Abstract
The Gene Ontology (GO) (http://www.geneontology.org) is a community bioinformatics resource that represents gene product function through the use of structured, controlled vocabularies. The number of GO annotations of gene products has increased due to curation efforts among GO Consortium (GOC) groups, including focused literature-based annotation and ortholog-based functional inference. The GO ontologies continue to expand and improve as a result of targeted ontology development, including the introduction of computable logical definitions and development of new tools for the streamlined addition of terms to the ontology. The GOC continues to support its user community through the use of e-mail lists, social media and web-based resources.
INTRODUCTION
The Gene Ontology (GO; http://www.geneontology.org) project is a bioinformatics resource that provides the scientific community with information about gene-product function (1) through the use of domain specific ontologies. The project consists of a collaborative effort to ‘annotate’ gene products (e.g. proteins) with terms that describe their functions and cellular location of action. A ‘GO annotation’ is an association, supported by evidence, between a gene product and a term from one of the structured, controlled vocabularies that describe how and where gene products act. Founded in 1998, the GO has grown to become an integrated resource containing functional information for over 11 million gene products from over 350 000 species (including strains) covering plants, animals and the microbial world. The GOC makes all annotations, vocabularies and tools freely available. Recent improvements to the GO resource include: expansion and refinement of the gene annotation set, further development of the ontology into key areas of biology, improved formalization of ontology structure and enhancements for biological investigation by researchers using the GO.
EXPANDED AND REFINED GENE-PRODUCT ANNOTATIONS
Increased annotation breadth and depth
Table 1 shows a summary of annotations available from the GO resource.
Table 1.
Status of the Gene Ontology as of 7 September 2011
| Biological process terms | 21 394 |
| Molecular function terms | 9062 |
| Cellular component terms | 2896 |
| Species with annotation (includes strains) | 367 887 |
| Total annotated gene products | 11 855 555 |
| Manually annotated gene products | 437 164 |
A major collaborative effort within the GOC has focused on providing a set of comprehensive experimental GO annotations for all gene products for human and 11 reference genomes of major model organisms, as well as tools for using these annotations to infer GO annotations for all fully sequenced genomes (Table 2). Through this project, GOC member databases have continued their efforts to provide a better annotation resource (2). Coordination through the reference genome project allows annotator interaction that ensures consistent annotation practice and allows for simultaneous development of the ontology as annotation progresses. The reference genome annotation project has been greatly enhanced by the use of the PAINT tool to infer functional information across closely related genes in a wide variety of organisms (3).
Table 2.
Twelve model organisms selected for targeted curation and their respective databases
| Arabidopsis thaliana | The Arabidopsis Information Resource (TAIR) |
| Caenorhabditis elegans | WormBase |
| Danio rerio | Zebrafish Information Network (Zfin) |
| Dictyostelium discoideum | Dictybase |
| Drosophila melanogaster | FlyBase |
| Escherichia coli | EcoliHub |
| Gallus gallus | AgBase |
| Homo sapiens | Human UniProtKB-Gene Ontology Annotation [UniProtKB-GOA] @ EBI |
| Mus musculus | Mouse Genome Informatics (MGI) |
| Rattus norvegicus | Rat Genome Database (RGD) |
| Saccharomyces cerevisiae | Saccharomyces Genome Database (SGD) |
| Schizosaccharomyces pombe | GeneDB S. pombe |
Introduction of GAF2.0
GO annotations are used both internally for GOC-developed tools and are provided to external developers for use in independently developed data analysis software. The GOC uses and provides annotation data in a standardized, tab-delimited format called a gene association file or GAF. Each line in the GAF includes information about the gene product being annotated, evidence supporting the annotation, the group making the annotation and the GO term associated with the annotation. One line represents one assertion about a gene product and includes information about the original reference on which the assumption is based as well as the evidence supporting that assumption. Since gene products can be involved in more than one process, carry out more than one function or be located in more than one cellular component, there may be many annotation lines in a GAF for a single gene product. In March 2010, the GOC began officially using an enhanced file format: GAF2.0 (http://www.geneontology.org/GO.format.gaf-2_0.shtml). In the GAF 2.0 format, there are 17 tab-delimited columns. GAF 2.0 improves and expands upon the GAF1.0 format by better capturing information about the identity of the specific gene products being annotated and by allowing annotations to contain contextual data thus enhancing the annotation specificity. Contextual data are captured using other biomedical ontology terms to narrow the meaning of an annotation. For example, the use of a Cell Type (4) ontology term as contextual data can be used to represent a process in a specific cell type if the base annotation represents a generic cellular process.
