Abstract
Gene Expression Atlas (http://www.ebi.ac.uk/gxa) is an added-value database providing information about gene expression in different cell types, organism parts, developmental stages, disease states, sample treatments and other biological/experimental conditions. The content of this database derives from curation, re-annotation and statistical analysis of selected data from the ArrayExpress Archive and the European Nucleotide Archive. A simple interface allows the user to query for differential gene expression either by gene names or attributes or by biological conditions, e.g. diseases, organism parts or cell types. Since our previous report we made 20 monthly releases and, as of Release 11.08 (August 2011), the database supports 19 species, which contains expression data measured for 19 014 biological conditions in 136 551 assays from 5598 independent studies.
INTRODUCTION
Two years ago, the European Bioinformatics Institute (EBI) launched a new database called the Gene Expression Atlas (the Atlas). The Atlas is a value-added database for querying differential gene expression across tissues, cell types and cell lines under various biological conditions, including developmental stages, physiological states, phenotypes and disease states, for multiple organisms. Data in the Atlas come directly from the ArrayExpress Archive of Functional Genomics Experiments, including data imported from GEO (1). We also now include data on microRNA expression as well as next-generation sequencing RNA-Seq data from the European Nucleotide Archive [ENA (2)].
Data sets imported in the Atlas are curated: microarray probes and quantified RNA-seq transcripts are mapped to the latest Ensembl genome builds (3), while sample attributes are systematized and mapped to the Experimental Factor Ontology (EFO) (4). Automatic statistical computations are performed, providing P-values and t-statistics linking each gene to each experimental condition for all studies. The Atlas query interface allows querying on gene and sample attributes, with an advanced interface for complex queries. A number of visual user interface improvements have been made since the last release, and a completely new interactive online training course is now available at http://www.ebi.ac.uk/training.
Gene Expression Atlas can now be installed locally; all content and source code is provided freely without restrictions and without requirements to register.
As of September 2011, the EBI's Gene Expression Atlas contains data for over 370 000 genes from nearly 6000 different independent studies, including more than 136 000 samples representing nearly 20 000 different biological conditions. Nineteen different species, including human and model organisms, are included. Overall, this represents a nearly 6-fold data volume increase compared with results reported when Atlas was launched (5). The database is updated monthly and is continuing to grow constantly, both in terms of curated data sets, downloads and hit rates.
RESULTS
Data and curation improvements
Global map of human gene expression
Work by Lukk et al. (6) on a large meta-analysis of human gene expression, consisting of re-processing and detailed re-analysis of nearly 6000 microarray samples performed on the Affymetrix U133A GeneChip™ platform has been loaded and integrated into the Atlas. The Functional Genomics group has now produced two more data sets of this nature, including a global mouse data re-analysis (7) and an order-of-magnitude expansion of the human Affymetrix data set (unpublished data). These new data sets will be made available in the Atlas in the course of 2012 as well.
MicroRNA curation
As planned in our previous report (5), we have focused on microRNA data curation in the Atlas. We re-annotate all microRNA chip designs in order to avoid incorrect or incomplete annotations of probes and incorrect probe-matching between different platforms. All probes are matched, by exact sequence comparison, to the miRBase database latest version (8) and are re-annotated with miRBase identifiers. This also allows us to ensure that different platforms are maximally comparable in the Atlas. Original manufacturers’ probe identifiers are then replaced with matched miRBase identifiers that are used uniformly in the Atlas.
RNA-Seq data processing
Early in 2011, we published our R-based pipeline, ArrayExpressHTS, for processing RNA-seq data sets (9). We integrated this pipeline in the Atlas. All new submissions with RNA-seq data into ArrayExpress Archive and European Nucleotide Archive are put through ArrayExpressHTS. At the moment, we are able to process automatically most RNA-seq experiments on human, mouse and fruit fly samples. From 45 human, 32 mouse and 64 fly RNA-Seq data sets in ArrayExpress in August 2011, 14 experiments have been processed and loaded into Atlas, with 20 more to be loaded before 2012. The pipeline uses bowtie (10) to align the reads and cufflinks (11) to quantify transcript isoform expression levels.
