Abstract
The Encyclopedia of DNA Elements (ENCODE) Consortium is entering its 5th year of production-level effort generating high-quality whole-genome functional annotations of the human genome. The past year has brought the ENCODE compendium of functional elements to critical mass, with a diverse set of 27 biochemical assays now covering 200 distinct human cell types. Within the mouse genome, which has been under study by ENCODE groups for the past 2 years, 37 cell types have been assayed. Over 2000 individual experiments have been completed and submitted to the Data Coordination Center for public use. UCSC makes this data available on the quality-reviewed public Genome Browser (http://genome.ucsc.edu) and on an early-access Preview Browser (http://genome-preview.ucsc.edu). Visual browsing, data mining and download of raw and processed data files are all supported. An ENCODE portal (http://encodeproject.org) provides specialized tools and information about the ENCODE data sets.
INTRODUCTION
Following a 4-year pilot phase aimed at identifying functional elements in selected regions comprising 1% of the human genome (1–2), the Encyclopedia of DNA Elements (ENCODE) project expanded to a whole-genome scope in September 2007 (3). Now beginning the 5th year of its mission to explore the ‘dark matter’ of the human genome, ENCODE contains an unprecedented range of diverse genomic data. With additional NHGRI support from the federal American Recovery and Reinvestment Act of 2009, complementary study of the mouse genome by ENCODE groups is underway. Previous manuscripts in this publication (4–5) have described the overall project and how the ENCODE Data Coordination Center at the University of California, Santa Cruz works with ENCODE labs worldwide to import their data sets, supporting documentation and metadata, and to make the data accessible to the broader biomedical community. A companion paper in this issue, ‘The UCSC Genome Browser database: Extensions and updates 2012’, provides background information about the UCSC Genome Browser database and infrastructure (6–7) that underlies ENCODE support at UCSC. This article focuses on ENCODE data and access tools introduced in 2011.
NEW DATA AVAILABILITY
With the increasing flood of ENCODE data production and the inevitable delays during quality review of submitted data, there arose a demand for an early access site for pre-reviewed data. In February 2011 UCSC deployed a Preview Browser (http://genome-preview.ucsc.edu) to serve this function. The Preview Browser is a weekly mirror of the UCSC internal development server. Data is made available on this site with the caveat that it is subject to change and has undergone only cursory review.
The year 2011 marked the first release of Mouse ENCODE data to the public. The Mouse ENCODE project serves to complement the Human ENCODE project, furthering the understanding of human functional elements through comparative analysis. Mouse experiments aim to be analogous to those in the Human ENCODE project, as well as address experimental conditions not feasible in human, such as genetic knockouts and embryonic tissues. On the public UCSC server this year, we released mouse ENCODE results identifying transcription factor binding sites and histone marks by ChIP-seq, regions of transcription by RNA-seq, and open chromatin by DNase-seq. Data sets representing these functional elements in additional cell and tissue types, developmental stages and treatment conditions are hosted on the Preview Browser in preparation for quality review.
During the previous year the ENCODE Consortium undertook a coordinated effort to remap and re-analyze all data sets from the initial phase of data production (referenced to the March 2006 NCBI36/hg18 human genome assembly) to the current standard human reference genome (February 2009 GRCh37/hg19). At the same time, data file formats were transitioned to newer standards [BAM (8) and bigWig/bigBed (9)]. The hg19 versions of all ENCODE data are now available at UCSC.
The ENCODE human data repertoire expanded with the addition of 90 additional cell types (for a total of 235) and 57 additional transcription factor and histone modifications assayed (for a total of 177). Table 1 shows how data sets are distributed across the most intensively studied cell types.
Table 1.
