Abstract
FlyRNAi (http://www.flyrnai.org), the database and website of the Drosophila RNAi Screening Center (DRSC) at Harvard Medical School, serves a dual role, tracking both production of reagents for RNA interference (RNAi) screening in Drosophila cells and RNAi screen results. The database and website is used as a platform for community availability of protocols, tools, and other resources useful to researchers planning, conducting, analyzing or interpreting the results of Drosophila RNAi screens. Based on our own experience and user feedback, we have made several changes. Specifically, we have restructured the database to accommodate new types of reagents; added information about new RNAi libraries and other reagents; updated the user interface and website; and added new tools of use to the Drosophila community and others. Overall, the result is a more useful, flexible and comprehensive website and database.
INTRODUCTION
RNA interference (RNAi) has become a method-of-choice for interrogating gene function at genome-wide scale (1). Among the most popular RNAi screening approaches is high-throughput screening of Drosophila cultured cells, an approach that has already led to new insights into a wide variety of cellular processes. To perform genome-wide screens in Drosophila cells requires a library of gene-specific screening reagents (i.e. double-stranded RNAs or dsRNAs) targeting the full set of approximately 14 600 Drosophila genes, as well as all of the equipment, data management and data analysis tools necessary for performing and interpreting the results of high-throughput cell-based assays. The Drosophila RNAi Screening Center (DRSC) was established in 2003 to provide a full-genome Drosophila dsRNA library and screening platform, enabling the community to perform genome-wide screens in Drosophila cells. Since then, the DRSC has provided libraries and screen support for a large number of projects by researchers from many institutions. Management of information about DRSC reagents, assay plates and experimental results presents a significant challenge.
The DRSC database, FlyRNAi (www.flyrnai.org), was initially designed around gene-specific primers used to amplify dsRNAs for screening, and has subsequently grown to track information about all stages of dsRNA production and RNAi screening [see Figure 1 and (2)]. RNAi screens at the DRSC are performed in 384-well micro-well plates, which are typically screened in duplicate. The type of biological processes examined; the form, number and characterization of phenotypes; and the choice among various whole-well or visual assay readouts vary from screen to screen. All of these factors influence the volume and type of data generated. Managing reagents and results in a single database has many advantages; for example, we can associate results with the full quality-analysis history of the reagents. In addition to storing in-house generated data, we also store information from other sources such as FlyBase (3), allowing us to display gene information alongside reagents and results. Additionally, we maintain a current list of Drosophila gene names, identifiers, symbols and synonyms, allowing us to provide intelligent and flexible searches. Moreover, we use our website not just as a platform for user interfaces with the database but also to provide protocols, software tools, links to other resources, and more, so that we can better communicate information to the community (Table 1).
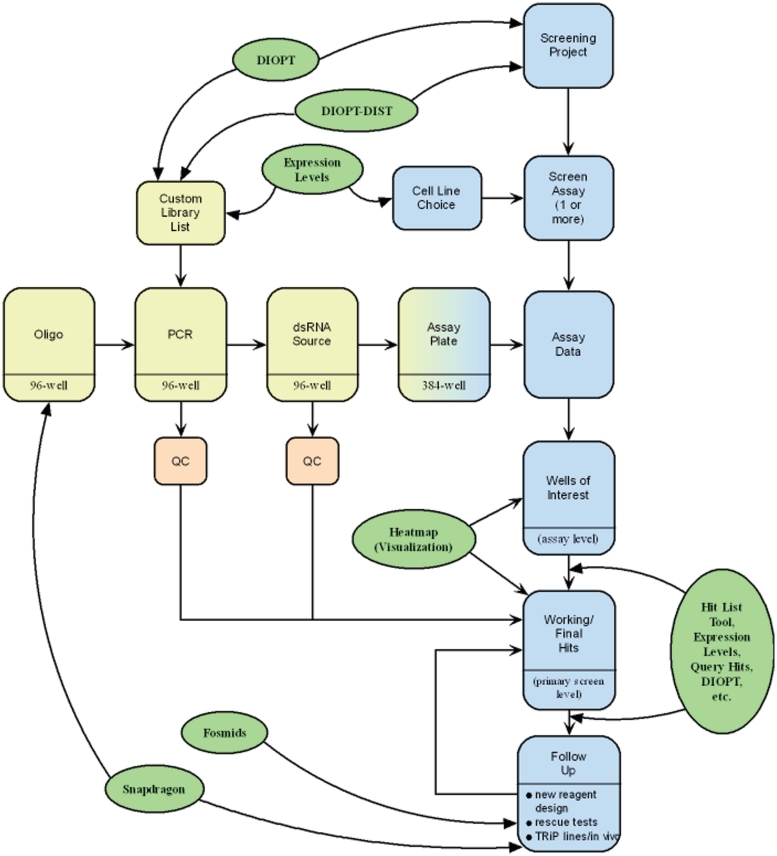
Figure 1.
