Abstract
Capturing how the structures of interacting partners evolved at their binding interfaces is a fundamental issue for understanding interactomes evolution. In that scope, the InterEvol database was designed for exploring 3D structures of homologous interfaces of protein complexes. For every chain forming a complex in the protein data bank (PDB), close and remote structural interologs were identified providing essential snapshots for studying interfaces evolution. The database provides tools to retrieve and visualize these structures. In addition, pre-computed multiple sequence alignments of most likely interologs retrieved from a wide range of species can be downloaded to enrich the analysis. The database can be queried either directly by pdb code or keyword but also from the sequence of one or two partners. Interologs multiple sequence alignments can also be recomputed online with tailored parameters using the InterEvolAlign facility. Last, an InterEvol PyMol plugin was developed to improve interactive exploration of structures versus sequence alignments at the interfaces of complexes. Based on a series of automatic methods to extract structural and sequence data, the database will be monthly updated. Structures coordinates and sequence alignments can be queried and downloaded from the InterEvol web interface at http://biodev.cea.fr/interevol/.
INTRODUCTION
Major insights into protein interaction networks have been brought through the physical mapping of protein interactions by a combination of high throughput techniques. Large databases such as Biogrid (1) and Intact (2) are now gathering several thousands of interactions for a number of model organisms. At a lower but significant rate, high resolution structures of protein complexes keep on expanding, providing a wealth of information for capturing the molecular logic underlying these interaction networks. Synergies, competitions, specificities can be best understood through the precise identification of residues contacting at the interface. Understanding how these interactions were preserved or altered through evolution remains an important challenge in both fields of network and structural biology (3–5). The important concept of interolog was introduced to define a conserved interaction between two binding partners (6) and, as an extension, when the structures of the corresponding complexes are known, they can be defined as ‘structural interologs’. The scope of the InterEvol database is to provide an integrated environment to explore coevolution processes in complexes of known structures. From the database, structural interologs between closely and distantly related homologs can be retrieved providing key insights into the fate of compensatory mutations within the structures of interfaces. Furthermore, an interactive PyMol plugin (The PyMol Molecular Graphics System, http://pymol.sourceforge.net) was developed to combine the structural exploration of interfaces with the targeted inspection of interologs multiple sequence alignments. Pre-calculated alignments are available in the InterEvol database and can also easily be recomputed using different options from the InterEvolAlign tool.
Large-scale analyses of the structural interactomes were tackled by a number of studies (7–9) and databases, including PSIBASE (10), PIBASE (11), PRISM (12), 3D complex (13), SCOPPI (14), PRINT (15), SCOWLP (16), JAIL (17), 3D interologs (18), IBIS (19), 3did (20) and ProtCID (21). Different strategies for clustering the interfaces were proposed depending on whether the entire chains (as in PRISM, IBIS) or the domains (as in SCOWLP, PIBASE, 3did, ProtCID) defined by either SCOP (22), CATH (23) or PFAM (24) were considered in the comparison process. A number of important insights for the understanding of interactomes were gained from these works. Thanks to the 3D-complex database (13), unanticipated distribution of symmetries among homo-oligomers revealed key rules in the evolution of protein assemblies (25). Providing more global perspectives about the structural organization of interactomes, the 3did database (20) clustered both domain–domain and domain–linear motifs complex structures, and provides a nicely interactive platform to travel through the networks of interactions made by a given domain superfamily. More focused on the details of the interfaces, databases such as SCOWLP are useful for grasping the types of physico-chemical contacts occurring at interfaces especially for water mediated interactions (26) while PRINT integrates predictions for hot spot residues (27).
Among the databases cited above several can provide useful insights into the evolution of interfaces, such as PRISM (12), IBIS (19), 3D interologs (18) and ProtCID (21). PRISM (12) database was among the first databases performing large-scale clustering of interface structures allowing for the extraction of similar interfaces with evolutionary relationships. IBIS (19) maps and infers interaction sites in proteins by inspecting the structures of protein complexes formed by homologous partners and, as PRISM, can be used to retrieve structural interologs. In 3D interologs, sequences from UniProt were blasted against a collection of structures of heterodimers offering the possibility to map the query sequence onto the structure of an interface and to align the sequences of interologs that matched the same template. ProtCID clustered and superimposed interacting proteins sharing the same PFAM architectures and generated a structural database of homologous interfaces. The primary goal of the database was to help discriminating crystal contacts from biological ones because biological interfaces tend to be conserved across different interologous structures. As an extension, ProtCID can also be used to explore the evolutionary properties of interologs provided they are not too distantly related. The InterEvol database pushes forward the exploitation of interology in two directions, not only it includes very remotely related interologs but also it combines structures and multiple sequence alignments of interologs so that they can be analysed in an interactive manner. Most of the numerous distant structural orthologs collected in InterEvol actually exhibit similar binding modes, significantly enhancing our perception of interfaces plasticity.