ENCODE experiments in the human genome are focused on a set of cell lines selected by the Consortium for intensive study
| Cell lines | Karyo | Tissue | Description | Datasets |
|---|---|---|---|---|
| Tier 1 | ||||
| GM12878 | Normal | Blood | Lymphoblastoid | 166 |
| H1-hESC | Normal | Embryonic stem | Embryonic stem | 89 |
| K562 | Cancer | Blood | Leukemia | 253 |
| Tier 2 existing | ||||
| HeLa-S3 | Cancer | Uterine cervix | Cervical carcinoma | 118 |
| HepG2 | Cancer | Liver | Liver carcinoma | 135 |
| HUVEC | Normal | Umbilical endothelium | Umbilical vein endothelial | 54 |
| Tier 2 added in 2011 | ||||
| A549 | Cancer | Lung | Lung carcinoma | 35 |
| CD14+ | Normal | Blood | Monocyte | 2 |
| IMR90 | Normal | Lung | Lung fibroblast | 3 |
| MCF-7 | Cancer | Breast | Breast carcinoma | 33 |
| SK-N-SH | Cancer | Brain | Neuroblastoma | 25 |
| Tier 3 | ||||
| 219 additional | 928 total |
All assays are performed in Tier 1; Tier 2 cell types are designated as the next level of priority.
New types of data available provided by UCSC this year include chromatin interaction maps by 5C (10) and ChIA-PET (11), nucleosome positioning by Mnase-seq, deep-sequenced DNAseI hypersensitive sites, SNP data for cell lines assayed for copy number variation, and three additional assays of RNA-binding proteins.
The Gencode Gene set (12) has been updated to version 7 (May 2011). This version features 25% more manual annotation, along with improved organization and display of the annotation to make it more intuitive to biologists. Details pages for the annotated elements show evidence used to build the annotation such as UniProt (13), CCDS (14), RefSeq (15) and GenBank (16) sequences, and PubMed IDs for published experimental evidence.
A notable addition this year was the first proteomics data within ENCODE. The new proteogenomics track features mappings of tandem mass spectrometry peptide profiles to the genome (17), complementing transcriptional evidence from RNA-based assays. The scope of DNA-binding site identification has been expanded by the introduction of epitope tagging of proteins (18) where antibodies suitable for chromatin immunoprecipitation are not available.
This year also featured two new integrative tracks provided by ENCODE analysts: a segmentation of the genome into 15 states based on the chromatin state in 9 cell lines (19) and a synthesis of multiple sources of the open chromatin state in 7 cell lines. As integrative analysis is now a major focus of Consortium efforts, more analysis tracks integrating function across primary data sets are expected in the coming year.
Table 2 lists the number of data sets currently available for each ENCODE data type.
Table 2.
ENCODE encompasses a diverse set of assays
| Data type | No. of experiments |
|---|---|
| Chromatin Interactions | |
| 5C | 4 |
| ChIA-PET | 6 |
| DNA methylation | |
| Methyl array | 63 |
| Methyl RRBS | 93 |
| Methyl-seq | 20 |
| Histone modifications | |
| ChIP-seq | 221 |
| ChIP-seq (MOUSE) | 28 |
| Open chromatin | |
| DNase-DGF | 19 |
| DNase-seq | 135 |
| Dnase-seq (MOUSE) | 27 |
| FAIRE-seq | 27 |
| RNA profiling | |
| CAGE | 45 |
| Exon array | 120 |
| RNA-chip | 26 |
| RNA-PET | 22 |
| RNA-seq | 151 |
| RNA-seq (MOUSE) | 27 |
| Transcription factor binding sites | |
| Epitope-tag ChIP-seq | 12 |
| ChIP-seq | 745 |
| ChIP-seq (MOUSE) | 92 |
| Other | |
| Bi-directional promoters | 1 |
| DNA cleavage | 1 |
| DNA-PET | 6 |
| Gencode genes | 5 |
| Genotype | 64 |
| Negative regulatory elements | 2 |
| Nucleosome positioning | 2 |
| Proteogenomics | 5 |
| RNA binding proteins | 49 |
| Short read mapability | 13 |
Descriptive overviews along with methods and references are included in the description page that accompanies all datasets.
Validation data sets to accompany primary data sets are now available for open chromatin and transcription factor binding site experiments.
