Abstract
MitoMiner (http://mitominer.mrc-mbu.cam.ac.uk/) is a data warehouse for the storage and analysis of mitochondrial proteomics data gathered from publications of mass spectrometry and green fluorescent protein tagging studies. In MitoMiner, these data are integrated with data from UniProt, Gene Ontology, Online Mendelian Inheritance in Man, HomoloGene, Kyoto Encyclopaedia of Genes and Genomes and PubMed. The latest release of MitoMiner stores proteomics data sets from 46 studies covering 11 different species from eumetazoa, viridiplantae, fungi and protista. MitoMiner is implemented by using the open source InterMine data warehouse system, which provides a user interface allowing users to upload data for analysis, personal accounts to store queries and results and enables queries of any data in the data model. MitoMiner also provides lists of proteins for use in analyses, including the new MitoMiner mitochondrial proteome reference sets that specify proteins with substantial experimental evidence for mitochondrial localization. As further mitochondrial proteomics data sets from normal and diseased tissue are published, MitoMiner can be used to characterize the variability of the mitochondrial proteome between tissues and investigate how changes in the proteome may contribute to mitochondrial dysfunction and mitochondrial-associated diseases such as cancer, neurodegenerative diseases, obesity, diabetes, heart failure and the ageing process.
INTRODUCTION
Mitochondria are far more than the ‘powerhouses’ of eukaryotic cells producing the adenosine triphosphate necessary for life (1). Their metabolism is dictated by their proteome, which is dynamic, responding to signals and the physiology of the cell, and their metabolic role includes supplying intermediates for anabolic pathways that produce essential metabolites, and catabolic pathways for disposal or recycling of excess or toxic metabolites. Their role in signalling is now evident and they participate in cellular growth, differentiation and death. Given these pivotal roles, mitochondrial dysfunction is associated with many diseases (2) including cancer, neurodegenerative diseases (3), obesity, diabetes, heart failure and the ageing process (4,5). As a marker for the health of mitochondria and cells (6), understanding the state of the mitochondrial proteome is necessary for further understanding mitochondrial-related disease.
The ability to isolate purified fractions of mitochondria promoted studies to catalogue the mitochondrial proteome in different species by using new proteomics techniques, particularly mass spectrometry (7). However, the isolation of pure mitochondria is not easy and contamination remains a serious problem (8). Conversely, even abundant membrane proteins are often absent or under-represented as most purification procedures are biased towards extracting soluble proteins (9). In recent years, mitochondrial proteomics has expanded to identifying post-translational modifications of mitochondrial proteins (10–12) and comparisons between mitochondria under different conditions (13,14). These proteomics studies are producing sizeable data sets, although many analyses are only possible if these studies are combined and can be compared among themselves and with other data. However, the use of these proteomics data and their inclusion in public protein sequence databases is hampered by their large size, high false positive rate and a prior lack of common data standards. Although the development of standards (15) and establishment of public repositories for proteomics data such as the Proteomics Identification (PRIDE) database (16) has attempted to address these issues, the repositories do not have the specific focus and advanced querying required for many types of analysis.
To enable better analysis of the mitochondrial proteome, resources cataloguing mitochondrial proteins have been published of varying data content and query capabilities, including the mitochondrial proteome database (MitoP2) (17), MitoRes (18) and MiGenes (19). Although it appears that many are no longer actively maintained and the majority are compiled by searching RefSeq or UniProt for proteins annotated as mitochondrial and so contain only well-characterized mitochondrial proteins. Proteomics studies are a source of previously unidentified mitochondrial proteins, but many resources incorporating such data are limited to a single species or have no protein homology data, which prevents cross-species comparisons and annotating proteins by using homology. We developed the MitoMiner mitochondrial proteomics data warehouse (20) to address four major limitations of other resources. First, to ease long-term maintenance and continuity of MitoMiner, the open source InterMine data warehouse system (21) was adopted for data import, management, querying and analysis. Second, to include data of many species and experimental types, MitoMiner uses an extensible data model that includes protein homology, which enables cross-species comparisons. Third, to enable evaluation of the evidence for mitochondrial localization, MitoMiner stores the provenance of the evidence for a protein being mitochondrial, which can be browsed and used as a constraint in queries. Fourth, to provide enhanced querying ability, the MitoMiner data warehouse can be searched by using an application programming interface (API) or a web interface with simple text searches, predefined template queries or custom queries built by using InterMine's interactive QueryBuilder.
