Abstract
The Orientations of Proteins in Membranes (OPM) database is a curated web resource that provides spatial positions of membrane-bound peptides and proteins of known three-dimensional structure in the lipid bilayer, together with their structural classification, topology and intracellular localization. OPM currently contains more than 1200 transmembrane and peripheral proteins and peptides from approximately 350 organisms that represent approximately 3800 Protein Data Bank entries. Proteins are classified into classes, superfamilies and families and assigned to 21 distinct membrane types. Spatial positions of proteins with respect to the lipid bilayer are optimized by the PPM 2.0 method that accounts for the hydrophobic, hydrogen bonding and electrostatic interactions of the proteins with the anisotropic water-lipid environment described by the dielectric constant and hydrogen-bonding profiles. The OPM database is freely accessible at http://opm.phar.umich.edu. Data can be sorted, searched or retrieved using the hierarchical classification, source organism, localization in different types of membranes. The database offers downloadable coordinates of proteins and peptides with membrane boundaries. A gallery of protein images and several visualization tools are provided. The database is supplemented by the PPM server (http://opm.phar.umich.edu/server.php) which can be used for calculating spatial positions in membranes of newly determined proteins structures or theoretical models.
INTRODUCTION
More than half of all proteins in cells associate with biological membranes permanently or temporarily. This includes integral monotopic and transmembrane (TM) proteins, which are encoded by 20–30% of sequenced genomes (1), and more numerous peripheral proteins and peptides that can form transient complexes with membrane lipids or proteins. Recent progress in structure determination techniques (2) have led to a significant growth of the number of membrane proteins with known three-dimensional (3D) structures. Currently, there are approximately 1200 and 10 000 entries in the Protein Data Bank (PDB) (3) related to TM and peripheral proteins, respectively, which corresponds to 1.6 and 13% of the PDB content. Many PDB entries represent different complexes, conformations, mutants or crystal forms of the same protein, so the set of distinct proteins is approximately 3-fold smaller.
Integral membrane proteins with known 3D structures can be found in several specialized databases, such as Stephen White's list (4), the Membrane Proteins Data Bank (MPDB) (5) and the transporter classification database (TCDB) (6). These resources provide some complementary information, including bibliography, crystallization and solubilization conditions (5) or classification and phylogenetic analysis of membrane transporters (6). More specialized resources cover membrane-targeting domains [MeTaDoR (7)], and antimicrobial peptides with non-standard amino acids [Peptaibol (8)].
The critical information missing in these databases is the exact position of membrane boundaries, which is not obvious from the protein 3D structure, even if the protein was crystallized with phospholipids. Spatial positions of membrane-associated proteins with respect to the lipid bilayer can be determined by experimental techniques or computationally. Experimental methods, including chemical modification, spin-labeling, X-ray scattering, neutron diffraction, infrared spectroscopy, electron-cryomicroscopy and NMR, are very laborious and, therefore, have been applied for a limited set of proteins and peptides (9,10). On the other hand, development of a fast and reliable computational approach would allow positioning of proteins in membranes in a timely manner, following the expanding flow of experimentally determined structures.
Several theoretical methods have been applied for positioning of proteins in membranes, which are based on molecular dynamics (MD) simulations (11), coarse-grained MD (12), optimization of electrostatic energy (13–16), energy minimization with implicit solvent models (17–21) or membrane depth-dependent scoring functions (22–24). The results for TM proteins have been collected in two databases, Protein Data Bank of TransMembrane proteins (PDBTM) (25), and Coarse-Grained DataBase (CGDB) (12,26). PDBTM includes an up-to-date set of 1441 PDB structures (release as of 24 June 2011) of α-helical and β-barrel proteins arranged in the lipid bilayer by the TMDET algorithm (23). CGDB holds pseudo-atoms models of approximately 370 TM proteins and around a dozen of monotopic proteins generated by the Coarse Grained MD simulations (12). Both databases are focused on integral membrane proteins but do not include peripheral proteins, because the prediction of their weak association with lipid bilayers would require a significantly higher precision in calculation of membrane binding affinities than can be provided by the underlying methods.