The FlyRNAi Database Information Tracking Pipeline. The database, website and tools support design and tracking of double-stranded RNA (dsRNA) reagent production (horizontal workflow), as well as design and tracking of cell-based RNAi screen assays, screens and follow-up (vertical workflow). Capture of quality control (QC) analysis information associated with reagent production is a critical step, as is capture of screen results. Yellow shading, steps related to dsRNA production; blue shading, steps related to cell-based screening; green shading, software tools.
Table 1.
Common questions to DRSC informatics staff and corresponding database or other resources
| Question | DRSC resource | URL |
|---|---|---|
| Is Gene X expressed in Drosophila cultured cells? | Cell Line Expression Levels | http://www.flyrnai.org/cellexpress |
| Where can I design dsRNAs against Gene X? | SnapDragon | http://www.flyrnai.org/snapdragon |
| Where can I find if past DRSC screens identified Gene X? | Gene Lookup | http://www.flyrnai.org/genelookup |
| Where can I view DRSC reagents for Gene X? | Gene Lookup | http://www.flyrnai.org/genelookup |
| Where can I view protocols for cell-based RNAi? | RNAi Protocols Page | http://www.flyrnai.org/DRSC-PRR.html |
| Have similar screens been performed at the DRSC? | Screen Summary Table | http://www.flyrnai.org/screensummary |
| How can I filter screen ‘hits’ based on expression data? | Cell Line Expression Levels | http://www.flyrnai.org/cellexpress |
| Where can I upload and view my own plate-based data? | Public Heat Map Tool | http://www.flyrnai.org/heatmap |
| Have genes identified in my screen been conserved? | DIOPT | http://www.flyrnai.org/diopt |
| Are orthologs of the genes I found linked to disease? | DIOPT-DIST | http://www.flyrnai.org/diopt-dist |
| Where can I access information about in vivo RNAi fly stocks? | TRiP Pages | http://www.flyrnai.org/TRiP-HOME.html |
| How do I find genomic fragments for RNAi rescue? | RNAi Rescue | http://www.flyrnai.org/RNAi-rescue |
| Where can I access information on published screens? | Publications Page | http://www.flyrnai.org/DRSC-PRY.html |
| Where can I view all public screens and access data? | Screen Summary Table | http://www.flyrnai.org/screensummary |
| How can I download all public DRSC screen data? | Power User: Link to All Hits | http://www.flyrnai.org/DRSC-TOO.html |
IMPROVEMENTS TO THE FlyRNAi DATABASE ORGANIZATION
The underlying database structure has been altered since our previous publication (2) to accommodate tracking other types of reagents (e.g. in vivo RNAi fly stocks). Specifically, instead of storing information primarily about dsRNAs, we now store information about ‘reagents’ that can be associated with specific reagent types (e.g. dsRNA, UAS-miRNA, or fly stock). This has allowed us to accommodate new reagents within the existing tracking infrastructure. The database is implemented in MySQL on redundant servers hosted by the Harvard Medical School Research Information Technology Group. The interface is presented as a collection of CGI scripts, primarily written in Perl and Javascript. Batch scripts, primarily written in Perl, C and Java, handle background data collection and processing. FlyBase sequence information is used for off-target effects (OTEs) prediction (see below).
IMPROVEMENTS AND ADDITIONS TO RNAi REAGENT LIBRARIES
FlyRNAi uses up-to-date gene information to calculate the risk of sequence-specific OTEs. As our understanding of the underlying cases of OTEs has improved, so, too, has our ability to help prevent them through changes to reagent design. Shortly after our previous database publication (2), our analysis of OTEs was updated to check for 19 bp matches and display information about CAN or CAR repeats (4–6). The set of dsRNAs included in the full-genome library also underwent a major update to reduce the chance for OTEs and the collection is perpetually updated to reflect updated gene annotations at FlyBase. Since our previous database publication (2), 6449 dsRNAs have been removed and 7572 newly designed dsRNAs have been added, improving coverage and quality of the library (7). To further increase confidence in screen results at the gene level, we have introduced a follow-up library of dsRNAs with independent designs as compared with the set of dsRNAs in the full-genome screening library. We have also added bioinformatically defined smaller libraries targeting kinases and phosphatases (DRSC-KP); transcription factors and related proteins (DRSC-TF); ubiquitin pathway-associated proteins (NYU-DRSC UBIQ); and transmembrane domain-containing proteins (NYU-DRSC TM; see http://www.flyrnai.org/DRSC-SUB.html). Furthermore, we added information about Transgenic RNAi Project (TRiP) fly stocks for in vivo RNAi (8); reagents for miRNA or protein over-expression (see http://www.flyrnai.org/DRSC-OEX.html); and fosmids for cross-species rescue (9).