Adopting InterMine offers a large number of advantages to biological resource providers, besides minimising development time. These include efficient construction and querying of large databases, an extensible core biological data model to describe and integrate its biological data, data import and export in a number of formats, creating and using lists of biological objects in queries with analysis tools to summarize their properties, a configurable user interface and personal accounts for users to store queries and results. As such, InterMine is used by a number of other biological resources for storing a variety of biological data, including FlyMine (21), modENCODE (22), RatMine, TargetMine (23) and YeastMine (24).
Since MitoMiner was first published (20), we have added new mitochondrial proteomics data sets, upgraded to the current version of InterMine with its new tools and functionality, and updated its integrated biological resources. MitoMiner currently contains data sets from proteomics studies on Homo sapiens, Mus musculus, Rattus norvegicus, Bos taurus, Drosophila melanogaster, Schizosaccharomyces pombe, Saccharomyces cerevisae, Neurospora crassa, Arabidopsis thaliana and the protists Tetrahymena thermophila and Giardia lamblia. Most importantly, we have now used the functionality of MitoMiner to define reference sets of mitochondrial proteins that are based on experimental proteomics data. These publicly available reference sets are available for further analyses in MitoMiner. Furthermore, we have recently demonstrated how MitoMiner provides the framework to build system biology models of mitochondrial metabolism (25).
DATA WAREHOUSE IMPLEMENTATION
This release of MitoMiner was built by using the InterMine open source data warehouse system (21) version 0.97. InterMine functionality depends upon a data model that describes biological data and the relationships between them. This data model is formalized by using the eXtensible Markup Language (XML) and is used by the InterMine system to generate the relational database schema and the Java classes that implement the data model as objects and coordinate data exchange with the data warehouse. The core data model of InterMine defines genes, proteins, publications and gene ontology terms. For MitoMiner, this data model was adapted and extended to include cellular sublocalization, protein homologs, metabolic pathways, genetic phenotypes, post-translational modifications and details of methods and results from green fluorescent protein (GFP) tagging and mass spectrometry experiments.
DATA IMPORT AND DATA WAREHOUSE CONTENT
The import of data into MitoMiner was achieved by using scripts that parse the original data, tabulated data or XML formatted files. Full details of the MitoMiner data conversion and import process are described in the first MitoMiner article (20). For each protein reported in a proteomics publication was recorded the type of experiment, the organism and the tissue or cell line analysed and the PubMed identifier of the publication. For mass spectrometry proteomics studies were recorded also the original protein identifier, subcellular localization, sequence of identified peptides, sequence coverage and details of the techniques used for protein purification, separation and identification. For species with mitochondrial localization data sets, all their UniProt entries were downloaded and the protein sequences and annotation imported plus their associated PubMed entries and annotations from the GO. Disease genes and proteins were imported from Online Mendelian Inheritance in Man (OMIM), and metabolic pathways from the Kyoto Encyclopaedia of Genes and Genomes (KEGG).
HomoloGene clusters (26) were used to define homologous proteins among species. However, HomoloGene does not include the protists T. thermophila and G. lamblia, which have mitochondrial proteome data sets in MitoMiner. To enable these protists to be compared with other species, protist proteins in mitochondrial proteome data sets were assigned to a HomoloGene cluster in MitoMiner if they had a BLASTP (27) expect value better than 10−35 to a member of the cluster.
AVAILABILITY
The current version of MitoMiner is freely accessible from the Medical Research Council Mitochondrial Biology Unit website (http://mitominer.mrc-mbu.cam.ac.uk/). It includes mitochondrial proteome data sets from 46 publications and 11 species (Table 1) and is cross-referenced with data from UniProt, GO, OMIM, HomoloGene, KEGG and PubMed.
Table 1.
Summary of mitochondrial proteomics studies and mitochondrial proteins in MitoMiner
| Species | Number of publications | Number of data entriesa |
Reference setb (number of HomoloGene clusters) | |
|---|---|---|---|---|
| Mass spectrometry | GFP | |||
| H. sapiens | 10 | 2482 | 273 | 1093 |
| M. musculus | 11 | 38 686 | 52 | 1075 |
| B. taurus | 1 | 28 | 0 | 850 |
| R. norvegicus | 5 | 1161 | 0 | 637 |
| D. melanogaster | 1 | 54 | 0 | 700 |
| S. cerevisiae | 9 | 2781 | 919 | 836 |
| A. thaliana | 5 | 696 | 0 | 551 |
| N. crassa | 1 | 293 | 0 | 590 |
| T. thermophila | 1 | 320 | 0 | 66 |
| G. lamblia | 1 | 1212 | 0 | 35 |
| P. falciparum | 0 | 0 | 0 | 143c |
aThe number of unique data entries from mass spectrometry or GFP tagging mitochondrial localization studies.
bThe number of HomoloGene clusters for a species that have member proteins that have been reported as mitochondrial in one GFP tagging study or more than two mass spectrometry studies.
cIn the absence of mitochondrial proteomics studies, the mitochondrial proteins of P. falciparum were predicted on the basis of their homology to proteins from other species with experimental evidence.