Improved annotation quality control
As part of the GOC's ongoing effort to standardize and improve annotation quality, we have also introduced a set of ‘hard’ and ‘soft’ quality control checks on annotations submitted by the participating groups. ‘Hard’ quality control checks identify incorrect annotations that will not be loaded into the GO database, but rather returned to the contributing resource for revision. These represent errors in annotation procedure such as annotating using an obsolete GO term/ID or annotating to the term 'protein binding' (GO:0005515) with an evidence code other than ‘inferred by physical interaction’.
Soft quality control checks identify annotations that are not necessarily incorrect, but that might be expected to have additional supporting evidence information and therefore should be subject to review. For example, annotation to the term ‘response to stress’ could likely be improved by specifying the type of stress. Another example of a soft check is the taxon constraint where a given annotation would be expected to be valid within certain taxonomic groups. We have continued to use and expand taxon restraints as a guide for identifying annotation errors (5). For example an annotation to ‘chloroplast’ should never be made for a mouse gene product. These taxon checks are considered as soft checks.
Summary of these and other error checking rules are available: http://www.geneontology.org/GO.annotation_qc.shtml.
The hard checks are implemented via a filtering script which removes offending annotations from the gene-association files (GAF) and the cleaned up GAF files are made available to users, loaded into the GO database and AmiGO. For the soft checks, a rule engine (GAF validator) allows curators to identify annotations that need to be reviewed.
NEW FEATURES OF THE ONTOLOGIES
We have continued to improve the ontologies themselves. A full list of projects to enhance the ontology is available at: http://wiki.geneontology.org/index.php/Ontology_Development
Our improvements have focused on three critical areas: making the ontology more useful for data aggregation, increasing biological content and improving the structure of the ontology to better reflect our current best understanding of biology.
New generic GO slim
GO Slims are predetermined sets of GO terms that are used to aggregate gene product information (http://www.geneontology.org/GO.slims.shtml). Since the terms in a given GO slim are manually chosen, they can be engineered to have a broad coverage of biology, or specific coverage of a limited subject area or a distribution of coverage based on experimental parameters such as stage of development. We have recently redesigned the generic GO Slim. The generic GO Slim is used for a broad categorization of the biological processes in which a set of gene products is involved. This GO Slim consists of 104 terms from the biological process portion of GO. The new generic GO Slim does not contain molecular function terms since these terms are necessarily very specific and only represent the action of individual gene products within a given biological process; however, we are currently working on a separate generic GO slim for molecular function grouping. Users can create custom GO slims with the OBO-Edit tool. Instructions can be found in the OBO-Edit help documentation.
Expanded biological content
The GOC has continued to work with community experts to expand and refine certain areas of the ontology. This work usually includes a face-to-face meeting between community experts and ontology developers where the structure and content of the ontology is discussed. After the meeting, ontology developers rearrange the ontology and add new terms to the ontology with review so that it reflects the most up to date views of the research community.
One area of intense focus over the last year has been the representation of transcription in GO (http://wiki.geneontology.org/index.php/Transcription). This work focused mainly on problematic terms in the molecular function ontology, particularly in the area of transcription factor function. The portion of the ontology describing transcription factors has been split into those transcription factors that act primarily as protein binding agents and those that act as DNA binding agents. We took advantage of the new has_part relationships in the ontology as well as the recent introduction of part_of relationships between molecular functions and biological processes so that the new structure reflects the complex nature of the activity of these molecules (6). For example, the molecular function ‘sequence-specific DNA binding transcription factor activity’ (GO:0003700) has as part of its activity ‘transcription regulatory region sequence-specific DNA binding’ (GO:0000976), indicating that binding to the regulatory region is necessary for the action of the gene product. GO:0003700 is part_of ‘regulation of transcription, DNA dependent’ (GO:0006355) (Figure 1). These relationships show that the action of a gene product annotated to this term controls whether or not transcription will take place.