Interfaces coevolution bears intriguing features related to their conservation and their sequence versatility which greatly complicate their analyses. Based on a large-scale analysis, a majority of domain pairs forming intermolecular contacts were found to interact in the same way with identities as low as 30–40% (28). At the interface itself, core positions were shown to evolve more slowly than the rest of the surface (29,30) and the modular organization was found to be conserved among a set of homologous complexes (31). However, prediction of contacting residues from pairwise covariation analyses was found difficult to extract (32) unless a large number of sequences was available (33). Reasons for such paradox may be that interfaces did not coevolve in a residue pairwise manner but rather through compensatory changes distributed within micro-environments of several residues (34). Complex mechanisms involving molecular epistasis were recently proposed to explain how such versatility may arise (35,36).
To facilitate the analysis of such micro-environments, the InterEvol database implemented several tools to generate multiple sequence alignments and visualize specific columns of these alignments in a structural context. Hence, it can be used to pinpoint local plasticity at an interface and help uncovering the rules for interface coevolution. From a user perspective, the InterEvol database provides a number of applications besides large-scale coevolution studies. For modelling usage, users can take advantage of the HHsearch profile–profile comparison program (37) running on the server, submit one or two sequences and retrieve all the structures of complexes involving a close or distant homolog of their queries. Multiple sequence alignments of interologs which can be optimized by changing the parameters in the InterEvolAlign web interface can provide important information to understand the specificity of an interaction, the deleterious effect of a mutation or to guide the design of compensatory mutations. All these analyses are greatly facilitated through the interactive PyMol plugin that can easily be installed to dive into interface coevolution properties. These developments represent key steps in tackling the complexity of interface coevolution from the analysis of sequence alignments and in generating enhanced statistics about the details of interface physico-chemistry throughout evolution. We believe that the way structures and sequences are combined together within the InterEvol database will provide new insights into the evolutionary analyses of interactomes.
CLUSTERING CLOSE AND REMOTE STRUCTURAL INTEROLOGS
The InterEvol database is built combining the PDB (38) coordinate files and the information in the xml header files (38). A general flow chart of the processing step is provided in Supplementary Figure S1. In the case of X-ray structures, the biological unit was generated by applying the transformation provided in the <PDBx:pdbx_struct_assembly_gen> xml tag under for the assembly_id ‘1’. Biological units were either defined by author, software or both as specified in the <PDBx:details> xml tag. This information is useful to rely as much as possible on authors expertise and only if required on assembly prediction methods such as PISA (39). We found this preferable in the case of some large complexes [such as the proteasome (pdb:1fnt)] or small interfaces [such as Hsp90-Sgt1 complex (pdb:2jki)] for which PISA returned improper assemblies predictions. As regards NMR structures, the model defined by the tag <PDBx:pdbx_nmr_representativeCategory> <PDBx:conformer_id> was used. All the entries containing multiple chains after reconstruction were further considered. A number of steps was performed to prepare the dataset and allow proper comparisons between the chains. The sequences of the different PDB chains were extracted from the <PDBx:entity_poly entity_id= ‘ENTITY’> tag and mapped on the sequences of the swissprot database using Blastp (40). For a given PDB, homomeric or heteromeric character was assigned depending on whether one or different sequences were mapped, respectively. Absence of overlap between the chains matching on the same sequence indicated that the chains were fragments of the same protein and chains were not considered further. To facilitate subsequent mapping with their multiple sequence alignments, chains were renumbered and relabelled if required so that the first residue index of every chain is 1 and every subsequent residue index is incremented by 1 (no gaps in the index). So far, only chains containing more than 30 amino acids were considered so that protein–peptide complexes are not taken into account in InterEvol.
To cluster close and remote structural interologs, we applied a two-step procedure first reducing the redundancy below 70% sequence identity and then using profile–profile alignments together with structural superposition. Chains sharing more than 70% sequence identity over more than 70% of their length were clustered using Uclust (41) and gathered in the ‘CHAIN>70’ subdatabase. Complexes sharing the same composition of ‘CHAIN>70’ groups were defined as redundant and the complex of best resolution was set as ‘reference complex’. All the binary complexes contained in every ‘reference complex’ were further processed provided at least 10 different pairs of residues have at least one atomic contact (inter-atomic threshold distance set at 5 Å). Structural interologs above 70% sequence identity were defined for pairs of binary complexes AB and A′B′ made of the same chains in ‘CHAIN>70’. To ensure that similar interfaces be clustered together, an additional constraint was that the positions involved in the interface of A and B overlapped with more than 40% of the positions involved in the interface of A′ and B′, respectively. Further details justifying the choice of this threshold are provided in Supplementary Figure S2A.