USER INTERFACE AND SERVICES
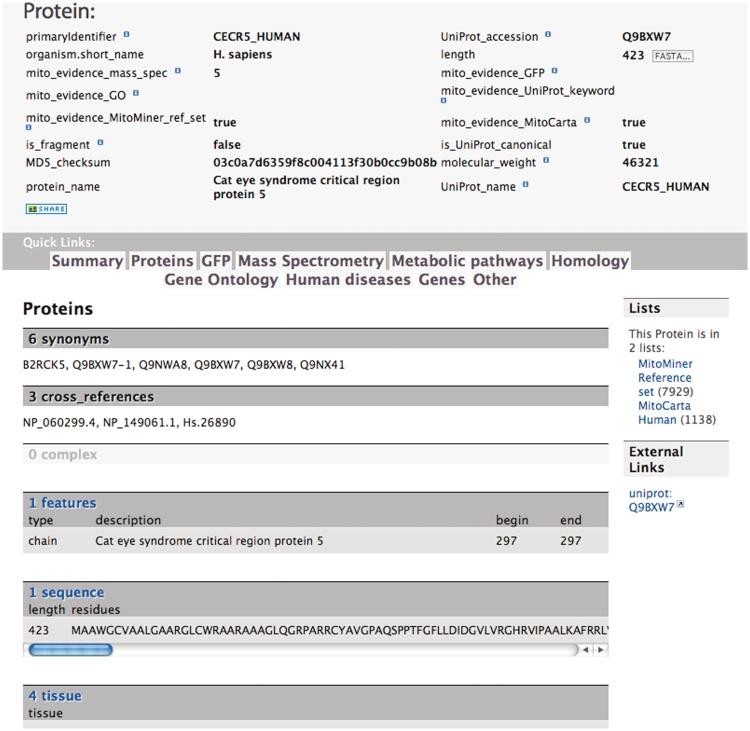
To provide convenient access to the data, the front page of MitoMiner shows the most popular queries organized into the data categories: mass spectrometry data, GFP tagging data, protein homology, protein annotation, metabolic pathways, proteomics publications and genetic phenotypes and diseases. A more detailed view of each data category page is available on selecting the data tab and provides information on the data sources, links to download datasets, relevant template queries to search these data and starting points for building custom queries with the QueryBuilder. For example, the protein data category page describes what protein annotation is available and from where it was taken; a link to download all proteins with experimental evidence of mitochondrial localization; and template queries for common searches of protein data. Data from entries in the database are presented in report pages that provide the information stored in the data warehouse and cross-references to external resources. For example, the protein report page lists protein attributes, a link to the UniProt entry, and tables of information about the protein, including mass spectrometry, GFP tagging, publications, tissue distribution, homologs, functional annotation and disease and phenotype and whether the protein is included in the MitoMiner mitochondrial protein reference set (Figure 1).
Figure 1.
The protein report page for the Cat eye syndrome critical region protein 5 (UniProt accession number Q9BXW7). This page reports that there is evidence for its mitochondrial localization in four tissues from five mass spectrometry studies including the human MitoCarta set and that the protein is included in the human MitoMiner mitochondrial reference set. However, a mitochondrial localization is not annotated in the UniProt keywords.