To fill this gap, we have proposed and recently advanced a method for Positioning of Proteins in Membrane (PPM 2.0) by optimizing free energy of protein transfer from water to the membrane environment that implements an anisotropic solvent model of the lipid bilayer (9,27). The method was thoroughly verified for several dozens of TM and peripheral proteins and peptides, whose arrangements in membranes have been experimentally studied (9,10,27). High computational efficiency of PPM 2.0 allows its application for the large-scale analysis of proteins from the PDB. The results are deposited in our Orientation of Proteins in Membranes (OPM) database that includes both TM and peripheral proteins (28). Hence, it covers a significantly larger number of membrane-associated macromolecules (1255 proteins and peptides) and PDB entries (3766 structures) than PDBTM and CGDB. It also provides a four-level protein classification system together with information about protein topology, type of intracellular membrane, source organism and comparison with experimental publications on arrangement of the corresponding proteins in membranes.
DATABASE CONTENT
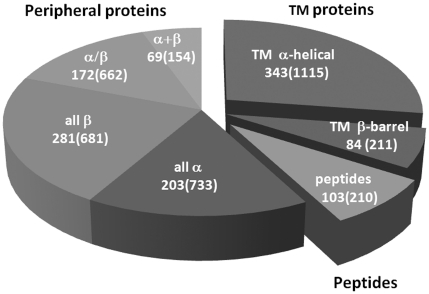
The OPM database was established in December of 2005 at the College of Pharmacy of the University of Michigan. The database currently holds 427 TM, 725 peripheral proteins and 103 membrane-active peptides related to 3766 PDB entries (Figure 1).
Figure 1.
Current distribution of different OPM entry types (as of 20 July 2011). Numbers of representative structures are indicated, as well as numbers of related PDB entries (in parenthesis).
The database includes only protein structures whose spatial positions in membranes can be computationally predicted, rather than a complete set of all membrane-associated proteins from the PDB. The positions of many peripheral proteins in membranes cannot be calculated because their membrane-anchoring structures (amphiphilic helices or loops, lipidated residues or specifically bound lipids) are disordered or missing in the experimental structure. In addition, peripheral proteins may adopt alternative conformations, some of which are not membrane associated.
All data are organized in pages associated with every protein class, superfamily, family or an individual protein. To deal with significant redundancy of PDB data, we select a ‘representative’ PDB entry for each protein. This entry represents the most complete protein structure that includes maximal number of protein domains and fewer disordered segments. Several ‘representative’ structures of the same protein are selected if they correspond to distinct conformational states or alternative quaternary complexes of the protein. All other available PDB entries of the same protein are included as ‘related’ structures linked to the ‘representative’ structure.