IMPROVED ACCESS TO REAGENT INFORMATION AND SCREEN RESULTS
We provide several routes for search and view of reagents and screen results (Table 1). The recently updated Gene Lookup (http://www.flyrnai.org/genelookup) allows users to view information online about cell-based or in vivo RNAi reagents, other types of reagents, screen results, etc. corresponding to a given query gene. Screen Summary (http://www.flyrnai.org/screensummary) facilitates view and download of data from all public cell-based RNAi screen datasets in tab-delimited text format. The Publications web pages list publications resulting from screens done at the DRSC or using DRSC reagents, organized by topic (http://www.flyrnai.org/DRSC-PTO.html) or year (http://www.flyrnai.org/DRSC-PRY.html), as well as our own publications (http://www.flyrnai.org/DRSC-PDR.html). As applicable, citations are linked to the corresponding PDF file, PubMed citation, FlyRNAi hits list, Supplemental data, and/or PubChem entry. Full data for DRSC reagents and results can be accessed from the Power User section of our tools page (http://www.flyrnai.org/DRSC-TOO.html). The power user section includes a tool for viewing or downloading a list of DRSC dsRNA designs in FASTA format, and a link to a tab-delimited table that shows in which screens each dsRNA was tested and/or was a hit. DRSC data has already enabled several meta-studies impacting our understanding of reagent design, screen design and interpretation, and specific biological topics (4,10–15).
INTERACTION WITH OTHER RESOURCES
Several external resources also facilitate search and view of FlyRNAi data. We deposited DRSC reagent information into NCBI PubChem Probe and Sequence, and we are uploading public screen data into PubChem BioAssay (16). The data can be searched and accessed at PubChem. In addition, PubChem records for specific screens are linked from our Publications and Screen Summary pages. Additionally, DRSC reagent information is linked from gene pages at FlyBase (3) and DRSC screen results are included in FlyMine (17). Moreover, FLIGHT (18) and GenomeRNAi (19) support search and display of DRSC and other RNAi screen datasets. Gene annotation undergoes constant update at FlyBase (3). As a result, gene identifiers such as FBgn numbers, CG numbers and gene symbol/names are retired or added over time. To facilitate accurate searches at FlyRNAi and keep our gene records up-to-date, we have implemented automatic algorithms for weekly upload of FlyBase changes.
NEW OR UPDATED SOFTWARE TOOLS FOR CELL-BASED RNAi
The DRSC has developed a number of software tools since our previous database publication, in particular for the design and analysis of Drosophila RNAi reagents and results. Several of these are of specific use for Drosophila RNAi screening or follow-up studies. SnapDragon (http://www.flyrnai.org/snapdragon) facilitates the design of primer pairs that will amplify regions predicted to confer effective and on-target RNAi knockdown. When given a DNA sequence or a gene identifier (e.g. FBgn, CG or gene symbol) via the user interface, SnapDragon searches for sequence regions suitable for dsRNA design (i.e. free of matches to genes other than the intended target) using an index-based algorithm developed in house and returns one or more pairs of primers suitable for PCR amplification of a template for in vitro transcription. The user can then rely on the default settings or define an OTE sequence match length to be considered (16–50 bp). Users also have the option to only consider regions shared by all isoforms (i.e. to target all forms or specific isoforms), as well as to define a maximum and minimum length for the dsRNA design. Cell Line Expression (http://www.flyrnai.org/cellexpress) allows users to check for evidence for expression of a given gene or set of genes in various Drosophila cultured cell lines, which is useful for assay development and filtering screen data (11,20). Fosmid Rescue (http://www.flyrnai.org/RNAi-rescue) allows users to identify genomic fragments in related Drosophila species likely to be useful for cross-species RNAi rescue (9).