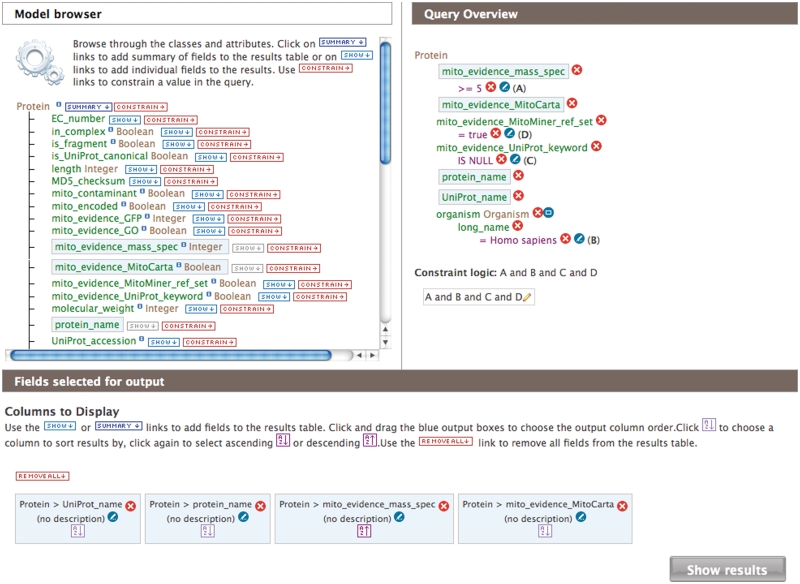
Querying of the MitoMiner data warehouse is provided by the InterMine system. There are three ways to search MitoMiner. First and simplest, the search text box in the top right corner of each page allows searching of identifiers, keywords or descriptions of entries in MitoMiner. Second, template queries for common searches are available from the front page, templates page or data category pages. They provide popular searches such as finding proteins with a specific GO annotation that have mitochondrial evidence or annotation. The template queries are form-based and users specify one or more search terms of the query, for example specifying a particular organism or metabolic pathway. Third, the interactive QueryBuilder tool allows users to view the data model, then select and apply search constraints on any combination of the data types and its attributes, and finally, select data for output in a results page (Figure 2). Template queries can be modified by using the QueryBuilder, and this provides a good introduction to using this interactive tool. Queries built with the QueryBuilder may be saved as XML files or stored for later use in a MyMine personal account. All query methods return their results as a table, where identifiers are linked to the report page for that entry. The results can be exported in a variety of formats, including those compatible with Microsoft Excel. The results of a query may also be saved as a list and used as the input to further queries.
Figure 2.
The web page of the QueryBuilder for building bespoke queries. The Web page has three components: the model browser (top left) from which data classes and attributes of the object model are selected for inclusion in the query; the data classes included in the query, the constraints on their attributes and the Boolean logic used to combine them (top right) and the data columns to display and sort the output of the results (bottom). The query displayed is to ‘find all human proteins that are in the MitoMiner reference set and reported in five or more mass spectroscopy studies, but do not have a mitochondrial localization annotated in UniProt.’ The query returns 21 proteins and also specifies if the protein is in the MitoCarta reference set.
List management is a valuable feature of InterMine that enables sets of identifiers to be uploaded or saved from queries and used as input to further queries or analyses. A list can also be merged, subtracted or intersected with another list. Combining results lists from different queries is an alternative way to compose and execute complex queries and build analysis pipelines. For example, a user can download the MitoP2 human mitochondrial reference set (17) as UniProt identifiers and upload this as a new list in MitoMiner. This list can be used as an input for a template or custom-built query, for example, to find the homologous proteins in species not present in MitoP2, or it can be compared with another list of proteins, for example, to compare and contrast it with the mitochondrial protein reference sets of MitoCarta (8) or MitoMiner. For user convenience, public lists were created and added to MitoMiner of the subunits of the respiratory complexes, the MitoCarta human and mouse mitochondrial protein reference sets, and the new MitoMiner mitochondrial protein reference sets.
UPDATES AND NEW FEATURES: DATA, SERVICES AND MITOCHONDRIAL PROTEIN REFERENCE SETS
There have been several significant advances and improvements to MitoMiner since it was first described in 2009. The data sets from 13 new publications have been added, including data sets from five new organisms for MitoMiner: S. pombe, T. thermophila, G. lamblia, N. crassa and A. thaliana. This greatly increases the phylogenetic diversity represented in MitoMiner and should give greater insight into the history and evolution of the mitochondrion. The data model has been improved and simplified to conform better to the requirements of the MitoMiner extended data model and to make the building of queries in the QueryBuilder more user-friendly.