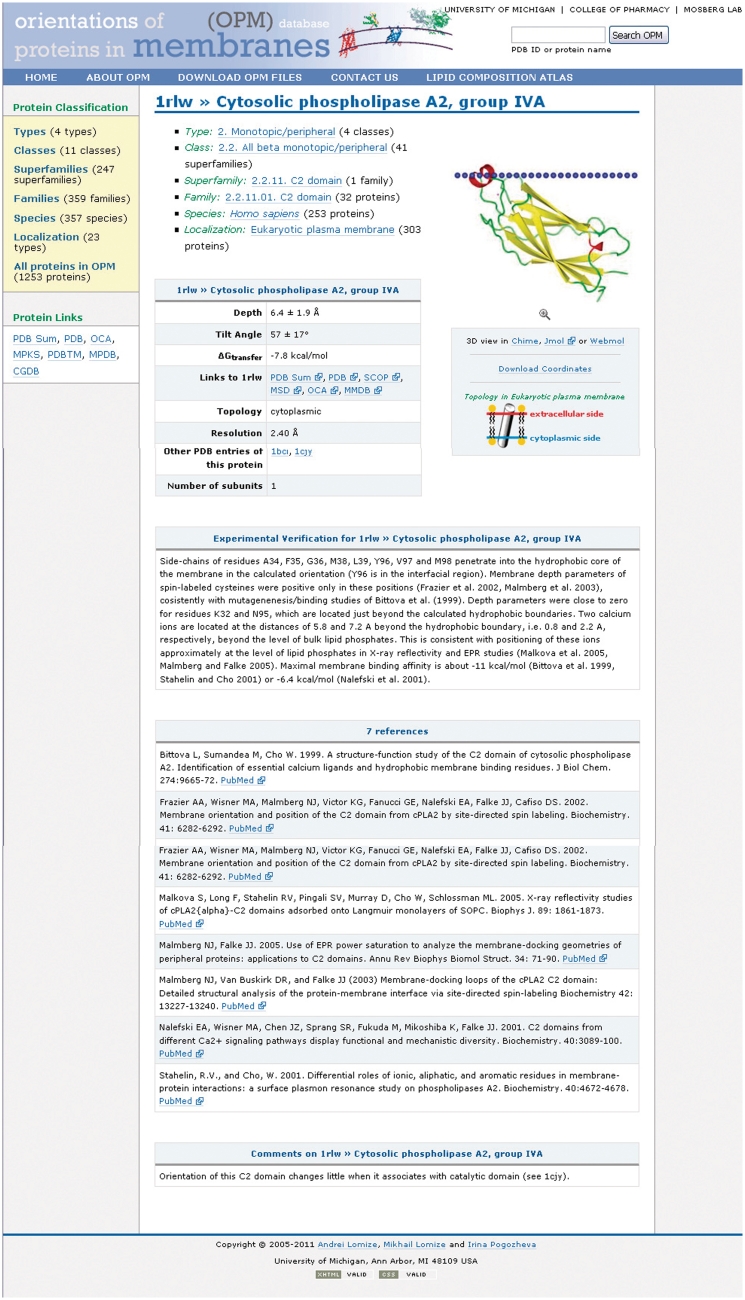
A ‘representative’ protein page (Figure 2) displays protein name, classification, subcellular localization (or destination membrane), source organism, protein topology (membrane side associated with protein N-terminus), number of subunits and links to other web resources. Another set of data describes the arrangement of a protein in the lipid bilayer as calculated by PPM 2.0. It includes: (i) downloadable atomic coordinates of a protein with lipid bilayer boundaries (located at the level of lipid carbonyls) that are indicated by dummy atoms; (ii) orientational parameters (tilt angle, hydrophobic thickness or membrane penetration depth); (iii) membrane binding energies; and (iv) list of TM segments.
Figure 2.
Example of entry page for C2 domain of peripheral protein phospholipase A2.
Data visualization is provided by static images and dynamic images generated by freely available interactive tools. Oligomeric states are taken from the PDBe (29) or generated by PISA (30), excluding a number of cases in which biological units were chosen in accordance with publications. For example, secretory phospolipases A2 and cytochromes P450 were taken in the physologically relevant monomeric state, even though PISA identifies some of them as stable dimers. Topology and intracellular localization of proteins were usually taken from the corresponding publications on protein structure determination, though for some peripheral proteins topology data from UniProt (31) were used and compared for homologous proteins in the database to minimize potential errors. Annotation and experimental verification of the calculated arrangement in membrane (with PubMed links) is provided for some well-studied proteins.
A ‘related’ protein page provides downloadable atomic coordinates of a protein with lipid bilayer boundaries presented by dummy atoms, protein static and dynamic images, and links to related web resources.
PROTEIN CLASSIFICATION
The classification has four-level hierarchy: type (TM, peripheral/monotopic protein and peptides), class (α-helical polytopic, α-helical bitopic, β-barrel TM proteins; and all-α, all-β, α+β, α/β peripheral/monotopic proteins), superfamily (evolutionarily related proteins) and family (proteins with clear sequence homology). Multi-domain proteins and their complexes are classified based on Pfam (32), SCOP (33) and TCDB (6) classification of their largest membrane-associated domain. OPM superfamilies usually correspond to Pfam clans and SCOP superfamilies, whereas OPM families correspond to Pfam, SCOP and TCDB families.
