Abstract
The polymorphism in microRNA target site (PolymiRTS) database aims to identify single-nucleotide polymorphisms (SNPs) that affect miRNA targeting in human and mouse. These polymorphisms can disrupt the regulation of gene expression by miRNAs and are candidate genetic variants responsible for transcriptional and phenotypic variation. The database is therefore organized to provide links between SNPs in miRNA target sites, cis-acting expression quantitative trait loci (eQTLs), and the results of genome-wide association studies (GWAS) of human diseases. Here, we describe new features that have been integrated in the PolymiRTS database, including: (i) polymiRTSs in genes associated with human diseases and traits in GWAS, (ii) polymorphisms in target sites that have been supported by a variety of experimental methods and (iii) polymorphisms in miRNA seed regions. A large number of newly identified microRNAs and SNPs, recently published mouse phenotypes, and human and mouse eQTLs have also been integrated into the database. The PolymiRTS database is available at http://compbio.uthsc.edu/miRSNP/.
INTRODUCTION
A primary, and often elusive, goal of genetics studies is identifying the specific genetic variants that cause individual variation in complex traits. One class of genetic variants that have been shown to impact gene expression and higher order traits is DNA polymorphisms that alter microRNA targeting. Polymorphisms in microRNA target sites (PolymiRTSs) have been associated with a wide range of diseases (1,2), including cancers (3–5), Parkinson disease (6), osteoporosis (7), diabetes (8) and hypertension (9). MicroRNAs are short (∼22 nt) non-coding RNAs that function as post-transcriptional regulators of genes, repressing mRNA translation and causing mRNA decay (10). It has been estimated that miRNAs may regulate ∼60% of all human genes. While the mechanism through which the specific mRNA targets of a miRNA are selected has yet to be completely understood, one common feature of many miRNA–mRNA pairs is sequence complementarity between the 5′-end, or seed region, of the mature form of the miRNA and a target site in the 3′-UTR of the mRNA. Because of this complementarity in miRNA binding, genetic variants, such as single-nucleotide polymorphisms (SNPs), in these sites can disrupt miRNA binding sites or create new binding sites, resulting in variation in both the levels of gene expression and, potentially, higher order traits across individuals.
We introduced the PolymiRTS database (11) (http://compbio.uthsc.edu/miRSNP/) to collect SNPs in putative miRNA target sites in human and mouse genomes and identify their possible implications in transcriptional and phenotypic variation. Specifically, the original version of the database integrated SNPs in predicted miRNA target sites in the 3′-UTRs of mRNAs with quantitative trait loci (QTLs) for both mRNA expression traits and higher-order phenotypes. The database is therefore organized to provide links between SNPs in mRNA target sites, cis-acting expression QTLs (eQTLs) and physiological QTLs. Since the introduction of the PolymiRTS database, several advances have been made in identifying both the targets of miRNAs and the polymorphisms that impact complex traits. In particular, the rapid expansion of genome-wide association studies (GWAS) in recent years has made it increasingly possible to link polymorphisms, such as those contained in the PolymiRTS database, with human diseases and traits. Additionally, new sequencing approaches, such as cross-linked immunoprecipitation sequencing (CLIP-Seq), have enabled the determination of the mRNA sequences that interact with miRNAs (12,13). In light of these advances, we have updated the PolymiRTS database to improve its utility as a tool for understanding how miRNA-related polymorphisms impact complex traits.
NEW FEATURES
We have both expanded the PolymiRTS database and added several new features, with a focus on incorporating information from experimental data sources, including GWAS and CLIP-Seq (Figure 1 and Table 1). New content in this update of the PolymiRTS database includes:
Polymorphic miRNA targets that have been linked to human diseases and traits in GWAS. The PolymiRTS database includes over 3000 genes that have been linked to human diseases and traits in GWAS and contain polymorphisms in predicted and experimentally supported miRNA binding sites.
Polymorphisms in experimentally supported miRNA target sites. Recently, new techniques, such as CLIP-Seq (12,13) and allelic imbalance sequencing (14), have been introduced and enabled more efficient experimental identification of microRNA targets. SNPs in miRNA target sites discovered using these techniques, as well as experimentally supported miRNA target sites in several databases (15–17), have been identified and are available for searching and browsing in the database.