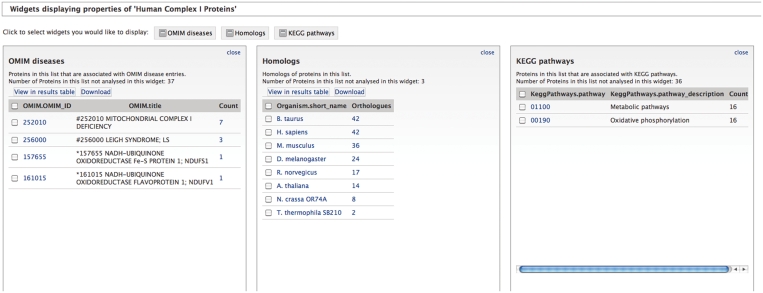
A new InterMine feature that has been deployed are widgets that analyse lists and display their results as tables, histograms or bar charts. These widgets are generated either by writing Java classes or from XML configuration files that define their behaviour. Currently, MitoMiner has XML-configured table widgets that summarize protein lists in terms of the frequency of disease genes in OMIM, homologous proteins in HomoloGene, and enzymes in KEGG metabolic pathways (Figure 3). Furthermore, Java-based widgets are in development that will perform statistical analyses and provide graphical views. For example, enrichment widgets that will calculate terms that are statistically over-represented (using the p- value) for a list of genes or proteins, such as KEGG metabolic pathways or tissue types.
Figure 3.
Table widgets in MitoMiner summarizing properties of a list of human complex I proteins. The three widgets calculate: the number of proteins associated with diseases reported in OMIM (left), the number of homologs in other species as defined by HomoloGene (middle) and the metabolic pathways as defined by KEGG (right).
The new InterMine system enables programmatic access via a RESTful API and web services. This enables developers to connect their applications directly to the MitoMiner data warehouse and automate complex data workflows. Client libraries are currently provided for Perl, Java and Python languages.
The most significant advance in MitoMiner is the creation of reference sets of mitochondrial proteins for many species based purely on proteomics data by analysing the data sets in MitoMiner from mass spectrometry and GFP tagging. This provides data sets that can be integrated and compared with mitochondrial data sets compiled in different ways, such as from localization annotation in UniProt or GO. As UniProt contains a large amount of sequence redundancy and protein fragments, we defined and counted mitochondrial proteins in terms of HomoloGene clusters, which group together homologous proteins and protein fragments. Thus, all the proteins from all species in a HomoloGene cluster were included in species-specific reference sets if mitochondrial localization had been reported among any of the cluster members in either one GFP tagging publication or three or more separate mass spectrometry publications. Only a report in one GFP tagging publication was required to define mitochondrial localization as it has a much higher accuracy compared with mass spectrometry of purified mitochondrial fractions. The threshold of three or more mass spectrometry publications was chosen as about half of the corresponding proteins were annotated as mitochondrial in UniProt. The MitoMiner mitochondrial protein reference sets are shown in Table 1. However, mitochondrial proteins may be present only in specific tissues or development stages, and the mass spectrometry techniques used often fail to detect membrane and low-abundance proteins (9). So the threshold for classifying a protein as mitochondrial as three or more reports in mass spectrometry publications eliminates some true positives as well as contaminants. Although there is no mitochondrial proteomics data for Plasmodium falciparum, the integration of HomoloGene in MitoMiner enabled the prediction of P. falciparum proteins in 143 HomoloGene clusters as mitochondrial on the basis of their homology with proteins from species that have experimental evidence (Table 1). The MitoMiner mitochondrial protein reference sets are available as public lists for further analysis. Using the lists features of MitoMiner, the different lists of mitochondrial proteins defined by MitoCarta, MitoMiner and UniProt can be compared and combined. For example, the number of H. sapiens mitochondrial proteins rises from 1093 to 1400 HomoloGene clusters when proteins annotated as mitochondrial in UniProt are combined with the MitoMiner reference set, and includes many known mitochondrial membrane proteins that may be missed in proteomics studies. Proteins from about 600 HomoloGene clusters of the MitoMiner reference set are not annotated as mitochondrial in UniProt. Furthermore, proteins of 680 HomoloGene clusters are in both the MitoMiner reference set and MitoCarta human data set, whereas 413 are unique to the MitoMiner reference set and 280 are unique to MitoCarta. Figure 2 shows a query to find human proteins in the MitoMiner mitochondrial protein reference set that have been reported in five or more mass spectrometry studies, but not annotated in UniProt as mitochondrial. The query returns 21 proteins, of which five are also present in the human MitoCarta set. Although many of these proteins are likely contaminants from other organelles (e.g. endoplasmin) or glycolytic proteins likely associated with the outer mitochondrial membranes (e.g. glyceraldehyde-3-phosphate dehydrogenase and fructose-bisphosphate aldolase A) (28), the list includes novel proteins such as cat eye syndrome critical region protein 5 (Figure 1) as well as stomatin-like protein 2, which has been reported as mitochondrial (29), although this is not reported in UniProt.