POSITIONING OF PROTEINS IN MEMBRANES
The spatial positions of proteins in membranes are calculated by the advanced version of our method, PPM 2.0, which combines all atom representation of a solute, an anisotropic solvent representation of the lipid bilayer and universal solvation model (27). This is a general physical method, which does not require a parameter adjustment for different classes of molecules. The anisotropic properties of the lipid bilayer are described by transbilayer profiles of dielectric constant and hydrogen bonding acidity and basicity parameters. We use polarity profiles of 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) bilayer derived from experimental distributions of quasi-molecular segments of lipids determined by neutron and X-ray scattering (34), and transbilayer distribution of water in DOPC bilayer determined in spin-labeling experiments (35). The location of a protein in the membrane coordinate system is obtained by optimization of protein transfer energy from water to the lipid bilayer (ΔGtransf). The transfer energy is calculated as a sum of two terms: (i) a solvent accessible surface area-dependent term that accounts for van der Waals and H-bonding solvent–solute interactions and entropy of solvent molecules in the first solvation shell; and (ii) an electrostatic term that includes solvation energy of dipoles and ions, and deionization penalty of ionizable groups in non-polar environment. The method also accounts for the preferential solvation by water of protein groups and for the hydrophobic mismatch for TM proteins.
The PPM 2.0 method automatically discriminates TM and peripheral/monotopic proteins based on their membrane penetration depth, transfer energy (ΔGtransf) and the detection of only one or two membrane boundary planes. For integral membrane proteins and peptides ΔGtransf is usually between −400 and −10 kcal/mol. For peripheral protein the calculated ΔGtransf varies between −15 and −1.5 kcal/mol. Proteins with marginal ΔGtransf values (between −1.5 and −5 kcal/mol) are in the gray zone and their potential membrane binding sites should be treated with caution because some of them might represent hydrophobic spots involved in protein–protein interactions. To distinguish membrane-bound proteins, additional criteria are needed: (i) similar membrane-binding modes are found for proteins from the same superfamily; (ii) calculated membrane boundaries are spatially close to potential binding sites for lipids or other hydrophobic ligands, to lipidated residues or to TM helices that are missing in the crystal structure; and (iii) some experimental indications of protein–membrane interaction are found in the literature for this or a closely homologous protein. Proteins from the gray zone, which do not satisfy at least two of these additional criteria cannot be reliably positioned in membranes and, therefore are not included in OPM. For the same reason some structures of short-protein fragments that miss membrane-anchoring elements, Cα-atom models, some NMR models with poorly defined disordered loops, and theoretical models are not included in the database.
The accuracy of PPM predictions was thoroughly tested for a large set of TM and peripheral proteins, peptides and small molecules whose membrane penetration depths, orientations with respect to the lipid bilayer or membrane binding affinities have been experimentally studied (9,10,27). The method was always able of reproducing the sets of residues penetrating to the lipid bilayer according to spin-labeling, fluorescence and chemical modification studies. The accuracy of determination of membrane-binding energy, which was assessed as RMSE between experimental and calculated values, was found to be 0.74 kcal/mol for small molecules and 1.13 kcal/mol for peripheral proteins (27). However, proteins are highly dynamic, rather than occupying a fixed spatial position in the membrane. To evaluate the uncertainty in the protein orientation, we calculated fluctuations of tilt angle, membrane penetration depth and hydrophobic thickness within 1 kcal/mol around the global minimum of energy for every protein structure. The values of the fluctuations are provided in OPM. The uncertainties in spatial positions can also be estimated from the comparison of different structures of the same protein. They are relatively small for TM proteins (1 Å for the hydrophobic thickness and approximately 5°C for the tilt angle), but larger for peripheral proteins, especially for NMR models with poorly defined conformations of membrane-interacting loops, where the uncertainty in tilt angle may reach 50°. Large differences in orientations may be observed for alternative conformations of proteins. For example, distinct conformations of Ca2+-ATPase, a TM α-helical protein, differ in protein tilt by 17° (PDB IDs: 1su4, 3b8c) and membrane thickness by 3 Å (PDB IDs: 2zbd, 3ar8), which may be of functional importance.
DATABASE ACCESS
Access to the OPM database is through the web site at http://opm.phar.umich.edu/. Pages are dynamically generated for every level of hierarchical classification including superfamilies, families and individual protein pages. The ‘representative’ protein pages can be accessed from any higher hierarchy page or using database search by PDB code or protein name, while the ‘related’ protein pages can be accessed through internal links from the ‘representative’ protein pages or using search by PDB code.