Figure 1.
Graphical view of the term ‘sequence-specific DNA binding transcription factor activity’ (GO:0003700). The grey arrows represent has_part relationships. The blue arrows represent is_a relationships. The purple arrow represents a regulates relationship. The gold arrows represent part_of relationships.
We have also begun to standardize the representation of signaling in the ontology (http://wiki.geneontology.org/index.php/Signaling). In particular, we have begun to define the starting points and stopping points of signaling processes. Clarifying the definitions is a great aid for both annotators who are looking for the right term to use, as well as for researchers looking at gene products associated with specific points in a signaling process. The functions of signaling ligands are represented as integral parts of the signal transduction process. The consequence of signaling is represented as a regulation of a cellular process. We have disentangled the processes that represent the complexities of ligand–receptor interactions where a single ligand can activate multiple transduction pathways and multiple ligands can activate the same pathway.
Kidney development is an area of biology that has important clinical relevance. As a follow-up to our targeted work on heart development (7), we have also met with community experts to vastly improve the representation of kidney development in GO. The meeting and subsequent work resulted in the addition of over 450 terms to improve the ontology. Renal system development now covers the renal systems of flies and vertebrates down to a cellular level. The structure of the graph represents similarities and differences that are reflected in major model organisms used to study renal development.
Improved ontology structure
The GO contains complex terms, particularly in the biological process ontology. In some cases the terms are internally referential, such as ‘regulation of cell growth’ (GO:0001558), which refers to both the process of ‘biological regulation’ (GO:0065007) as well as the process of ‘growth’ (GO:0040007). We have introduced formal descriptions of these properties into the OBO stanzas of compound terms (http://wiki.geneontology.org/index.php/Category:Cross_Products). ‘Regulation of cell growth’ is formally defined as a ‘biological regulation’ that regulates ‘growth’ (8). The formal descriptions are used to computationally analyze the placement of a term in the ontology. In this example, computational reasoning can be used to infer that ‘regulation of cell growth’ is_a ‘regulation of growth’ (GO:0040008) because ‘cell growth’ (GO:0016049) is_a ‘growth’ (GO:0040007). Compound logical definitions for terms that express regulates, occurs_in and part_of relationships now reside in the live version of the full ontology.
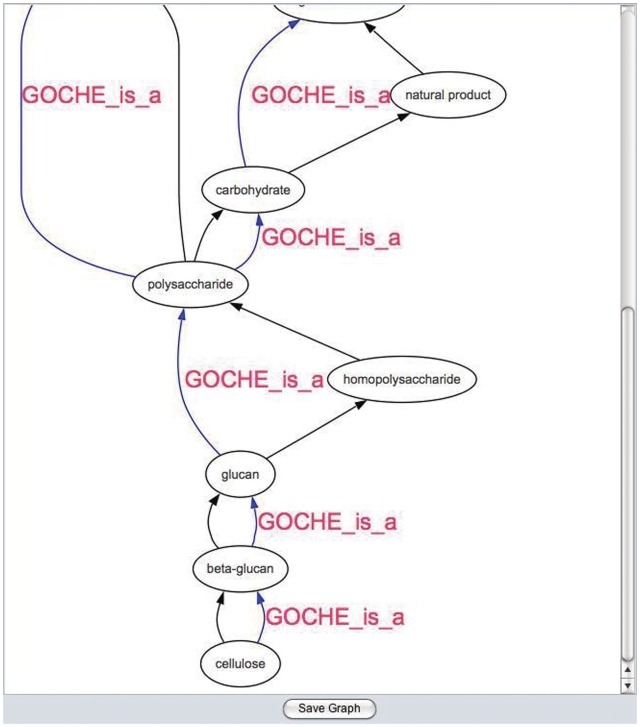
Complex terms in GO can reference both other terms within GO and terms from other biomedical ontologies that are outside the scope of GO. In particular, many biological process terms reference anatomical structures, cell types and chemicals. For example, the term ‘epithelial cell differentiation’ refers to the term ‘epithelial cell’ (CL:0000066) from the cell type ontology (4). Formally cross-referencing terms from external ontologies is a powerful way to integrate expertise from different specialist communities into an existing ontology (8,9). To begin the formal representation of an external ontology within GO, we have been deconstructing GO terms that refer to chemicals and cross-referencing those term to the Chemicals of Biological Interest (ChEBI) ontology (10). GO developers have worked closely with ChEBI developers to assign ChEBI IDs to GO terms that refer to chemicals. The chemical references are arranged into a structure representing the intrinsic chemical hierarchy within GO (GOChe). Ontology developers use the GOChe to check alignment of the representation of chemicals in GO with the representation of chemicals in ChEBI (Figure 2). When misalignments of the two ontologies are found, GO curators work with ChEBI curators to resolve the discrepancy.