DISCUSSION
Among the available resources cataloguing mitochondrial proteins, only MitoP2, MitoCarta and MitoMiner include mitochondrial proteomics data from more than one species. However, the MitoCarta data sets are lists of M. musculus and H. sapiens mitochondrial proteins produced from an analysis of several data sets, rather than a database. The MITOP database for mitochondria-related proteins, genes and diseases was first published in 1999 (30) and the current version, MitoP2 (17) includes mitochondrial data sets for H. sapiens, M. musculus, S. cerevisiae, N. crassa and A. thaliana from proteomics, cellular sublocalization, mutant screens, disease genes, expression profiling and protein–protein interactions. In comparison, MitoMiner has imported data sets of 11 different species and is up-to-date with UniProt, OMIM, PubMed, GO and KEGG. MitoP2 and MitoMiner have many similarities and several differences, aside from in MitoMiner the greater number of imported recent mitochondrial proteome data sets and different species. Both resources import many of the same early mitochondrial proteome data sets, use OMIM and UniProt as a source of information, incorporate protein homology, enable queries and provide reference sets of mitochondrial proteins. The main differences are that (i) MitoMiner is built using the open source InterMine data warehouse that has an active community for its development and documentation and that has been adopted by several other biological resources, (ii) MitoMiner provides an interactive QueryBuilder for building bespoke queries over all aspects of the data warehouse, and enables the construction of analysis pipelines, which can be saved for future use, (iii) MitoMiner provides a list functionality to enable results to be saved and used as input to further queries as well as the MitoMiner and MitoCarta reference sets of mitochondrial proteins, (iv) MitoMiner uses the GO for functional annotation of proteins, whereas MitoP2 defines its own categories of mitochondrial function, (v) MitoMiner uses HomoloGene for its definition of homologous proteins, whereas MitoP2 uses the similarity matrix of proteins (SIMAP) (31), (vi) The MitoMiner mitochondrial protein reference set is generated automatically from proteomics data sets, whereas the MitoP2 set was automatically extracted from Swiss-Prot, then verified and complemented by manual annotation, (vii) MitoMiner includes the KEGG compound and metabolic pathways data that define functional links between the proteins as well as details about proteomics studies that are not present in other mitochondrial resources, such as the techniques used for purification, separation and identification of the proteins, and which are available to query, and (viii) MitoP2 includes computational predictions of subcellular localization and data sets of protein–protein interactions and expression profiles, whereas MitoMiner does not.
To assess the mitochondrial proteome, we developed mitochondrial protein reference sets for different species. The MitoMiner mitochondrial protein reference set for M. musculus has 1075 HomoloGene clusters, which requires the proteins to have been reported in one GFP tagging or more than two mass spectrometry publications. By comparison, the MitoMiner reference set of H. sapiens has 1093 HomoloGene clusters. The difference between M. musculus and H. sapiens mostly arises due to some UniProt accessions not being mapped by the UniProt mapping service to RefSeq proteins that are in HomoloGene. The number of predicted mitochondrial proteins is lower than may be expected for other species with few proteomics data sets, such as D. melanogaster and T. thermophila (Table 1), because of the stringent clustering threshold of HomoloGene, which only clusters near identical proteins. In particular, HomoloGene may not cluster orthologous short proteins, including transmembrane proteins, if they have low similarity. Two further limitations of HomoloGene are that, first, HomoloGene does not use phylogenetic analyses to distinguish between orthologs and paralogs. Thus, if a gene has duplicated and one copy is not targeted to the mitochondrion, HomoloGene may cluster them together leading to false positives in the data set. Second, HomoloGene is limited to a set of model species, which required the use of BLAST to incorporate T. thermophila and G. lamblia. The BLAST cutoff for inclusion in a cluster was very conservative to prevent the misclustering together of non-orthologous proteins, such as from different subfamilies of the mitochondrial carriers. However, this implies that diverged and short orthologous proteins may not have been clustered either. Because of these caveats in the use of HomoloGene, future versions of MitoMiner will include other definitions of protein homology for comparison.