To facilitate data retrieval and analysis, higher hierarchy pages are organized in protein lists and tables supplemented by protein images, internal and external links. For example, to compare membrane interaction modes of evolutionarily related proteins from the database, one can navigate to a protein superfamily page, which simultaneously displays images of all proteins from the superfamily with calculated membrane boundaries. Tables are automatically generated for every protein type, class, superfamily, family, membrane localization and source organism. Tables allow sorting of proteins based on the content of different fields: protein family code, protein name, PDB ID, biological source, destination membrane, number of TM α-helices or β-strands, number of subunits, transfer energy and orientational parameters of proteins in membranes.
All coordinate files of protein structures with hydrocarbon core boundaries marked by dummy atoms can be downloaded individually for each protein or as a single file for various protein sets: α-helical polytopic proteins, α-helical bitopic proteins, β-barrel proteins, monotopic and peripheral proteins and peptides. Lists of PDB codes for every protein family, superfamily, class and type are automatically generated at the beginning of every table for the corresponding protein set. Semiannual updated releases of the database will be provided as downloadable sql files.
Visualization is provided by static images and dynamic visualization tools. Static molecular images in PNG format are automatically generated using scripts for PyMOL molecular graphic software (36). Proteins with calculated membrane boundaries can be interactively displayed in Chime, Jmol (37) or WebMol (38), which allows the orientation from both membrane sides and packing through the membrane to be readily visible. The whole gallery of protein images can be retrieved separately. The database provides links to TCDB (6), Pfam (32) from family and superfamily pages and to SCOP (33), PDB (3), PDBsum (39), PDBe (29), OCA (40), MMDB (41) from protein pages. Links to CGDB (12), MPKS (4), PDBTM (25) and MPDB (5) are also provided. Links to the OPM database are currently integrated in several widely used resources including PDBsum, OCA, Wikipedia, Membrane Builder (42), and Cell Microcismos Membrane Editor (43).
MAINTAINANCE AND UPDATES
OPM was developed with PHP, MySQL and the Smarty engine, which separates the program logic (PHP, MySQL) and presentation (XHTML, CSS, JavaScript), and enables caching. The database is populated by experimental structures of proteins and their complexes extracted from the PDB. Some of the structures were modified using PPM 2.0 to reconstruct missing side chain atoms and optimize side chain conformers at the membrane interface. The database curation includes selection of ‘representative’ PDB entries, identification of topology, localization and oligomeric state using available informatics resources, classification of proteins to families and superfamilies and verification of the predicted arrangement in membrane, as described above.
The OPM content is updated using queries and online forms, which we have developed. The data for TM proteins are normally updated on a biweekly basis. The newly released TM structures are regularly retrieved from the PDB by PDBTM, or by combined PDBe/Uniprot/Interpro keyword search implemented in PDBe (29). Update of peripheral proteins is significantly more time-consuming and, therefore, is conducted on a yearly basis. To identify peripheral proteins, we perform an automatic screening of PDB entries using PPM 2.0 and selection criteria mentioned above, which is followed by the automatic comparison with lists of proteins that are indicated as membrane-associated by Pfam, PDBe, Uniprot or InterPro databases, the manual analysis of the results and examination of related publications.
PPM SERVER
To provide a web tool for calculation of spatial positions of proteins in the lipid bilayer we designed a PPM server that implements our PPM 2.0 method. The PPM server can be used for positioning in membranes of newly determined experimental structures or theoretical models of TM, peripheral proteins or peptides prior to their deposition in the PDB. The majority of TM proteins (1326 entries) and a large part of peripheral membrane proteins (2230 entries) from the PDB has already been pre-calculated by our method and can be found in the OPM database.
On the web interface of the PPM server the user can upload the atomic coordinates of a protein or a peptide, whose arrangement in the lipid bilayer will then be evaluated by PPM 2.0. The protein structure should have a biologically relevant oligomeric state and all side-chain atoms that may interact with lipids. The user has an option to specify topology of the protein and include ligands (lipids, cofactors, etc.) in the calculation.