Figure 2.
Graphical view showing the inherent GO-chemical (GOChe) ontology and ChEBI. Black arrows represent CHEBI is_a relationships. Blue arrows represent GOChe is_a relationships. Note that the term ‘homopolysaccharide’ only exists in ChEBI.
Addition of logical definitions into the GO permits the use of automated reasoning tools to check the logical consistency of the ontology. Reports resulting from these reasoning tools are used periodically by ontology developers to add missing relationships to the ontology and to identify incorrect relationships that should be modified or removed.
With formal, computable definitions of GO terms now represented in the ontology we can add new terms that fit standard term formats to the ontology without adding relationships manually. For example many terms such as ‘X involved in Y’ fit into the ontology in a consistent way where ‘X’ is part_of ‘Y’. Ontology developers use a web-based tool called TermGenie to add these stereotypical terms into the ontology. When using TermGenie ontology editors are prompted to select a template such as ‘all regulates’, the editor can then choose if they want all three types of regulation and search for a target term such as ‘transcription’. Once the term is chosen the request can be completed and the proper ‘regulation of transcription’ terms are created with the appropriate relationships to other terms in the ontology. TermGenie is currently capable of handling terms in several standard formats.
IMPROVEMENTS FOR COMMUNITY ACCESS
AmiGO is the GOC's primary web application that provides access to annotations and the ontology (http://amigo.geneontology.org) using the GO database. AmiGO allows users to browse the ontology and search the annotation corpus. Over the past year, several improvements were made to the AmiGO resource (Table 3). Term views are now more informative; displaying the term name, ID, definition and subsets of GO in which the term is included. The term view also has a link to the GONUTS wiki for users to contribute to information about the usage of the term (http://gowiki.tamu.edu/wiki/index.php/Main_Page). At the bottom of the term-view page, there are several tabbed options for viewing the term in the context of the rest of the ontology. In particular, there is an inferred tree view of the term that gives a compact view of the term in the context of its parents and children, a view that lists the parents and children of a given term, and a graphical view of the term using the QuickGO graphical utility. Additionally, an on-going rewrite of the software that underlies ‘GOOSE’, the GO online SQL environment (http://berkeleybop.org/goose) has been undertaken. Users can also access new software tools that are under development by the GOC through a link to AmigoLabs (http://wiki.geneontology.org/index.php/AmiGO_Labs).
Table 3.
Enhancements made to the AmiGO tool
| GOOSE | GO Online SQL Environment | http://berkeleybop.org/goose |
|---|---|---|
| Visualization | Create custom graphical representations of the ontology | http://amigo.geneontology.org/cgi-bin/amigo/amigo?mode=visualize |
| Live Search | Search annotations or terms and obtain results automatically in an embedded frame | http://amigo.geneontology.org/cgi-bin/amigo/amigo?mode=live_search |
| Homology Set Summary | Browse gene product annotation summaries from homology sets coordinately curated by the GOC | http://amigo.geneontology.org/cgi-bin/amigo/amigo?mode=homolset_summary |
We have also been improving our community outreach by continuously modifying and enhancing documentation available through the main web site and the GO wiki. To keep users and members of the GOC up to date with respect to changes that are made to the ontologies, we now provide a weekly report of changes and modifications to terms and/or their definitions (http://www.geneontology.org/internal-reports/ontology/).