The continued interest and recognition of important mitochondrial-associated diseases together with advances in protein purification and mass spectrometry techniques, particularly for detection of post-translational modifications, means that mitochondrial proteomes of healthy and diseased samples will continue to be published. It is desirable that the results of these studies are archived in public repositories upon publication so that they are accessible to researchers. It remains to be seen how the new proteomics repositories such as PRIDE will compete with dedicated resources such as MitoMiner. But currently there remains a need for dedicated resources that can add further value and analysis tools to subsets of these data. In particular, a mitochondrial proteome catalogue forms the basis for understanding the physiology of the mitochondrion and the unique integration of metabolic pathway data with proteomics data in MitoMiner gives a physiological context for a protein as well as supporting systems biology and modelling approaches. An important application of MitoMiner has been in the development of a systems biology model of human cardiomyocyte mitochondria (25). The tissue and species-specific proteomics data in MitoMiner were essential to reconstruct the metabolic network described in the iAS253 model, which simulates normal mitochondrial physiology and predicts the effects of metabolic disorders, as well as how these perturbations may be rectified. During the modelling process, MitoMiner was used to define the list of reactions to be included in the model that represent mitochondrial metabolism (25). This was done by first identifying all proteins associated with a reaction in KEGG that had annotation for localization to the mitochondrial matrix. MitoMiner was then used to evaluate all proteins with any level of evidence or annotation as mitochondrial that were associated with a reaction, in conjunction with information from other resources. Reactions with convincing evidence and whose metabolites could be used elsewhere in the model were included. In addition MitoMiner was used to determine whether the protein had been identified and reported by mass spectrometry or GFP in human heart tissue to ensure the model only included reactions occurring in this tissue. By using the data in MitoMiner, detailed models of mitochondrial metabolism can be developed for other species, organs, tissues and developmental and disease states. Thus, the collection of proteomics data on mitochondrial proteins remains an important endeavour for understanding mitochondrial physiology and a resource such as MitoMiner that can integrate and serve these data.
FUNDING
Funding for open access charge: Medical Research Council, UK.
Conflict of interest statement. None declared.
REFERENCES
- 1.McBride HM, Neuspiel M, Wasiak S. Mitochondria: more than just a powerhouse. Curr. Biol. 2006;16:R551–R560. doi: 10.1016/j.cub.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 2.Wallace DC. Mitochondrial diseases in man and mouse. Science. 1999;283:1482–1488. doi: 10.1126/science.283.5407.1482. [DOI] [PubMed] [Google Scholar]
- 3.Schapira AH. Mitochondrial disease. Lancet. 2006;368:70–82. doi: 10.1016/S0140-6736(06)68970-8. [DOI] [PubMed] [Google Scholar]
- 4.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–1252. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 5.Ladiges W, Wanagat J, Preston B, Loeb L, Rabinovitch P. A mitochondrial view of aging, reactive oxygen species and metastatic cancer. Aging Cell. 2010;9:462–465. doi: 10.1111/j.1474-9726.2010.00579.x. [DOI] [PubMed] [Google Scholar]
- 6.Zhang J, Liem DA, Mueller M, Wang Y, Zong C, Deng N, Vondriska TM, Korge P, Drews O, Maclellan WR, et al. Altered proteome biology of cardiac mitochondria under stress conditions. J. Proteome Res. 2008;7:2204–2214. doi: 10.1021/pr070371f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rabilloud T, Kieffer S, Procaccio V, Louwagie M, Courchesne PL, Patterson SD, Martinez P, Garin J, Lunardi J. Two-dimensional electrophoresis of human placental mitochondria and protein identification by mass spectrometry: toward a human mitochondrial proteome. Electrophoresis. 1998;19:1006–1014. doi: 10.1002/elps.1150190616. [DOI] [PubMed] [Google Scholar]
- 8.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carroll J, Fearnley IM, Walker JE. Definition of the mitochondrial proteome by measurement of molecular masses of membrane proteins. Proc. Natl Acad. Sci. USA. 2006;103:16170–16175. doi: 10.1073/pnas.0607719103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 11.Carroll J, Fearnley IM, Skehel JM, Runswick MJ, Shannon RJ, Hirst J, Walker JE. The post-translational modifications of the nuclear encoded subunits of complex I from bovine heart mitochondria. Mol. Cell. Proteomics. 2005;4:693–699. doi: 10.1074/mcp.M500014-MCP200. [DOI] [PubMed] [Google Scholar]