NEW ACCESS INFORMATION AND TOOLS
The ENCODE portal (http://encodeproject.org), which is the centralized resource for accessing the information and tools described in this section, was extensively upgraded this year. An entire section for Mouse ENCODE resources has been added. The experimental guidelines and data standards developed by the ENCODE Consortium this year for a broad range of whole-genome assays (RNA-seq, ChIP-seq, DNase-seq, DNA methylation assays) are hosted on a dedicated portal Data Standards page, along with platform characterization summaries and references.
A key resource for learning about ENCODE data is the OpenHelix ENCODE tutorial (openhelix.com/ENCODE), a free Online resource released in November 2010. This tutorial provides an overview of the ENCODE project, summarizes the types of data available through ENCODE, and details methods for accessing ENCODE data via the UCSC Genome Browser. The tutorial, and accompanying instructional material, is free to the public and is sponsored by the DCC. Other resources for learning about ENCODE data usage can be found on the new ENCODE portal Education and Outreach page.
The DCC devoted considerable engineering effort this year to developing tools to enable users to easily locate data of interest within the overwhelming set of ENCODE data tracks and subtracks. For an overview of ENCODE data, the DCC now provides a Data Summary page on the ENCODE portal. This page includes a spreadsheet in multiple formats itemizing ENCODE experiments by lab, data type, cell type and other experimental variables.
The premier methods for locating ENCODE data are the new Track Search and File Search tools, available from the ENCODE portal and Genome Browser web pages. Both of these tools allow free-text searching by keyword, coupled with an advanced search feature that provides selectable lists of terms from the ENCODE controlled vocabulary (described below) to guide the search. Multiple terms can be applied in both ‘and’ and ‘or’ combinations. For example, in a single advanced search, a user can locate tracks showing evidence of the enhancer-associated histone modifications ‘H3K4me1’ and ‘H3K27Ac’ in either ‘NHLF’ or ‘IMR90’ lung cell lines. The Track Search tool is described more fully in the companion Genome Browser paper in this issue. The File Search tool locates downloadable files for analysis across the full range of ENCODE data sets, and the related track File Downloads tool (available from the track configuration page) selects files within a single track. The Downloads page of many ENCODE tracks include hundreds and even thousands of files. Using controlled vocabulary terms relevant for each experiment set, the files are now listed in a sortable and filterable table.
In a related effort, the DCC this year implemented an accessioning scheme to group related files and tracks within logical experiments. These accessions make it easier to relate associated files and provide a short, stable identifier for citations. Each experiment groups a set of data from a single providing laboratory for a single assay in a single cell type and set of experimental conditions. All replicates and levels of data (raw sequence files and mappings to multiple genome assemblies, processed data such as peak calls or putative transcription isoforms) associated with a single logical experiment are assigned the same accession. The DCC accession is visible everywhere metadata for a track or file appears. As of this writing, ENCODE comprises 1861 experiments in human and 174 experiments in mouse.
The ENCODE DCC controlled vocabulary (CV) is a mechanism for associating metadata with ENCODE experiments. Metadata terms are added as needed, and the metadata controlled vocabularies have been expanded this year for both human and mouse. There are currently 23 metadata controlled vocabularies. The largest vocabularies are ‘Antibody’ (199 terms) and ‘Cell Line’ (235 human and 34 mouse cell types). The CV has received extensive curation and quality review this year to ensure completeness and eliminate duplicate and confusing terms. This effort has led to a more informative set of metadata associated with each track, including links to term descriptions and supporting documents. Two specific areas where the CV was improved are the cell type karyotype and lineage terms. The karyotype term has been simplified to describe cell lines that are derived from normal or cancerous tissues. At present 72 cell lines have been annotated as normal and 47 cell lines as cancerous. The lineage term has been used to describe the progenitor tissue type from which the source tissue type has differentiated. The values ectoderm, endoderm, mesoderm and inner cell mass are associated with 36, 45, 90 and 12 cell lines, respectively.