Polymorphisms in miRNA seed regions. SNPs in miRNA seed regions have potentially wide-spread effects, as the change in seed sequence may disrupt the binding of the miRNA to all of its original targets and redirect it to new target genes, and have been linked with complex traits. For example, polymorphisms in the seed region of miR-96 have been associated with hearing loss in humans (18) and mice (19) and
A significantly expanded number of expression QTL datasets. Variation in gene expression caused by a polymiRTS may result in detection of a cis-eQTL in microarray or RNA-Seq data. We have added cis-eQTLs from several studies in mouse and human, including those in the GTEx eQTL browser, to the polymiRTS database to identify the genetic variants that may cause these cis-eQTLs.
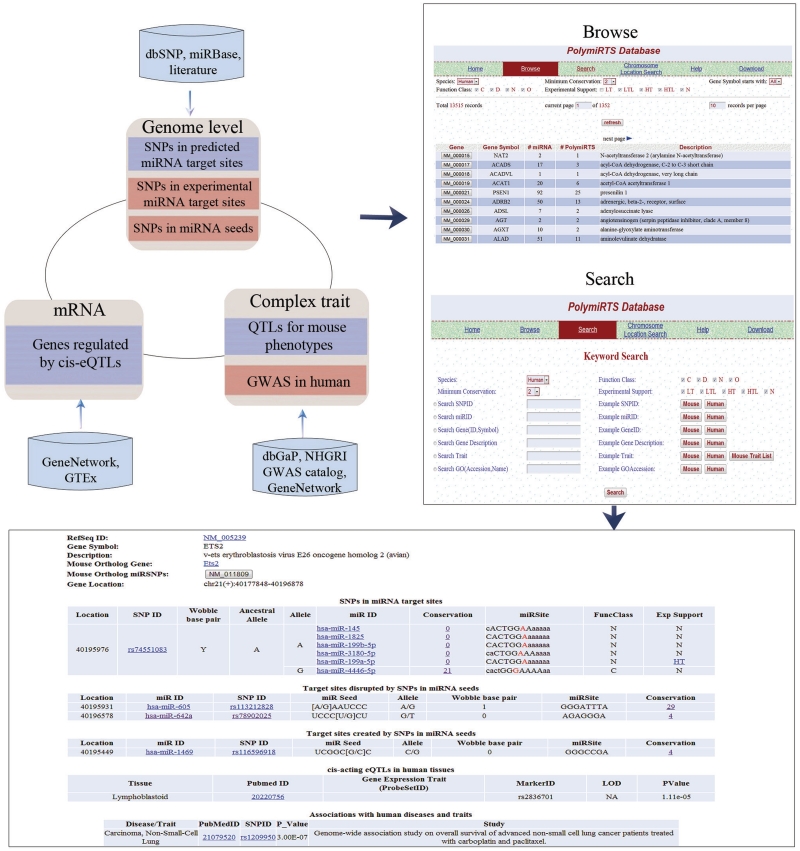
Figure 1.
Overview of PolymiRTS 2.0. The database integrates genomic, expression and complex trait data which can be accessed through a variety of browse and search options. New features in the database have a red background. A sample entry in the database is also shown.
Table 1.
Summary of data available in PolymiRTS database
| Type of data | Number of records |
|
|---|---|---|
| Human | Mouse | |
| miRNAs | 1733 | 1111 |
| SNPs in predicted miRNA target sites | 117 167 | 112 763 |
| SNPs in experimentally supported miRNA target sites | 1117 | 426 |
| SNPs in miRNA seeds | 20 | 5 |
| Expression datasets used to determine cis-eQTLs | 12 | 9 |
| cis-eQTLs | 24 000 | 27 862 |
| Genes associated with human traits in GWAS | 3509 | NA |
| Genes associated with BXD phenotype QTLs | NA | 2331 |
DATABASE CONTENT
SNPs in predicted and experimentally validated miRNA target sites
The main table in the original version of the PolymiRTS database contained SNPs in predicted miRNA target sites for human and mouse. We have updated this table following the same general workflow outlined in our previous paper (11), utilizing the latest versions of publically available databases (as of August 2011). We first identified all SNPs in dbSNP build 132 (20,21) that were in 3′-UTRs of mRNAs and used Galaxy (22–24) to obtain the sequences surrounding these SNPs in the human and mouse genomes (hg19 and mm9). Mature miRNA sequences were downloaded from miRBase 17 (25). The seed regions of the miRNAs were compared to the 3′-UTR sequences to determine if the SNPs affected complementarity between the seed and potential target sequences. We used the criteria of TargetScan (26) in the prediction of miRNA sites, which requires a perfect Watson–Crick match to the seed nucleotides 2–7 of miRNA, and either a perfect match to the 8th nucleotide of the miRNA or an anchor adenosine immediately downstream of the 2–7 seed in the target. We then classified predicted target sites containing SNPs into one of four functional classes. The four functional classes were defined as follows: (i) O: the ancestral allele could not be determined, (ii) D: the derived allele disrupts a target site that was conserved in two or more other vertebrate genomes based on the 46-way Multiz alignment, (iii) N: the derived allele disrupts a non-conserved target site and (iv) C: the derived allele creates a new miRNA site. The ancestral allele for each SNP was determined through comparison with the rat (rn4) and chimpanzee (panTro2) genomes for mouse and human, respectively.