Automation: mapping to EFO with Zooma
Zooma is a bespoke application designed to automate the problem of associating sample or assay annotations submitted by data providers to ontology classes in EFO and other ontologies. This process involves mapping the annotations, which consist of a type (e.g. disease) and a value (e.g. leukemia), to the appropriate ontology class (in this case, EFO_0000565). Zooma automates this process by searching for previously mapped instances of the same annotation in the Atlas, exact text matches to EFO, and also performs a search against the BioPortal (12) and OLS (13) web services using OntoCAT (14). It uses a simple ranking-based heuristic to evaluate the optimal match and automatically writes any new mappings to the Atlas database prior to each new data release.
Auto-updating genome annotations
Ensembl BioMart (15) provides detailed annotations for genomic elements, as well as regularly updated mappings for probes of most microarray designs from major platforms, e.g. Affymetrix and Illumina, to reference Ensembl genome builds. We developed a subsystem within Atlas that automatically checks the BioMart Central Portal for new data releases and updates Atlas annotations. This subsystem also enables the Atlas to support alternative probe mappings and genomic annotations, e.g. custom Affymetrix CDFs from the Bioconductor project (16), or NCBI genome build annotations.
Improving Atlas statistics
The core statistical engine in the Atlas remains the same: making use of the Bioconductor package limma in combination with structured curation of sample annotations to identify experimental variables and automatically compute per-factor contrasts. New to the Atlas is the reporting of computed t-statistics together with P-values and sample sizes on experiment pages and via API access routes.
User-interface features
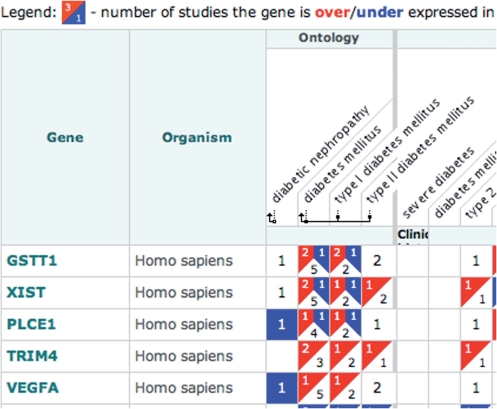
Non-differentially expressed genes
The Atlas allows searching for non-differentially expressed genes, i.e. those with multiple testing-adjusted P values above the significance threshold of 0.05 in simultaneous t-test comparisons with global factor means. These ‘non-differential expression’ results can be searched in the same way as others: by gene and sample attributes from the Atlas homepage, specifying ‘non-d.e.’ or ‘up/down/non-d.e.’ in the query dropdown. Whereas over- and under-expression results appear on red and blue backgrounds, respectively, non-differential expression results are on white background (Figure 1).
Figure 1.
Example of non-differential expression results display.
Advanced interface features
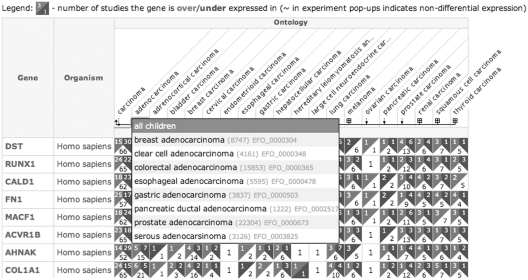
We have introduced the ability to filter results by specifying the number of experiments in which a result is found, e.g. one can search for all genes that appear as over-, under- or non-differentially expressed in leukemia samples in at least five experiments. Integration of Experimental Factor Ontology sample attribute mappings has been improved: a more compact ontology tree is displayed for large queries, with functionality to expand collapsed parts of the hierarchy and to traverse upward. For instance, a search for ‘carcinoma’ shows results aggregated for top-level ontology entries such as ‘adenocarcinoma’ and ‘melanoma’, which in turn can be further expanded. Also, one can navigate up from ‘carcinoma’ to show results for other cancer types (Figure 2).
Figure 2.
Example of compact ontology display for carcinoma results.
Genome browser integration
Our RNA-Seq pipeline ArrayExpressHTS saves short read alignments in as BAM files. These files are incorporated into the Atlas and are integrated into Atlas Experiment Pages: for every transcript isoform identified by the pipeline, a link is provided to the Ensembl Genome Browser, where aligned reads are displayed as one of the tracks. Upon new releases of the pipeline, its underlying tools and reference genome builds, the alignments are rebuilt.