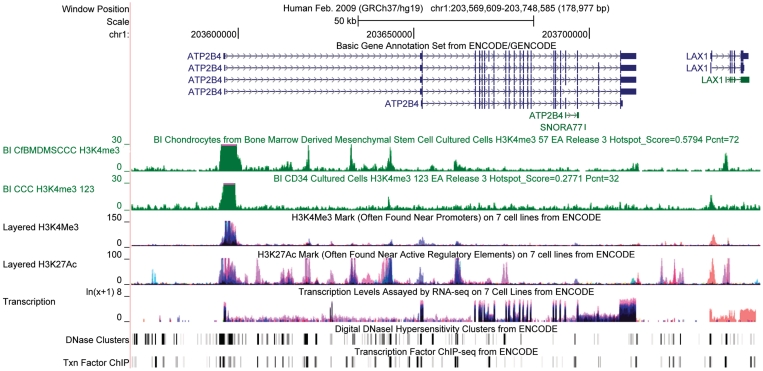
A new Genome Browser feature, Data Hubs, supports display of off-site annotations alongside ENCODE data. The first publicly provided hub presents the Roadmap Epigenomics (20) catalog of data sets, enabling close comparison of the voluminous and complementary results from these two consortia. Figure 1 shows a Genome Browser screen showcasing ENCODE and Roadmap Epigenomics data together. For more information about the Data Hubs feature, see the Genome Browser update in this issue.
Figure 1.
ENCODE data displayed in the UCSC Genome Browser together with two annotations from the Roadmap Epigenomics Release III data hub. The genomic region contains two protein coding genes, plasma membrane calcium ATPase 4a (ATP2B4) and lymphocyte transmembrane adaptor 1 isoform a (LAX1). The GENCODE Genes track shows multiple variant transcripts for both genes as well as a snoRNA in the region. The Epigenomics Roadmap tracks just below the GENCODE track show H3K4me3, a histone mark associated with promoters, in two cell lines not assayed by the ENCODE project. These tracks show support for the short, non-coding form of LAX1 in mesenchymal stem cells, and support for the longer isoform in CD34 cells, based on peaks at likely promoter regions. The next three tracks are transparent overlays from seven cell lines assayed by the ENCODE project showing the H3K4me3 mark again, the H3K27Ac mark associated with active regulatory regions, and a log plot of transcription levels in the same cell lines. The histone marks and pattern of transcription show coordinated, cell-type-specific activity; the ATP2B4 gene is most active in NHEK (purple) and K562 (blue) cells, while LAX1 is most active in GM12878 (orange) cells. The DNAse and Transcription Factor ChIP-seq clusters shown in the last two tracks summarize data from a much wider range of cell lines and indicate a large number of regulatory regions. Additional details for these annotations are available on click-through.
The DCC effort to pass quality-reviewed ENCODE data to the NCBI Gene Expression Omnibus (GEO) (21) and Short Read Archive (SRA) as an auxiliary data repository has made considerable progress in the past year. Since September 2010 we have accessioned 916 GEO Samples, in 15 GEO Series in human and mouse over 3 assemblies (NCBI36/hg18, GRCh37/hg19 and NCBI37/mm9). To further organize the data and facilitate access, NCBI BioProjects have been created for ENCODE.
ACCESSING ENCODE DATA
ENCODE data availability is summarized in Tables 1–3 in this article, and a comprehensive spreadsheet of experiments available from the ENCODE portal Data Summary page. Data sets marked as having ‘released’ status are available from the UCSC public server, http://genome.ucsc.edu. Data sets marked ‘displayed’ or ‘reviewing’ can be viewed at the preview site, http://genome-preview.ucsc.edu. Human ENCODE data is available on two human genome assemblies: NCBI36/hg18 and GRCh37/hg19. Mouse ENCODE data is provided on the mouse NCBI37/mm9 assembly.
Table 3.