GO keeps its community informed through two email lists (go-consortium@lists.stanford.edu and go-friends@lists.stanford.edu), RSS feeds and social media like LinkedIn, Facebook and Twitter. We continue to support our users by responding to queries and data requests sent to: go-helpdesk@lists.stanford.edu or http://www.geneontology.org/GO.contacts.shtml.
FUNDING
National Human Genome Research Institute (NHGRI) (P41 grant 5P41HG002273-09 to Gene Ontology Consortium) and European Union RTD Programme ‘Quality of Life and Management of Living Resources’ (QLRI-CT-2001-00981 and QLRI-CT-2001-00015 to GO and UniProtKB-GOA groups at EMBL-EBI). Funding for open access charge: National Human Genome Research Institute (NHGRI) (P41 grant 5P41HG002273-09).
Conflict of interest statement. None declared.
APPENDIX 1
J.A. Blake, M. Dolan, H. Drabkin, D.P. Hill, L. Ni, D. Sitnikov (MGI, The Jackson Laboratory, Bar Harbor, ME, USA); S. Burgess, T. Buza, C. Gresham, F. McCarthy, L. Pillai, H. Wang (AgBase, Mississippi State University; MS, USA); S. Carbon, S.E. Lewis, C.J. Mungall, (BBOP, LBNL, Berkeley, CA, USA); P. Gaudet (CALIPHO group, SIB, Geneva, Switzerland); R.L. Chisholm, P. Fey, W.A. Kibbe, S. Basu (dictyBase, Northwestern University, Chicago, IL, USA); D.A. Siegele, B.K. McIntosh, D.P. Renfro, A.E. Zweifel and J.C. Hu (EcoliWiki, Departments of Biology and Biochemistry and Biophysics, Texas A&M Univ., College Station, TX, USA); N.H. Brown, S. Tweedie (FlyBase, Gurdon Institute and Department of Genetics, University of Cambridge, Cambridge, UK); Y. Alam-Faruque, R. Apweiler, A. Auchinchloss, K. Axelsen, G. Argoud-Puy, B. Bely, M.-C. Blatter, L. Bougueleret, E. Boutet, S. Branconi-Quintaje, L. Breuza, A. Bridge, P. Browne, W.M. Chan, E. Coudert, I. Cusin, E. Dimmer, P. Duek-Roggli, R. Eberhardt, A. Estreicher, L. Famiglietti, S. Ferro-Rojas, M. Feuermann, M. Gardner, A. Gos, N. Gruaz-Gumowski, U. Hinz, C. Hulo, R. Huntley, J. James, S. Jimenez, F. Jungo, G. Keller, K. Laiho, D. Legge, P. Lemercier, D. Lieberherr, M. Magrane, M.J. Martin, P. Masson, M. Moinat, C. O'Donovan, I. Pedruzzi, K. Pichler, D. Poggioli, P. Porras Millán, S. Poux, C. Rivoire, B. Roechert, T. Sawford, M. Schneider, H. Sehra, E. Stanley, A. Stutz, S. Sundaram, M. Tognolli, I. Xenarios (UniProtKB: EBI, Hinxton, UK and SIB, Geneva, Switzerland); R. Foulger, J. Lomax, P. Roncaglia (GO-EBI, Hinxton, UK); E. Camon, V.K. Khodiyar, R.C. Lovering, P.J. Talmud (Institute of Cardiovascular Science, University College London, London, UK); M. Chibucos, M. Gwinn Giglio (Institute for Genome Sciences, University of Maryland School of Medicine, Baltimore, MD, USA); K. Dolinski, S. Heinicke, M.S. Livstone (Lewis-Sigler Institute for Integrative Genomics, Princeton University, Princeton, NJ, USA); R. Stephan, (MTBBASE, Berlin); M.A. Harris, S.G. Oliver, K. Rutherford, V. Wood (PomBase, University of Cambridge, Cambridge, UK); J. Bahler, A. Lock (PomBase, University College, London UK); P.J. Kersey, M.D. McDowall, D.M. Staines (PomBase, EBI, Hinxton UK); M. Dwinell, M. Shimoyama, S. Laulederkind, T. Hayman, S.