ADDITIONAL NEW SOFTWARE TOOLS
Additional tools developed by our group or others are available on the website and are useful not just to screeners but also to other researchers. Public Heat Map (http://www.flyrnai.org/heatmap) is a free online statistical analysis and visualization tool for plate-based datasets. Any researcher can use the tool, including those without access to commercial software applications or licenses necessary for using many similar tools. DIOPT (http://www.flyrnai.org/diopt) combines results from a number of ortholog prediction tools published by previous groups, facilitating rapid identification of putative orthologs in human and model organism genomes. (21). The related tool DIOPT-DIST (http://www.flyrnai.org/diopt-dist) identifies putative human orthologs of model system genes based on DIOPT results and displays information about diseases or traits associated with those human genes (21). MinoTar (http://www.flyrnai.org/cgi-bin/DRSC_MinoTar.pl) is a look-up tool which provides data about microRNA coding region targets based on analysis performed by Bonnie Berger’s group at MIT (22). Lastly, we provide access to DRSC RNAi reagents and results as described above, as well as links to related external resources (http://www.flyrnai.org/DRSC-LIN.html).
FUTURE DIRECTIONS
Over the years, the FlyRNAi database of the DRSC has evolved from tracking information about a single, first-generation reagent library for cell-based Drosophila RNAi and a few full-genome screens to tracking information pertaining to an expanded number and variety of reagents and results. Our website additionally provides access to information about conducting screens and a number of different software tools useful to screeners and others. Based on user feedback we have identified two additional areas where further improvement would be beneficial. These are (1) collection and display of full raw or analyzed numerical datasets for all full-genome and smaller screens conducted using DRSC reagents, and (2), storage and public availability of image files associated with microscopy-based screens. To achieve these goals we require input and cooperation from other researchers and informatics experts. Storage and availability of image files currently presents a technical hurdle (i.e. as individual image-based screen datasets can be several terabytes in size) faced not just by our group but by the screening community more generally (23). As mentioned above, several other groups facilitate search of DRSC datasets in various contexts (3,16–19). Thus, we anticipate that in the next few years, our efforts regarding the database per se are best focused on continued tracking of reagent production, managing screen data during acquisition and analysis, and exporting raw and analyzed datasets to public repositories. Annotation of reagent quality, such as through annotation of in vivo RNAi fly stocks with phenotypic and/or validation information, is another area in which we plan to make significant additions. As a community-focused group, we welcome input from all researchers on how to define and prioritize further changes to the DRSC's FlyRNAi database, website and suite of tools.
FUNDING
National Institutes of Health [grant number R01 GM067761]; N.P. is a Howard Hughes Medical Institute Investigator; Dana Farber/Harvard Cancer Center and National Cancer Institute [grant number NIH 5 P30 CA06516] to S.E.M. Funding for open access charge: National Institutes of Health [grant number R01 GM067761].
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank the many researchers who have performed screens at the DRSC, used our libraries, and/or used our database and tools, for providing critical feedback that guides our development of the database, website and software tools. The authors extend our thanks also to R. DasGupta and colleagues at the NYU RNAi Core for collaboration on bioinformatically defined sub-library collections. Additionally the authors thank Liz Perkins and Laura Holderbaum for helpful discussions regarding the DRSC database and website.