ENCODE vital statistics, as of September 2011
| Category | Human | Mouse |
|---|---|---|
| Experiments | 1861 | 174 |
| Assay types | 29 | 3 |
| Cell and tissue types | 235 | 34 |
| ChIP antibodies | 179 | 30 |
All ENCODE data is subject to the Consortium data policy, which places some restrictions on use for the 9 months after the data becomes publicly available. Restriction timestamps for all experiments are prominently displayed on the track and file information pages, as well as being listed on the Data Summary spreadsheet. The data policy is described in detail on the Data Policy page of the ENCODE portal.
ENCODE GEO submissions are listed on the GEO ENCODE summary page, http://www.ncbi.nlm.nih.gov/geo/info/ENCODE.html. ENCODE has been assigned NCBI BioProject identifiers to further organize the data: PRJNA30707 for Human ENCODE (with the subproject PRJNA63443 for Production phase data) and PRJNA50617 for Mouse ENCODE. Data in each project is further categorized as epigenomic, functional genomics or transcriptome.
FUTURE WORK
Highlights of the fifth and final year of this phase of the ENCODE project will be the fruition of ongoing integrative analysis efforts and dissemination of the results to the DCC, promotion of an additional collection of cell types for Consortium-wide use (see Table 1), expansion of the transcription factor space based on community input, selected new experiment types in high-value areas such as single-cell assays, and additional validation data sets. The Mouse ENCODE project makes its future experiment planning publicly available on the ENCODE portal Mouse Data Summary page.
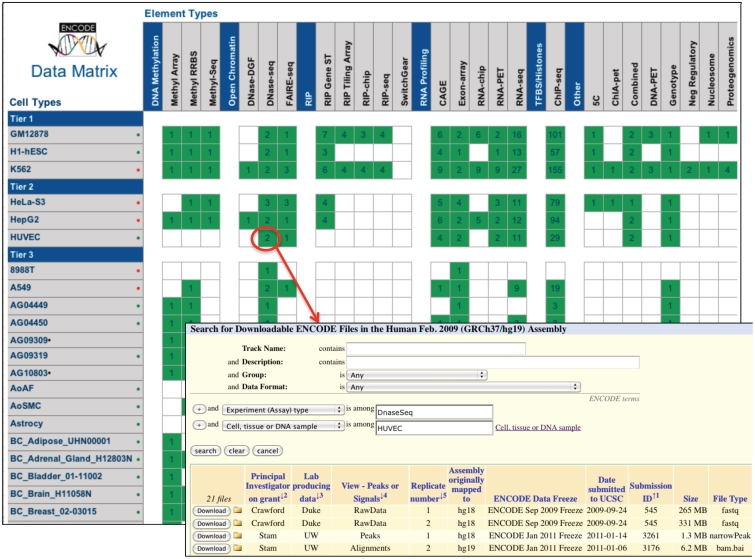
DCC efforts during the 5th year will continue to emphasize data accessibility and usability. We have scheduled an update to the OpenHelix ENCODE tutorial, and are contracting for the design and production of ENCODE Quick Reference Cards. A new Data Matrix web application on the portal will provide table and matrix-based display of the breadth of ENCODE data, with click-through access to search results for selected experiments. Figure 2 shows a snapshot as of September 2011. We expect to release this feature on the ENCODE portal by late fall 2011.
Figure 2.
Data matrix display and selection of files for download. This feature will be linked to the ENCODE portal, and will navigate to the Advanced Search features of File and Track Search.
In upcoming months we expect the new data hub feature will be adopted more widely, and we anticipate that the larger ENCODE production groups will migrate to hub-based hosting of much of their data. The DCC will be implementing search across data hubs to further enhance the synergy between UCSC-hosted and remote data sources.
CONTACT INFORMATION
General questions and feedback about ENCODE data at UCSC should be directed to the ENCODE mailing list: encode@soe.ucsc.edu. General questions about the Genome Browser should be sent to the UCSC browser mailing list: genome@soe.ucsc.edu. Specific questions about details of laboratory methods or data interpretation should be directed to the ENCODE laboratory contact listed on the description page for that data set. We announce releases of new ENCODE data via the ENCODE announcement list. To subscribe, visit https://lists.soe.ucsc.edu/mailman/listinfo/encode-announce.