-J. Wang, V. Petri, T. Lowry (RGD, Medical College of Wisconsin, Milwaukee, WI, USA); P. D'Eustachio, L. Matthews (Reactome, Department of Biochemistry, NYU School of Medicine, New York, NY, USA); C.D. Amundsen, R. Balakrishnan, G. Binkley, J.M. Cherry, K.R. Christie, M.C. Costanzo, S.S. Dwight, S.R. Engel, D.G. Fisk, J.E. Hirschman, B.C. Hitz, E.L. Hong, K. Karra, C.J. Krieger, S.R. Miyasato, R.S. Nash, J. Park, M.S. Skrzypek, S. Weng, E.D. Wong (SGD, Department of Genetics, Stanford University, Stanford, CA, USA); T.Z. Berardini, D. Li, E. Huala (TAIR, Department of Plant Biology, Carnegie Institution for Science, Stanford, CA, USA); D. Slonim, H. Wick (Tufts University, Medford, MA, USA); P. Thomas (USC, Los Angeles, CA, USA); J. Chan, R. Kishore, P. Sternberg, K. Van Auken (WormBase, California Institute of Technology, Pasadena, CA, USA); D. Howe, M. Westerfield (ZFIN, University of Oregon, Eugene, OR, USA). The GO also acknowledges the annotation effort from the annotators at: Gramene, Department of Botany and Plant Pathology, Oregon State University, Corvallis, OR, USA; The J. Craig Venter Institute, Rockville, MD, USA; PAMGO, Wells College, Aurora, NY, USA and PAMGO, Virginia Bioinformatics Institute, VA, USA; AspGD, Stanford, CA, USA; CGD, Stanford, CA, USA; Sanger GeneDB, Hinxton, UK; InterPro EBI, Hinxton, UK; IntAct, EBI, Hinxton, UK; seudoCAP, British Columbia, Canada; SGN, Ithaca, NY, USA.
REFERENCES
- 1.The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.The Reference Genome Group of the Gene Ontology Consortium. The Gene Ontology's Reference Genome Project: a unified framework for functional annotation across species. PLoS Comput. Biol. 2009;5:e1000431. doi: 10.1371/journal.pcbi.1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gaudet P, Livstone MS, Lewis SE, Thomas PD. Phylogenetic-based propagation of functional annotations within the Gene Ontology consortium. Brief Bioinform. 2011;12:449–462. doi: 10.1093/bib/bbr042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meehan TF, Masci AM, Abdulla A, Cowell LG, Blake JA, Mungall CJ, Diehl AD. Logical Development of the Cell Ontology. Bioinformatics. 2011;12:6. doi: 10.1186/1471-2105-12-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deegan née Clark JI, Dimmer EC, Mungall CJ. Formalization of taxon-based constraints to detect inconsistencies in annotation and ontology development. BMC Bioinformatics. 2010;11:530. doi: 10.1186/1471-2105-11-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Khodiyar VK, Hill DP, Howe D, Berardini TZ, Tweedie S, Talmud PJ, Breckenridge R, Bhattarcharya S, Riley P, Scambler P, et al. The representation of heart development in the gene ontology. Dev. Biol. 2011;354:9–17. doi: 10.1016/j.ydbio.2011.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mungall CJ, Bada M, Berardini TZ, Deegan J, Ireland A, Harris MA, Hill DP, Lomax J. Cross-product extensions of the Gene Ontology. J. Biomed. Inform. 2011;44:80–86. doi: 10.1016/j.jbi.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hill DP, Blake JA, Richardson JE, Ringwald M. Extension and integration of the gene ontology (GO): combining GO vocabularies with external vocabularies. Genome Res. 2002;74:121–128. doi: 10.1101/gr.580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Matos P, Alcántara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical entities of biological interest: an update. Nucleic Acids Res. 2009;38:D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]