To identify structural interologs below 70% sequence identity, including remotely related ones, the set of ‘CHAIN>70’ had to be further clustered to the superfamily level. The profile–profile comparison algorithm HHsearch is well suited for that purpose since it was calibrated against the SCOP database to detect superfamilies relationships between sequences with high sensitivity (37). For every ‘CHAIN>70’ referent chain, a sequence profile was generated using three iterations of Psi-blast (40) against the nr database. All-vs-all HHsearch comparisons were performed to group together ‘CHAIN>70’ referent chains matching with a probability higher than 90% (local alignment mode). To improve the specificity for short matches, superfamily assignment was further controlled by checking that both structures superimposed with Matras (42) with a fold similarity probability above 80%. The fold similarity probability was defined using the reliability score calculated by Matras (42) which was calibrated by all-vs-all comparison of protein domains in SCOP 1.59 database. Such combined use of both HHsearch and Matras scores was found optimal to retrieve remotely related domains while preventing false positive assignments. Structural interologs sharing <70% sequence identity were identified following the same procedure as described in the former paragraph except that we used the chains clustered at the superfamily level resulting in the ‘INTER<70’ database. For every pair of structural interologs AB and A′B′ in ‘INTER<70’, an interface RMSD (iRMSD) was computed inspiring from CAPRI metrics (43) with some variations to account for the fact that superimposed chains can be substantially different: residues having a difference in accessibility between the free and the bound states were defined as ‘interface residues’. Next, chains A and A′ were superimposed using Matras and ‘common interface residues for A and A′’ were defined as all the pairwise aligned positions involved in the interface of both AB and A′B′ complexes, respectively. The same calculation was repeated for chains B and B′ to define the ‘common interface residues for B and B′’. Two interface iRMSDs were computed using the coordinates of the backbone atoms, first, chains A and A′ were superimposed and an iRMSD_BB′ was computed between common interface residues of chains B and B′; second, chains B and B′ were superimposed and an iRMSD_AA′ between the common interface residues of chain A and A′ was calculated. The minimal value between both iRMSD_AA′ and iRMSD_BB′ was chosen as the representative iRMSD. In most cases, both iRMSD values lie in the same range. Typical exceptions are presented in Supplementary Figure S2B and illustrate why the choice of the minimal iRMSD was found a good compromise to represent the structural divergence between interfaces. Further details regarding the number of residues generally involved in the iRMSD calculation and the dependence between the iRMSD and this number of residues are provided in Supplementary Figure S3.
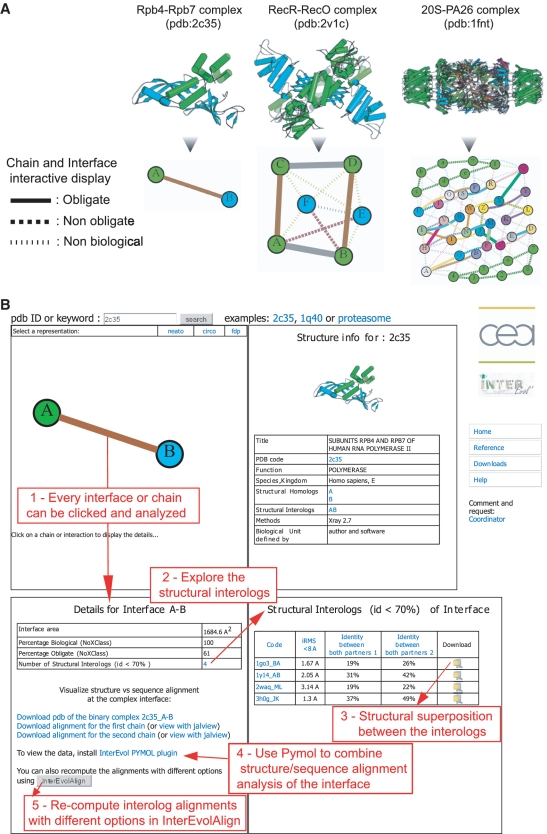
For every interface, we estimated the biological/non-biological and obligate/non-obligate characters relying on the NoXClass probabilities (44). This program was used with the two-stage support vector machine option trained with the three parameters, interface size, size ratio between interface and surface and amino acids composition. The NoXClass predictions are graphically represented by the type and thickness of the links connecting the chains in the networks of interactions displayed as in Figure 1A. From the InterEvol browser page, chains (nodes) and interfaces (edges) can be interactively clicked to display information panels about their respective homologs and interologs (Figure 1B). All in all, the InterEvol database contains nearly 12 000 non-redundant interfaces (below the 70% sequence identity threshold) predicted by NoXClass as biological interfaces (Table 1). They can be subdivided into 9309 homodimers and 2589 heterodimers. Focusing on the heterodimers set, 513 groups of structural interologs with sequence identity below 70% could be retrieved (distribution of the size of the groups is shown in Figure 2A). Certain groups exhibit very interesting features illustrated by the structural interologs of Mtr2-Mex67 heterodimer involved in nuclear transport (pdb: 1q40). Six structural interologs were identified with sequence identities for the pair of chains A–B ranging from 33–50% to 7–15% with corresponding iRMS ranging from 1.25 to 4.14 Å. Five out of the six complexes are also involved in nuclear transport while another is involved in the ubiquitination process. As a comparison, ProtCID (21) identified only one out of the six homologous interfaces likely due to the distant homology relationships between them. Querying at a large scale other databases such as ProtCID, IBIS or PRISM with all the interologs retrieved in InterEvol indicated that below 30% sequence identity, InterEvol interologs contained at least twice as many pairs of interologs (Supplementary Figure S4).