REFERENCES
- 1.Mohr S, Bakal C, Perrimon N. Genomic screening with RNAi: results and challenges. Annu. Rev. Biochem. 2010;79:37–64. doi: 10.1146/annurev-biochem-060408-092949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Flockhart I, Booker M, Kiger A, Boutros M, Armknecht S, Ramadan N, Richardson K, Xu A, Perrimon N, Mathey-Prevot B. FlyRNAi: the Drosophila RNAi screening center database. Nucleic Acids Res. 2006;34:D489–D494. doi: 10.1093/nar/gkj114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, Perrimon N, Mathey-Prevot B. Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat. Methods. 2006;3:833–838. doi: 10.1038/nmeth935. [DOI] [PubMed] [Google Scholar]
- 5.Ma Y, Creanga A, Lum L, Beachy PA. Prevalence of off-target effects in Drosophila RNA interference screens. Nature. 2006;443:359–363. doi: 10.1038/nature05179. [DOI] [PubMed] [Google Scholar]
- 6.Moffat J, Reiling JH, Sabatini DM. Off-target effects associated with long dsRNAs in Drosophila RNAi screens. Trends Pharmacol. Sci. 2007;28:149–151. doi: 10.1016/j.tips.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Horn T, Sandmann T, Boutros M. Design and evaluation of genome-wide libraries for RNA interference screens. Genome Biol. 2010;11:R61. doi: 10.1186/gb-2010-11-6-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ni JQ, Zhou R, Czech B, Liu LP, Holderbaum L, Yang-Zhou D, Shim HS, Tao R, Handler D, Karpowicz P, et al. A genome-scale shRNA resource for transgenic RNAi in Drosophila. Nat. Methods. 2011;8:405–407. doi: 10.1038/nmeth.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kondo S, Booker M, Perrimon N. Cross-species RNAi rescue platform in Drosophila melanogaster. Genetics. 2009;183:1165–1173. doi: 10.1534/genetics.109.106567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wiles AM, Ravi D, Bhavani S, Bishop AJ. An analysis of normalization methods for Drosophila RNAi genomic screens and development of a robust validation scheme. J. Biomol. Screen. 2008;13:777–784. doi: 10.1177/1087057108323125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Booker M, Samsonova AA, Kwon Y, Flockhart I, Mohr SE, Perrimon N. False negative rates in Drosophila cell-based RNAi screens: a case study. BMC Genomics. 2011;12:50. doi: 10.1186/1471-2164-12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DasGupta R, Nybakken K, Booker M, Mathey-Prevot B, Gonsalves F, Changkakoty B, Perrimon N. A case study of the reproducibility of transcriptional reporter cell-based RNAi screens in Drosophila. Genome Biol. 2007;8:R203. doi: 10.1186/gb-2007-8-9-r203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan H, Venkatesan K, Beaver JE, Klitgord N, Yildirim MA, Hao T, Hill DE, Cusick ME, Perrimon N, Roth FP, et al. A genome-wide gene function prediction resource for Drosophila melanogaster. PLoS One. 2010;5:e12139. doi: 10.1371/journal.pone.0012139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Muller P, Boutros M, Zeidler MP. Identification of JAK/STAT pathway regulators–insights from RNAi screens. Semin. Cell Dev. Biol. 2008;19:360–369. doi: 10.1016/j.semcdb.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Tu Z, Sun F. A network-based integrative approach to prioritize reliable hits from multiple genome-wide RNAi screens in Drosophila. BMC Genomics. 2009;10:220. doi: 10.1186/1471-2164-10-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Bolton E, Dracheva S, Karapetyan K, Shoemaker BA, Suzek TO, Wang J, Xiao J, Zhang J, Bryant SH. An overview of the PubChem BioAssay resource. Nucleic Acids Res. 2010;38:D255–D266. doi: 10.1093/nar/gkp965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyne R, Smith R, Rutherford K, Wakeling M, Varley A, Guillier F, Janssens H, Ji W, McLaren P, North P, et al. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007;8:R129. doi: 10.1186/gb-2007-8-7-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sims D, Bursteinas B, Jain E, Gao Q, Baum B, Zvelebil M. The FLIGHT Drosophila RNAi database: 2010 update. Fly. 2010;4:344–348. doi: 10.4161/fly.4.4.13303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilsdorf M, Horn T, Arziman Z, Pelz O, Kiner E, Boutros M. GenomeRNAi: a database for cell-based RNAi phenotypes. 2009 update. Nucleic Acids Res. 2010;38:D448–D452. doi: 10.1093/nar/gkp1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cherbas L, Willingham A, Zhang D, Yang L, Zou Y, Eads BD, Carlson JW, Landolin JM, Kapranov P, Dumais J, et al. The transcriptional diversity of 25 Drosophila cell lines. Genome Res. 2011;21:301–314. doi: 10.1101/gr.112961.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Y, Flockhart I, Vinayagam A, Bergwitz C, Berger B, Perrimon N, Mohr SE. An integrative approach to ortholog prediction for disease-focused and other functional studies. BMC Bioinformatics. 2011;12:357. doi: 10.1186/1471-2105-12-357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schnall-Levin M, Zhao Y, Perrimon N, Berger B. Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3'UTRs. Proc. Natl Acad. Sci. USA. 2010;107:15751–15756. doi: 10.1073/pnas.1006172107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang M, Instrell R, Saunders B, Berven H, Howell M. Tales from an academic RNAi screening facility; FAQs. Brief. Funct. Genomics. 2011;10:227–237. doi: 10.1093/bfgp/elr016. [DOI] [PubMed] [Google Scholar]