- 12.Chen R, Fearnley IM, Palmer DN, Walker JE. Lysine 43 is trimethylated in subunit c from bovine mitochondrial ATP synthase and in storage bodies associated with Batten disease. J. Biol. Chem. 2004;279:21883–21887. doi: 10.1074/jbc.M402074200. [DOI] [PubMed] [Google Scholar]
- 13.Hammer E, Goritzka M, Ameling S, Darm K, Steil L, Klingel K, Trimpert C, Herda LR, Dorr M, Kroemer HK, et al. Characterization of the human myocardial proteome in inflammatory dilated cardiomyopathy by label-free quantitative shotgun proteomics of heart biopsies. J. Proteome Res. 2011;10:2161–2171. doi: 10.1021/pr1008042. [DOI] [PubMed] [Google Scholar]
- 14.Hadsell DL, Olea W, Wei J, Fiorotto ML, Matsunami RK, Engler DA, Collier RJ. Developmental regulation of mitochondrial biogenesis and function in the mouse mammary gland during a prolonged lactation cycle. Physiol. Genomics. 2011;43:271–285. doi: 10.1152/physiolgenomics.00133.2010. [DOI] [PubMed] [Google Scholar]
- 15.Martens L, Chambers M, Sturm M, Kessner D, Levander F, Shofstahl J, Tang WH, Rompp A, Neumann S, Pizarro AD, et al. mzML—a community standard for mass spectrometry data. Mol. Cell. Proteomics. 2011;10:R110.000133. doi: 10.1074/mcp.R110.000133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vizcaino JA, Cote R, Reisinger F, Barsnes H, Foster JM, Rameseder J, Hermjakob H, Martens L. The Proteomics Identifications database: 2010 update. Nucleic Acids Res. 2010;38:D736–D742. doi: 10.1093/nar/gkp964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prokisch H, Andreoli C, Ahting U, Heiss K, Ruepp A, Scharfe C, Meitinger T. MitoP2: the mitochondrial proteome database—now including mouse data. Nucleic Acids Res. 2006;34:D705–D711. doi: 10.1093/nar/gkj127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Catalano D, Licciulli F, Turi A, Grillo G, Saccone C, D'Elia D. MitoRes: a resource of nuclear-encoded mitochondrial genes and their products in Metazoa. BMC Bioinformatics. 2006;7:36–42. doi: 10.1186/1471-2105-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Basu S, Bremer E, Zhou C, Bogenhagen DF. MiGenes: a searchable interspecies database of mitochondrial proteins curated using gene ontology annotation. Bioinformatics. 2006;22:485–492. doi: 10.1093/bioinformatics/btk009. [DOI] [PubMed] [Google Scholar]
- 20.Smith AC, Robinson AJ. MitoMiner, an integrated database for the storage and analysis of mitochondrial proteomics data. Mol. Cell. Proteomics. 2009;8:1324–1337. doi: 10.1074/mcp.M800373-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyne R, Smith R, Rutherford K, Wakeling M, Varley A, Guillier F, Janssens H, Ji W, McLaren P, North P, et al. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007;8:R129. doi: 10.1186/gb-2007-8-7-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen YA, Tripathi LP, Mizuguchi K. TargetMine, an integrated data warehouse for candidate gene prioritisation and target discovery. PLoS ONE. 2011;6:e17844. doi: 10.1371/journal.pone.0017844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Skrzypek MS, Hirschman J. Using the Saccharomyces Genome Database (SGD) for analysis of genomic information. Curr. Protoc. Bioinformatics. 2011;35:1–23. doi: 10.1002/0471250953.bi0120s35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smith AC, Robinson AJ. A metabolic model of the mitochondrion and its use in modelling diseases of the tricarboxylic acid cycle. BMC Syst. Biol. 2011;5:102–214. doi: 10.1186/1752-0509-5-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giege P, Heazlewood JL, Roessner-Tunali U, Millar AH, Fernie AR, Leaver CJ, Sweetlove LJ. Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell. 2003;15:2140–2151. doi: 10.1105/tpc.012500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hajek P, Chomyn A, Attardi G. Identification of a novel mitochondrial complex containing mitofusin 2 and stomatin-like protein 2. J. Biol. Chem. 2007;282:5670–5681. doi: 10.1074/jbc.M608168200. [DOI] [PubMed] [Google Scholar]
- 30.Scharfe C, Zaccaria P, Hoertnagel K, Jaksch M, Klopstock T, Lill R, Prokisch H, Gerbitz KD, Mewes HW, Meitinger T. MITOP: database for mitochondria-related proteins, genes and diseases. Nucleic Acids Res. 1999;27:153–155. doi: 10.1093/nar/27.1.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arnold R, Rattei T, Tischler P, Truong MD, Stumpflen V, Mewes W. SIMAP—the similarity matrix of proteins. Bioinformatics. 2005;21(Suppl. 2):ii42–ii46. doi: 10.1093/bioinformatics/bti1107. [DOI] [PubMed] [Google Scholar]