Anatomograms and mappings to the vertebrate bridging ontology
We have developed several diagrammatic vector representations of human, mouse and fruit fly, and propagated anatomical terms from the EFO into these images. This allows highlighting and marking up the images according to expression profiles. These anatomograms, with highlighted tissues, are displayed on individual gene pages in the Atlas, and are integrated in EBI-wide search. The Vertebrate Bridging Ontology (VBO) is an ontology of homologies between anatomical structures across vertebrates, developed in the Functional Genomics group (17). Cross-species homologous structure annotations were imported into the EFO and are used to enhance condition searches in the Atlas, expanding multi-organism queries along these axes.
Integration with GeneSigDB
GeneSigDB, Gene Signature Database is an expert-curated database of fully traceable, standardized, annotated gene signature from published research (18). We have collaborated closely with GeneSigDB and have imported the latest (GeneSigDB v4) set of signatures into the Atlas, enabling users to search not only by individual gene or gene attribute, but also by signature. For example, the famous van ‘t Veer breast cancer signature (19) is represented by ID 11823860-Figure2 in GeneSigDB, and one can see the expression profiles of these genes across all of Atlas by entering this identifier into the gene search box on the home page.
Atlas infrastructure developments
Standalone Atlas
Starting from August 2010, we made the Atlas into a standalone, installable software system for managing transcriptomics data sets. Every month as we make public releases of the EBI Gene Expression Atlas website, simultaneously, we release the Atlas data and software on the associated website (http://github.com/gxa/gxa), with release notes, download and installation instructions. The standalone Atlas is built with open-source components and requires an Oracle database.
Standalone Atlas software provides a comprehensive administration interface, which supports loading and unloading data from the Atlas and performing various maintenance tasks. The Atlas look and feel and some aspects of its behavior can be adjusted via UI templates and options in the Configuration tab.
Distributed Atlas
A separate version of Atlas called Distributed Atlas has been released, providing online query federation among multiple Atlas deployments and integration of results with conflict resolution. Distributed Atlas uses a system of simple, powerful rules for integrating distributed query results over multiple semantically aligned Atlas instances within a single Atlas interface, with natural extension and integration points.
We separated the Atlas into client and server parts, allowing the client to communicate with multiple Atlas servers, federating gene and condition queries simultaneously. Two core rules were introduced to control possible query result conflicts:
First Not Null: when assembling results from multiple servers, the first non-empty result is used. This rule is used to integrate ontology- and experiment-related queries.
Aggregation: multiple results of the same kind are combined according to a pre-defined statistical integration strategy. For over/under expression counts, results can be summed. Results with statistical significance measurements are combined using meta-analytical strategies, e.g. using Fisher's method.
The Distributed Atlas user interface looks nearly identical to the public EBI Atlas, except that the client can be configured to communicate with multiple servers and result pages include integrated results as well as subsections of query results from individual servers. The software is available at http://www.ebi.ac.uk/fg/gxa-distributed.
FUTURE DIRECTIONS
As Atlas data have grown >6-fold in the last year and half, we are seeing that for a growing number of experiments our statistical approach may not be best suited. We have been developing alternative methods of statistical analysis for complex multifactorial experiments and will work on releasing these into production in 2012.
While in prior Atlas releases we have focused on processed, normalized data as provided by original authors, we are gradually switching to using raw data where possible and reprocessing these data for best standardization and comparability. For microRNA data sets, specifically, we intend to combine data by platform and by organism, identify common microRNAs and appropriate low-signal intensity thresholds and renormalize the data using cyclic lowness or quantile normalization methods (20).
Lower costs and technological barriers have made possible two comparatively new kinds of experiments: large meta-analysis experiments, e.g. E-MTAB-62, the global map of human gene expression (6), and large reference experiments such as the Illumina Body Map 2.0 data set. These experiments call for different statistics and visualization approaches entirely, and work is underway to make this possible.
We have made good progress this year with RNA-Seq data processing and integration, but it is clear that we have only scratched the surface of what is possible. Our goal overall is to focus on NGS data in 2012. In particular, we plan to process more high-throughput experiments from ArrayExpress and the ENA and improve how we handle other types of sequencing experiments, such as small non-coding RNA-Seq and ChIP-Seq data.