The calculation of protein positions in the lipid bilayer may take from a few seconds to a few minutes, depending on the number of atoms. The output window displays orientational parameters: membrane penetration depth for peripheral proteins or hydrophobic thickness for TM proteins (Å), tilt angle (°), and water-to-membrane transfer energy (kcal/mol). The fluctuations of depth/hydrophobic thickness and tilt angle are defined within 1 kcal/mol around the global minimum of transfer energy and indicated by ± values. The output also contains TM segments of integral proteins and a list of membrane-embedded residues for all proteins. The downloadable atomic coordinates of the protein together with positions of hydrophobic core boundaries marked by dummy atoms are provided. The interactive visualization of the protein with calculated membranes borders is provided by Jmol (37). The server is hosted at the LAMP type (Linux, Apache, MySQL, Perl/PHP/Python) virtual server at the University of Michigan.
Comparison of the PPM-server with other existing web servers for positioning of proteins in membranes, EZ (22), TMDET (23), MAPS (24) and MAPAS (16), demonstrates that PPM clearly outperforms all of them in scope and accuracy and represents the only server that correctly predicts membrane-binding sites of peripheral proteins (see Supplementary Data).
CONCLUSIONS
The OPM database is the first comprehensive resource for membrane-associated peptides and proteins with known structures whose arrangement in membranes can be reliably assessed by the PPM 2.0 method, which is based on the evaluation of free energy of transfer of molecules from water to the anisotropic lipid environment. We also provide a web tool, PPM server, which enables the user to evaluate the membrane binding energy and parameters of spatial arrangement in the lipid bilayer of proteins not yet included into the OPM database.
OPM is highly accessed with more than 435 000 unique visits since its first release (from 4000 to 10 000 first time visitors and from 500 to 1200 returning visitors each month). The availability of the OPM database contributes to basic scientific research advances including understanding of the physics of protein–membrane interactions, determining the role of protein–lipid interactions in molecular transport, signal transduction, membrane transformations, formation of multi-proteins functional units and comparative analysis of mechanisms of insertion and translocation of proteins from different families into or across membranes. We are dedicated to incorporating new data in a timely manner as long as funding support is available.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The OPM database is sponsored by Division of Biological Infrastructure of the National Science Foundation (NSF) (Award #0849713 to A.L., I.P.). This work is also supported by National Institute of Health (National Institute on Drug Abuse, grant 5R01DA003910, to H.M.). Funding for open access charge: National Science Foundation (award #0849713); National Institutes of Health (grant 5R01DA003910).
Conflict of interest statement. None declared.
REFERENCES
- 1.Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998;7:1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bill RM, Henderson PJF, Iwata S, Kunji ERS, Michel H, Neutze R, Newstead S, Poolman B, Tate CG, Vogel H. Overcoming barriers to membrane protein structure determination. Nat. Biotechnol. 2011;29:335–340. doi: 10.1038/nbt.1833. [DOI] [PubMed] [Google Scholar]
- 3.Berman HM, Battistuz T, Bhat TN, Bluhm WF, Bourne PE, Burkhardt K, Iype L, Jain S, Fagan P, Marvin J, et al. The Protein Data Bank. Acta Crystallogr. D. 2002;58:899–907. doi: 10.1107/s0907444902003451. [DOI] [PubMed] [Google Scholar]
- 4. White, S. H. and Snaider, C. http://blanco.biomol.uci.edu/mpstruc/listAll/list (20 July 2011, date last accessed)