SNPs in experimentally supported target sites
While computationally predicted miRNA target sites have been successfully used for several applications, they still result in a large number of false positives (27,28), making experimentally determined and validated target sites of great interest. We have therefore added experimentally determined miRNA target sites from a variety of experimental sources to the PolymiRTS database. First, we used three databases, miRecords (16), TarBase (15) and miRTarBase (17), that contain collections of miRNA targets from both low- and high-throughput experiments. The large majority of the entries in these databases contain only the miRNA–mRNA pairs that interact, not the specific location of the miRNA target site. For these cases, we scanned the entire 3′-UTR region of the mRNA for predicted binding sites of the interacting miRNA that included SNPs, as described earlier. These experimentally supported entries in the PolymiRTS database, where the specific locations of the miRNA target sites were not provided by the experiment, are labeled with the classes HT or LT depending on whether miRNA-mRNA target pair was determined from high-throughput (e.g. microarray or pSILAC) or low-throughput (e.g. luciferase reporter assay or western blot) methods, respectively.
Additionally, several experimental techniques, such as the HITS-CLIP (13) and PAR-CLIP (12), have recently been developed and used to identify the specific mRNA sequences that bind with miRNAs in the RNA-inducing silencing complex (RISC). To include data from these experiments, we first obtained the mRNA sequences bound in RISCs from ago.rockefeller.edu, which is associated with the HITS-CLIP experiment, and the Supplementary Material from Ref. (12). We then determined if these sequences contained SNPs that would disrupt miRNA binding. The disrupted target sites identified from this method are labeled with the code HTL to indicate that the target site was identified using a high-throughput method that provides the specific location information about the target site. Finally, alleleic imbalance sequencing has recently been proposed as an experimental method to determine if SNPs in miRNA target sites impact expression of the target (14). The allelic imbalance for 67 predicted target sites of three miRNAs in mouse was measured, and it was determined that at least 16% of these target sites were functional. We included the 11 (67 × 0.16 = 10.7) target sites with the highest allelic imbalance ratios, and labeled them with the code LTL, indicating that the specific location of the miRNA target in the mRNA was confirmed through a low-throughput experimental method.
SNPs in miRNA seed regions
With the increasing number of SNPs entries in dbSNP and annotated miRNAs in miRBase, it has become practical to identify SNPs in miRNAs. A SNP in a miRNA seed region may impact the binding of several hundred targets. We identified 5 and 20 SNPs in miRNA seed regions for mouse and human, respectively. We extracted the entire 3′-UTR of all genes in Enesembl Biomart (29) for the mouse and human genomes and used the criteria discussed earlier to identify all predicted target sites that would be either disrupted or created by the SNPs in the miRNA seed regions. Disrupted sites are predicted targets of the miRNAs with the reference allele at the SNP location, while the created sites are predicted targets of the miRNA with the derived allele.
PolymiRTS in cis-acting eQTLs
Polymorphisms that disrupt miRNA binding may be the causal variants that underlie cis-acting expression QTLs (eQTLs). We have extensively increased the datasets used to identify gene regulated by cis-eQTLs in mouse and human. For mouse, eQTL mapping results for gene expression were accessed from publically available datasets in the GeneNetwork (www.genenetwork.org) browser. We included eQTLs in nine tissues (whole brain, cerebellum, eye, hippocampus, kidney, liver, nucleus accumbens, prefrontal cortex and retina) in the BXD recombinant inbred panel in PolymiRTS. Gene expression levels were treated as quantitative traits and were mapped onto genomic regions (eQTL) using standard marker regression. A gene is said to have a significant cis-acting eQTL if the QTL peak location is within 5 Mb from the gene's physical location and the genome-wide adjusted P-value was ≤0.05.