FUNDING
The project was funded by the European Molecular Biology Laboratory (EMBL), SLING, SYBARIS, GEUVADIS and GEN2PHEN grants from the European Commission, grant #BB/G022755/1 from the BBSRC, and the MAGE grant from the NHGRI. Funding for open access charge: EMBL.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
In addition to the invaluable help from the entire Functional Genomics Team, led by Ugis Sarkans, we would like to thank particularly Ekaterina Pilicheva for help with gene re-annotation and Nikolay Kolesnikov for help with ArrayExpress Archive interface. Many thanks to all our students, postdocs and visitors, especially Daniel Gusenleitner (DFCI), Markus Schroeder (DFCI) and Rodrigo Santamaria Vicente (U. of Salamanca). We are grateful to the Enright Group at the EBI, particularly Stijn van Dongen and Cei Abreu-Goodger. We would like to acknowledge the contribution of David Dean and Ketan Patel from Pfizer.
REFERENCES
- 1.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets—10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leinonen R, Akhtar R, Birney E, Bower L, Cerdeno-Tárraga A, Cheng Y, Cleland I, Faruque N, Goodgame N, Gibson R, et al. The European Nucleotide Archive. Nucleic Acids Res. 2011;39:D28–D31. doi: 10.1093/nar/gkq967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Malone J, Holloway E, Adamusiak T, Kapushesky M, Zheng J, Kolesnikov N, Zhukova A, Brazma A, Parkinson H. Modeling sample variables with an Experimental Factor Ontology. Bioinformatics. 2010;26:1112–1118. doi: 10.1093/bioinformatics/btq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kapushesky M, Emam I, Holloway E, Kurnosov P, Zorin A, Malone J, Rustici G, Williams E, Parkinson H, Brazma A. Gene expression atlas at the European bioinformatics institute. Nucleic Acids Res. 2010;38:D690–D698. doi: 10.1093/nar/gkp936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lukk M, Kapushesky M, Nikkilä J, Parkinson H, Goncalves A, Huber W, Ukkonen E, Brazma A. A global map of human gene expression. Nat. Biotechnol. 2010;28:322–324. doi: 10.1038/nbt0410-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng-Bradley X, Rung J, Parkinson H, Brazma A. Large scale comparison of global gene expression patterns in human and mouse. Genome Biol. 2010;11:R124. doi: 10.1186/gb-2010-11-12-r124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goncalves A, Tikhonov A, Brazma A, Kapushesky M. A pipeline for RNA-seq data processing and quality assessment. Bioinformatics. 2011;27:867–869. doi: 10.1093/bioinformatics/btr012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- 12.Whetzel PL, Noy NF, Shah NH, Alexander PR, Nyulas C, Tudorache T, Musen MA. BioPortal: enhanced functionality via new Web services from the National Center for Biomedical Ontology to access and use ontologies in software applications. Nucleic Acids Res. 2011;39:W541–545. doi: 10.1093/nar/gkr469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Côté R, Reisinger F, Martens L, Barsnes H, Vizcaino JA, Hermjakob H. The Ontology Lookup Service: bigger and better. Nucleic Acids Res. 2010;38:W155–160. doi: 10.1093/nar/gkq331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adamusiak T, Burdett T, Kurbatova N, Joeri van der Velde K, Abeygunawardena N, Antonakaki D, Kapushesky M, Parkinson H, Swertz MA. OntoCAT—simple ontology search and integration in Java, R and REST/JavaScript. BMC Bioinformatics. 2011;12:218. doi: 10.1186/1471-2105-12-218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guberman JM, Ai J, Arnaiz O, Baran J, Blake A, Baldock R, Chelala C, Croft D, Cros A, Cutts RJ, et al. BioMart Central Portal: an open database network for the biological community. Database. 2011;2011:bar041. doi: 10.1093/database/bar041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Travillian R, Malone J, Pang C, Hancock J, Holland PWH, Schofield P, Parkinson H. The Vertebrate Bridging Ontology (VBO) ISMB/ECCB Proceedings. 2011 Bio-Ontologies SIG. [Google Scholar]
- 18.Culhane AC, Schwarzl T, Sultana R, Picard KC, Picard SC, Lu TH, Franklin KR, French SJ, Papenhausen G, Correll M, et al. GeneSigDB—a curated database of gene expression signatures. Nucleic Acids Res. 2010;38:D716–725. doi: 10.1093/nar/gkp1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van 't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AAM, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 20.Yauk CL, Rowan-Carroll A, Stead JD, Williams A. Cross-platform analysis of global microRNA expression technologies. BMC Genomics. 2010;11:330. doi: 10.1186/1471-2164-11-330. [DOI] [PMC free article] [PubMed] [Google Scholar]