- 5.Raman P, Cherezov V, Caffrey M. The membrane protein data bank. Cell. Mol. Life Sci. 2006;63:36–51. doi: 10.1007/s00018-005-5350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saier MH, Yen MR, Noto K, Tamang DG, Elkan C. The transporter classification database: recent advances. Nucleic Acids Res. 2009;37:D274–D278. doi: 10.1093/nar/gkn862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhardwaj N, Stahelin RV, Zhao GJ, Cho W, Lu H. MeTaDoR: a comprehensive resource for membrane targeting domains and their host proteins. Bioinformatics. 2007;23:3110–3112. doi: 10.1093/bioinformatics/btm395. [DOI] [PubMed] [Google Scholar]
- 8.Whitmore L, Wallace BA. The peptaibol database: a database for sequences and structures of naturally occurring peptaibols. Nucleic Acids Res. 2004;32:D593–D594. doi: 10.1093/nar/gkh077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lomize AL, Pogozheva ID, Lomize MA, Mosberg HI. Positioning of proteins in membranes: a computational approach. Protein Sci. 2006;15:1318–1333. doi: 10.1110/ps.062126106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lomize AL, Pogozheva ID, Lomize MA, Mosberg HI. The role of hydrophobic interactions in positioning of peripheral proteins in membranes. BMC Struct. Biol. 2007:7. doi: 10.1186/1472-6807-7-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gumbart J, Wang Y, Aksimentiev A, Tajkhorshid E, Schulten K. Molecular dynamics simulations of proteins in lipid bilayers. Curr. Opin. Struct. Biol. 2005;15:423–431. doi: 10.1016/j.sbi.2005.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sansom MSP, Scott KA, Bond PJ. Coarse-grained simulation: a high-throughput computational approach to membrane proteins. Biochem. Soc. T. 2008;36:27–32. doi: 10.1042/BST0360027. [DOI] [PubMed] [Google Scholar]
- 13.BenTal N, Honig B, Miller C, McLaughlin S. Electrostatic binding of proteins to membranes. Theoretical predictions and experimental results with charybdotoxin and phospholipid vesicles. Biophys. J. 1997;73:1717–1727. doi: 10.1016/S0006-3495(97)78203-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Murray D, BenTal N, Honig B, McLaughlin S. Electrostatic interaction of myristoylated proteins with membranes: simple physics, complicated biology. Structure. 1997;5:985–989. doi: 10.1016/s0969-2126(97)00251-7. [DOI] [PubMed] [Google Scholar]
- 15.Murray D, Honig B. Electrostatic control of the membrane targeting of C2 domains. Mol. Cell. 2002;9:145–154. doi: 10.1016/s1097-2765(01)00426-9. [DOI] [PubMed] [Google Scholar]
- 16.Sharikov Y, Walker RC, Greenberg J, Kouznetsova V, Nigam SK, Miller MA, Masliah E, Tsigelny IF. MAPAS: a tool for predicting membrane-contacting protein surfaces. Nat. Methods. 2008;5:119–119. doi: 10.1038/nmeth0208-119. [DOI] [PubMed] [Google Scholar]
- 17.Feig M, Brooks CL. Recent advances in the development and application of implicit solvent models in biomolecule simulations. Curr. Opin. Struct. Biol. 2004;14:217–224. doi: 10.1016/j.sbi.2004.03.009. [DOI] [PubMed] [Google Scholar]
- 18.Ducarme P, Rahman M, Brasseur R. IMPALA: a simple restraint field to simulate the biological membrane in molecular structure studies. Proteins. 1998;30:357–371. [PubMed] [Google Scholar]
- 19.Lazaridis T. Implicit solvent simulations of peptide interactions with anionic lipid membranes. Proteins. 2005;58:518–527. doi: 10.1002/prot.20358. [DOI] [PubMed] [Google Scholar]
- 20.Efremov RG, Nolde DE, Konshina AG, Syrtcev NP, Arseniev AS. Peptides and proteins in membranes: what can we learn via computer simulations? Curr. Med. Chem. 2004;11:2421–2442. doi: 10.2174/0929867043364496. [DOI] [PubMed] [Google Scholar]
- 21.Lazaridis T. Effective energy function for proteins in lipid membranes. Proteins. 2003;52:176–192. doi: 10.1002/prot.10410. [DOI] [PubMed] [Google Scholar]
- 22.Senes A, Chadi DC, Law PB, Walters RFS, Nanda V, DeGrado WF. E-z, a depth-dependent potential for assessing the energies of insertion of amino acid side-chains into membranes: Derivation and applications to determining the orientation of transmembrane and interfacial helices. J. Mol. Biol. 2007;366:436–448. doi: 10.1016/j.jmb.2006.09.020. [DOI] [PubMed] [Google Scholar]