We have also added eQTLs identified in several human tissues to the database. The GTEx eQTL browser (http://www.ncbi.nlm.nih.gov/gtex/test/GTEX2/gtex.cgi) currently includes eQTL mapping results for seven data sets across six tissues and cell lines (liver, cerebellum, frontal cortex, temporal cortex, pons and lymphblastoid cells). These eQTLs from GTEx were downloaded and filtered to select cis-eQTLs by identifying the marker with the minimum P-value within 1 Mb of the gene. Additionally, cis-eQTLs in four studies (not in GTEx) of expression genetics in the human skin (30), cortex (31), monocytes (32) and lymphoblastoid cells (33) were also added to polymiRTS. The methods and significance criteria used in identifying cis-eQTLs in each of these four studies were maintained.
PolymiRTS in physiological QTLs and disease genes
The main goal of many genetic studies is to determine the polymorphisms that cause variation in complex traits and diseases. For mouse, we identified polymorphisms and genes that may impact complex traits by mapping QTLs (genome-wide adjusted P-value ≤0.1) for more than 2000 published BXD phenotypes (physiological and behavioral traits) available in GeneNetwork. For each QTL, we linked it with genes that are located in the QTL interval and have at least one PolymiRTS. These genes, together with genes with other types of potentially functional sequence variations, are candidate causal genes underlying the physiological and disease QTLs.
To associate genes containing polymorphisms in miRNA target sites with human diseases and traits, we utilized results from GWAS available in the NHGRI GWAS Catalog (34) and dbGaP (35). We first used these databases to collect the genes that contain or are located nearby the polymorphisms that have been linked to complex traits in the GWAS. Then, we found the intersection of this list of GWAS-identified genes with the list of genes containing SNPs in predicted and experimentally supported miRNA target sites. The polymiRTSs in these genes are potential causal genetic variants that underlie the GWAS results.
DATABASE ACCESS
The PolymiRTS interface has been modified to allow for quick access to polymorphisms that are of high interest and to the new features that have been included in this update. Specifically, we have added new tables to the database which are accessible via the Browse link on the PolymiRTS homepage. The database now includes the following browsable tables: (i) genes with SNPs in miRNA target sites, (ii) SNPs in miRNA seeds, (iii) human diseases and traits, which contains human diseases and traits that have been linked to genes with SNPs in miRNA target sites in GWAS and (iv) experimentally supported targets, which contains only the SNPs in miRNA target sites with experimental support. Users are also able to search the database by SNP, miRNA, gene, complex trait and gene ontology, as well as perform a chromosome location search to select all entries in PolymiRTS between two genomic locations, which may be used by researchers looking for functional polymorphisms that underlie a physiological QTL. Both the browsable tables and search results can be filtered to select only entries in the database with specific functional classes, experimental support or high levels of conservation across species. Tab-delimited text files containing all of the tables used in the database are also available for download.
The main method of viewing the records in PolymiRTS is a webpage page separated into five sections, which lists all data in the database pertaining to a particular gene (Figure 1). The five sections on this page are: (i) a gene description section listing the RefSeqID, gene symbol, gene description and genomic location, as well as, for genes with a human/mouse ortholog, the ortholog gene and a link to the record in PolymiRTS in the other species, if available; (ii) a table of SNPs in miRNA target sites in the 3′-UTR regions of the gene, which provides the genomic location and miRNAs that target the site, as well as the functional class and evolutionary conservation of the target site; (iii) tables of predicted targets sites that are either created or disrupted by SNPs in miRNA seeds; (iv) a table of tissues in which the gene is regulated by cis-acting eQTLs and (v) a table listing associations with complex traits. For human genes, the associations with complex traits section provide the title of the study establishing the link between the gene and the trait as well as a link to the study's PubMed entry.
DISCUSSION AND FUTURE DIRECTIONS
One of the limiting factors in understanding the impact of miRNAs on complex traits has been in identifying the mRNA sequences that are targeted by miRNAs, and, in response, several computational methods have been developed to predict these target sites (36–38). These methods have traditionally been based on complementarity between miRNA seeds and mRNA sequences and conservation of mRNA sequences across species; however, other criteria including thermodynamics, target site accessibility and features of the sequence surrounding the predicted binding site have been used. Because of the relatively small number of experimentally validated miRNA targets, it has been difficult to thoroughly evaluate the performance of these algorithms, and the existing comparisons of computational predictions with experimental results have shown that the predictions have high false positive rates, particularly for non-conserved sites (27,28). As there has yet to be a clear winner among target prediction algorithms, we have continued to use a simple method of predicting target sites in the PolymiRTS database that uses only seed-target complementarity and, if the user chooses, conservation, allowing users the choice of displaying all potential target sites or only those sites with high conservation.