FUNDING
National Human Genome Research Institute (grants 5P41HG002371-10 and 3P41HG002371-10S1 to the UCSC Center for Genomic Science, and grant 5U41HG004568-04 and 3U41HG004568-03S1 to the UCSC ENCODE Data Coordination Center); Howard Hughes Medical Institute (to D.H.). Funding for the open access charge: The Howard Hughes Medical Institute.
Conflict of interest statement. The authors receive royalties from the sale of UCSC Genome Browser source code licenses to commercial entities.
ACKNOWLEDGEMENTS
We would like to thank the systems administration staff at the Center for Biomolecular Science and Engineering: Jorge Garcia, Erich Weiler, Victoria Lin and Gary Moro, for their dedication and support, keeping high-volume ENCODE data flowing to our public site while assuring our servers are reliable and available. Thanks also to members of the ENCODE Consortium for providing these valuable data sets.
REFERENCES
- 1.ENCODE Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- 2.The ENCODE Project Consortium. Birney E, Stamatoyannopoulos J, Dutta A, Guigó R, Gingeras T, Margulies E, Weng Z, Snyder M, Dermitzakis E, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Myers RM, Stamatoyannopoulos J, Snyder M, Dunham I, Hardison RC, Bernstein BE, Gingeras TR, Kent WJ, Birney E, Wold B, et al. A user’s guide to the encyclopedia of DNA elements (ENCODE) PLoS Biol. 2011;9:e1001046. doi: 10.1371/journal.pbio.1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenbloom KR, Dreszer TR, Pheasant M, Barber GP, Meyer LR, Pohl A, Raney BJ, Wang T, Hinrichs AS, Zweig AS, et al. ENCODE whole-genome data in the UCSC Genome Browser. Nucleic Acids Res. 2010;38:D620–D625. doi: 10.1093/nar/gkp961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Raney BJ, Cline MS, Rosenbloom KR, Dreszer TR, Learned K, Barber GP, Meyer LR, Sloan CA, Malladi VS, Roskin KM, et al. ENCODE whole-genome data in the UCSC genome browser (2011 update) Nucleic Acids Res. 2011;39:D871–D875. doi: 10.1093/nar/gkq1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fujita PA, Rhead B, Zweig AS, Hinrichs AS, Karolchik D, Cline MS, Goldman M, Barber GP, Clawson H, Coelho A, et al. The UCSC Genome Browser database: update 2011. Nucleic Acids Res. 2011;39:D876–D882. doi: 10.1093/nar/gkq963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li G, Fullwood MJ, Xu H, Mulawadi FH, Velkov S, Vega V, Ariyaratne PN, Mohamed YB, Ooi HS, Tennakoon C, et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 2010;11:R22. doi: 10.1186/gb-2010-11-2-r22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, et al. GENCODE: producing a reference annotation for ENCODE. Genome Biol. 2006;7(Suppl. 1):S41–S49. doi: 10.1186/gb-2006-7-s1-s4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2011;39:D32–D37. doi: 10.1093/nar/gkq1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krug K, Nahnsen S, Macek B. Mass spectrometry at the interface of proteomics and genomics. Mol Biosyst. 2011;7:284–291. doi: 10.1039/c0mb00168f. [DOI] [PubMed] [Google Scholar]
- 18.Poser I, Sarov M, Hutchins JR, Heriche JK, Toyoda Y, Pozniakovsky A, Weigl D, Nitzsche A, Hegemann B, Bird AW, et al. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat Methods. 2008;5:409–415. doi: 10.1038/nmeth.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature. 2011;473:43–49. doi: 10.1038/nature09906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bernstein BE, Stamatoyannopoulos JA, Costello JF, Ren B, Milosavljevic A, Meissner A, Kellis M, Marra MA, Beaudet AL, Ecker JR, et al. The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 2010;28:1045–1048. doi: 10.1038/nbt1010-1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]