- 23.Tusnady GE, Dosztanyi Z, Simon I. TMDET: web server for detecting transmembrane regions of proteins by using their 3D coordinates. Bioinformatics. 2005;21:1276–1277. doi: 10.1093/bioinformatics/bti121. [DOI] [PubMed] [Google Scholar]
- 24.Cheema J, Basu G. MAPS: An interactive web server for membrane annotation of transmembrane protein structures. Ind. J. Biochem. Biophys. 2011;48:106–110. [PubMed] [Google Scholar]
- 25.Tusnady GE, Dosztanyi Z, Simon I. PDB_TM: selection and membrane localization of transmembrane proteins in the protein data bank. Nucleic Acids Res. 2005;33:D275–D278. doi: 10.1093/nar/gki002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chetwynd AP, Scott KA, Mokrab Y, Sansom MSP. CGDB: A database of membrane protein/lipid interactions by coarse-grained molecular dynamics simulations. Mol. Membr. Biol. 2008;25:662–669. doi: 10.1080/09687680802446534. [DOI] [PubMed] [Google Scholar]
- 27.Lomize AL, Pogozheva ID, Mosberg HI. Anisotropic solvent model of the lipid bilayer. 2. Energetics of insertion of small molecules, peptides, and proteins in membranes. J. Chem. Inf. Model. 2011;51:930–946. doi: 10.1021/ci200020k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: Orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 29.Velankar S, Best C, Beuth B, Boutselakis CH, Cobley N, Da Silva AWS, Dimitropoulos D, Golovin A, Hirshberg M, John M, et al. PDBe: Protein Data Bank in Europe. Nucleic Acids Res. 38:D308–D317. doi: 10.1093/nar/gkp916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 31.Consortium TU. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bateman A, Birney E, Cerruti L, Durbin R, Etwiller L, Eddy SR, Griffiths-Jones S, Howe KL, Marshall M, Sonnhammer ELL. The Pfam Protein Families Database. Nucleic Acids Res. 2002;30:276–280. doi: 10.1093/nar/30.1.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJP, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kucerka N, Nagle JF, Sachs JN, Feller SE, Pencer J, Jackson A, Katsaras J. Lipid bilayer structure determined by the simultaneous analysis of neutron and X-ray scattering data. Biophys. J. 2008;95:2356–2367. doi: 10.1529/biophysj.108.132662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marsh D. Membrane water-penetration profiles from spin labels. Eur.Biophys. J. Biophy. 2002;31:559–562. doi: 10.1007/s00249-002-0245-z. [DOI] [PubMed] [Google Scholar]
- 36. DeLano, W. L. (2003) The PyMOL molecular graphics system. DeLano Scientific LLC, San Carlos, CA, USA. http://www.pymol.org/. http://www.pymol.org/ (20 July 2011, date last accessed)
- 37.Jmol: an open-source Java viewer for chemical structures in 3D. http://jmol.sourceforge.net/ (20 July 2011, date last accessed) [Google Scholar]
- 38.Walther D. WebMol - A Java-based PDB viewer. Trends Biochem. Sci. 1997;22:274–275. doi: 10.1016/s0968-0004(97)89047-0. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, Chistyakov VV, Thornton JM. PDBsum more: new summaries and analyses of the known 3D structures of proteins and nucleic acids. Nucleic Acids Res. 2005;33:D266–D268. doi: 10.1093/nar/gki001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Prilusky J. OCA, a browser-database for protein structure/function. 1996 http://bip.weizmann.ac.il/oca (20 July 2011, date last accessed) [Google Scholar]
- 41.Wang YL, Addess KJ, Chen J, Geer LY, He J, He SQ, Lu SN, Madej T, Marchler-Bauer A, Thiessen PA, et al. MMDB: annotating protein sequences with Entrez's 3D-structure database. Nucleic Acids Res. 2007;35:D298–D300. doi: 10.1093/nar/gkl952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jo S, Lim JB, Klauda JB, Im W. CHARMM-GUI membrane builder for mixed bilayers and its application to yeast membranes. Biophys. J. 2009;97:50–58. doi: 10.1016/j.bpj.2009.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sommer B, Dingersen T, Gamroth C, Schneider SE, Robert S, Kruger J, Dietz KJ. CELLmicrocosmos 2.2 membrane editor: a modular interactive shape-based software approach to solve heterogeneous membrane packing problems. J. Chem. Inf. Model. 2011;51:1165–1182. doi: 10.1021/ci1003619. [DOI] [PubMed] [Google Scholar]