Due to the limitations of computational target predictions, we have added results from a large number of the existing experimentally supported miRNA–mRNA interactions in PolymiRTS. While many of these results rely on computational algorithms to determine the precise location on the mRNA that is the miRNA target site, recent experimental techniques, such as CLIP-Seq and allelic imbalance sequencing, can provide the specific miRNA target sequence. These experiments have the ability to improve the quality of databases such as PolymiRTS because they not only have the potential to greatly increase the number of experimentally validated miRNA targets, but they also provide a training set that can be used to improve computational prediction of target sites.
The expansion of the PolymiRTS database in this update increases its usefulness to researchers investigating the impact of polymorphisms on miRNA-mediated biological processes. The inclusion of experimentally supported target sites and SNPs associated with human traits and diseases in GWAS will enable users to select SNPs of high-interest to understanding the effect of these genetic variations on gene expression and complex traits. In the future, we expect PolymiRTS to continue to grow with the increasing availability of data in resources such as GTEx and dbGaP. One area that we can see PolymiRTS improving greatly in the future is in having improved methods for prediction and identification of miRNA target sites. We will perform quarterly searches for updates to databases such as miRTarBase and miRecords as well as for new data sets containing targets determined from CLIP-Seq or similar experimental methods in order to include new experimentally supported miRNA–mRNA interactions in PolymiRTS as they become available. Also, the improvements to computational prediction algorithms facilitated by these experiments will be used to enable selection of only high confidence targets sites in the PolymiRTS database.
FUNDING
UT Center for Integrative and Translational Genomics; National Institutes of Health (grants NR009270, AI081050, AI019782); Department of Defense (grant W81XHW-05-01-0227) and American Heart Association (grant 0830134N). Funding for open access charge: Department of Defense (grant W81XHW-05-01-0227).
Conflict of interest statement. None declared.
REFERENCES
- 1.Sethupathy P, Collins FS. MicroRNA target site polymorphisms and human disease. Trends Genet. 2008;24:489–497. doi: 10.1016/j.tig.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 2.Badiera S, Hatem E, Syonnet S, Henrion-Caude A. microRNAs in diseases: from candidate to modifier genes. Clin. Genet. 2010;77:306–313. doi: 10.1111/j.1399-0004.2010.01370.x. [DOI] [PubMed] [Google Scholar]
- 3.Chen K, Song F, Calin GA, Wei Q, Hao X, Zhang W. Polymorphisms in microRNA targets: a gold mine for molecular epidemiology. Carcinogenesis. 2008;29:1306–1311. doi: 10.1093/carcin/bgn116. [DOI] [PubMed] [Google Scholar]
- 4.Bao BY, Pao JB, Huang CN, Pu YS, Chang TY, Lan YH, Lu TL, Lee HZ, Juang SH, Chen LM, et al. Polymorphisms inside microRNAs and microRNA target sites predict clinical outcomes in prostate cancer patients receiving androgen-deprivation therapy. Clin. Cancer Res. 2010;17:928–936. doi: 10.1158/1078-0432.CCR-10-2648. [DOI] [PubMed] [Google Scholar]
- 5.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat. Rev. Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang G, van der Walt JM, Mayhew G, Li YJ, Zuchner S, Scott WK, Martin ER, Vance JM. Variation in the miRNA-433 binding site of FGF20 confers risk for Parkinson disease by overexpression of alpha-synuclein. Am. J. Hum. Genet. 2008;82:283–289. doi: 10.1016/j.ajhg.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lei SF, Papasian CJ, Deng HW. Polymorphisms in predicted miRNA binding sites and osteoporosis. J. Bone Mineral Res. 2010;26:72–78. doi: 10.1002/jbmr.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lv K, Guo Y, Zhang Y, Wang K, Jia Y, Sun S. Allele-specific targeting of hsa-miR-657 to human IGF2R creates a potential mechanism underlying the association of ACAA-insertion/deletion polymorphism with type 2 diabetes. Biochem. Biophys. Res. Commun. 2008;374:101–105. doi: 10.1016/j.bbrc.2008.06.102. [DOI] [PubMed] [Google Scholar]
