Wohlgemuth G, Roth MR, et al. PlantMetabolomics.org: a web portal for plant metabolomics experiments. Plant Physiol. 2010;152:1807–1816. doi: 10.1104/pp.109.151027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science. 2003;301:653–657. doi: 10.1126/science.1086391. [DOI] [PubMed] [Google Scholar]
- 3.Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee do Y, Lu Y, Moon S, Nikolau B. Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J. 2008;53:691–704. doi: 10.1111/j.1365-313X.2007.03387.x. [DOI] [PubMed] [Google Scholar]
- 4.Fiehn O, Sumner LW, Rhee SY, Ward J, Dickerson J, Lange BM, Lane G, Roessner U, Last R, Nikolau B. Minimum reporting standards for plant biology context information in metabolomics studies. Metabolomics. 2007;3:195–201. [Google Scholar]
- 5.Matsuda F, Hirai M, Sasaki E, Akiyama K, Yonekura-Sakakibara K, Provart N, Sakurai T, Shimada Y, Saito K. AtMetExpress development: a phytochemical atlas of Arabidopsis development. Plant Physiol. 2010;152:566–578. doi: 10.1104/pp.109.148031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Matsuda F, Nakabayashi R, Sawada Y, Suzuki M, Hirai MY, Kanaya S, Saito K. Mass spectra-based framework for automated structural elucidation of metabolome data to explore phytochemical diversity. Front. Plant Sci. 2011;2:40. doi: 10.3389/fpls.2011.00040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lopes CT, Franz M, Kazi F, Donaldson SL, Morris Q, Bader GD. Cytoscape web: an interactive web-based network browser. Bioinformatics. 2010;26:2347–2348. doi: 10.1093/bioinformatics/btq430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang P, Dreher K, Karthikeyan A, Chi A, Pujar A, Caspi R, Karp P, Kirkup V, Latendresse M, Lee C, et al. Creation of a genome-wide metabolic pathway database for populus trichocarpa using a new approach for reconstruction and curation of metabolic pathways for plants. Plant Physiol. 2010;153:1479–1491. doi: 10.1104/pp.110.157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collins TJ. ImageJ for microscopy. BioTechniques. 2007;43:S25–S30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
- 10.Fahy E, Subramaniam S, Murphy RC, Nishijima M, Raetz CRH, Shimizu T, Spener F, van Meer G, Wakelam MJO, Dennis EA. Update of the LIPID MAPS comprehensive classification system for lipids. J. Lipid Res. 2009;50:S9–S14. doi: 10.1194/jlr.R800095-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van den Berg RA, Hoefsloot HC, Westerhuis JA, Smilde AK, van der Werf MJ. Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genom. 2006;7:142. doi: 10.1186/1471-2164-7-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seber GAF. Multivariate Observations. Hoboken, NJ: John Wiley & Sons; 1984. [Google Scholar]
- 14.Spearman C. The proof and measurement of association between two things. Am. J. Psychol. 1904;15:72–101. [PubMed] [Google Scholar]
- 15.Scott IM, Vermeer CP, Liakata M, Corol DI, Ward JL, Lin W, Johnson HE, Whitehead L, Kular B, Baker JM, et al. Enhancement of plant metabolite fingerprinting by machine learning. Plant Physiol. 2010;153:1506–1520. doi: 10.1104/pp.109.150524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breiman L. Random forests. Mach. Learn. 2001 2001, 45, 5–32. [Google Scholar]
- 17.Díaz-Uriarte R, Alvarez de Andrés S. Gene selection and classification of microarray data using random forest. BMC Bioinform. 2006;7:3. doi: 10.1186/1471-2105-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liaw A, Wiener M. Classification and regression by randomForest. R. News. 2002;2/3:18–22. [Google Scholar]
- 19.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. PubChem Compound Database. PMID: 19933261.
- 21.Zhang P, Foerster H, Tissier CP, Mueller L, Paley S, Karp PD, Rhee SY. MetaCyc and AraCyc. Metabolic pathway databases for plant research. Plant Physiol. 2005;138:27–37. doi: 10.1104/pp.105.060376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Hemert JL, Dickerson JA. PathwayAccess: CellDesigner plugins for pathway databases. Bioinformatics. 2010;26:2345–2346. doi: 10.1093/bioinformatics/btq423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hummel J, Selbig J, Walther D, Kopka J. The Golm Metabolome Database: a database for GC-MS based metabolite profiling. In: Nielsen J, Jewett M, editors. Metabolomics. Berlin, Heidelberg, New York: Springer-Verlag; 2007. pp. 75–96. [Google Scholar]