- 9.Martin MM, Buckenberger JA, Jiang J, Malana GE, Nuovo GJ, Chotani M, Feldman DS, Schmittgen TD, Elton TS. The human angiotensin II type 1 receptor +1166 A/C polymorphism attenuates microrna-155 binding. J. Biol. Chem. 2007;282:24262–24269. doi: 10.1074/jbc.M701050200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Sayed D, Abdellatif M. MicroRNAs in development and disease. Physiol. Rev. 2011;91:827–887. doi: 10.1152/physrev.00006.2010. [DOI] [PubMed] [Google Scholar]
- 11.Bao L, Zhou M, Wu L, Lu L, Goldowitz D, Williams RW, Cui Y. PolymiRTS database: linking polymorphisms in microRNA target sites with complex traits. Nucleic Acids Res. 2007;35:D51–D54. doi: 10.1093/nar/gkl797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano J, Jungkamp M, Munschauer ACM, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chi SW, Zang JB, Mele A, Darnell RB. Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature. 2009;460:479–486. doi: 10.1038/nature08170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Bartel DP. Allelic imbalance sequencing reveals that single-nucleotide polymorphisms frequently alter microRNA-directed repression. Nat. Biotechnol. 2009;27:472–477. doi: 10.1038/nbt.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sethupathy P, Corda B, Hatzigeorgiou AG. TarBase: a comprehensive database of experimentally supported animal microRNA targets. RNA. 2006;12:192–197. doi: 10.1261/rna.2239606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA-target interactions. Nucleic Acids Res. 2009;37:D105–D110. doi: 10.1093/nar/gkn851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, Tsai WT, Chen GZ, Lee CJ, Chiu CM, et al. miRTarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011;39:D163–D169. doi: 10.1093/nar/gkq1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mencia A, Modamio-Hoybjor S, Redshaw N, Morin M, Mayo-Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- 19.Lewis MA, Quint E, Glazier AM, Fuchs H, De Angelis MH, Langford C, van Dongen S, Abreu-Goodger C, Piipari M, Redshaw N, et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat. Genet. 2009;41:614–618. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. 2010 doi: 10.1002/0471142727.mb1910s89. Chapter 19, Unit 19, 10.1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giardine B, Riemer C, Hardison RC, Burhans R, Elnitski L, Shah P, Zhang Y, Blankenberg D, Albert I, Taylor J, et al. Galaxy: a platform for interactive large-scale genome analysis. Genome Res. 2005;15:1451–1455. doi: 10.1101/gr.4086505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat. Method. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- 28.Baek D, Villen J, Shin C, Carmargo FD, Gygi SP, Bartel DP. The impact of microRNAs on protein output. Nature. 2008;455 doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinsella RJ, Kahari A, Haider S, Zamora J, Proctor G, Spudich G, Almeida-King J, Staines D, Derwent P, Kerhornou A, et al. Ensembl BioMarts: a hub for data retrieval across taxonomic space. Database. 2011;2011:bar030. doi: 10.1093/database/bar030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ding J, Gudjonsson JE, Liang L, Stuart PE, Li Y, Chen W, Weichenthal M, Ellinghaus E, Franke A, Cookson W, et al. Gene expression in skin and lymphoblastoid cells: refined statistical method reveals extensive overlap in cis-eQTL signals. Am. J. Hum. Genet. 2010;87:779–789. doi: 10.1016/j.ajhg.2010.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers AJ, Gibbs JR, Webster JA, Rohrer K, Zhao A, Marlowe L, Kaleem M, Leung D, Bryden L, Nath P, et al. A survey of genetic human cortical gene expression. Nat. Genet. 2007;39:1494–1499. doi: 10.1038/ng.2007.16. [DOI] [PubMed] [Google Scholar]
- 32.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, et al. Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS One. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pickrell JK, Marioni JC, Pai AA, Degner JF, Engelhardt BE, Nkadori E, Veyrieras JB, Stephens M, Gilad Y, Pritchard JK. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature. 2010;464:768–772. doi: 10.1038/nature08872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mendes ND, Freitas AT, Sagot MF. Current tools for the identification of miRNA genes and their targets. Nucleic Acids Res. 2009;37:2419–2433. doi: 10.1093/nar/gkp145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Xu J, Yang D, Tan X, Wang H. Computational approaches for microRNA studies: a review. Mamm. Genome. 2010;21:1–12. doi: 10.1007/s00335-009-9241-2. [DOI] [PubMed] [Google Scholar]
- 38.Hammell M. Computational methods to identify miRNA targets. Semin. Cell Dev. Biol. 2010;21:738–744. doi: 10.1016/j.semcdb.2010.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]