Figure 1.
(A) Three examples of network representation with clickable nodes and edges to navigate interactively through the InterEvol database. The biological and obligate properties of the interactions (edges) between chains (nodes) were predicted using the NoXClass (44) program with plain (obligate), dashed (non-obligate) or dotted lines (non-biological) as indicated in the figure. Thickness of the edges scales with the NoXClass probability (44) for an interaction to be obligate. Network representation was developed using the python library NetworkX. (B) Screen capture of the InterEvol browser page with five red caption boxes enumerating a typical search process from an edge request (step 1 which provides details about the corresponding interface) to the download of structural interologs superpositions and/or of interologs sequences alignments.
Table 1.
Statistics about the complexes, interfaces and structural interologs collected in InterEvol
| Number of complexes analysed | 33 472 |
| Homomers | 26 167 |
| Heteromers | 7305 |
| Number of non-redundant complexes (%id < 70%) | 12 095 |
| Number of non-redundant interfaces (%id < 70%) | 16 943 |
| Homodimers interfaces | 13 274 |
| Heterodimers interfaces | 3669 |
| Predicted as biological (NoXClass) | 11 898 |
| Homodimers predicted as obligate (NoXClass) | 7326 |
| Homodimers predicted as non-obligate (NoXClass) | 1983 |
| Heterodimers predicted as obligate (NoXClass) | 1148 |
| Heterodimers predicted as non-obligate (NoXClass) | 1441 |
| Number of interologs sequences alignments (more than 10 sequences) | 1230 |
| Heterodimers predicted as obligate (NoXClass) | 579 |
| Heterodimers predicted as non-obligate (NoXClass) | 651 |
| Number of groups of structural interologs (%id < 70%) | 1741 |
| Homodimers interfaces | 1129 |
| Heterodimers interfaces | 513 |
| Combination of homodimers and heterodimers | 99 |
Summary of the statistics of the number of complexes, interfaces and interologs collected and analysed in the initial version of the InterEvol database. Biological and obligate properties of the interfaces were predicted using the NoXClass method (44).
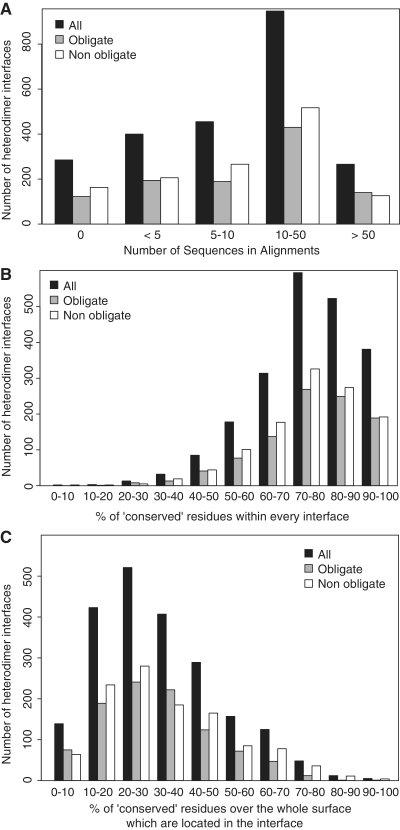
Figure 2.
(A) Histogram reporting the number of groups of structural interologs which contain from 2 to 10 different members. A vast majority of the groups is composed of a single pair of structural interologs underscoring the diversity of structures available in these groups. (B) Given a binary interaction between two chains A–B, the plot represents the interface RMSD (iRMSD) of this couple with its corresponding structural interologs noted A′–B′ versus the minimal percentage identity obtained from the structural alignment of A and A′ and from B and B′. Groups of structural interologs with more than 10 members were not represented to prevent that their large combinatorial pairwise comparisons bias the interpretation of the graph. The few points exhibiting >70% identity are due to chains which were not clustered together because they did not respect the coverage condition.
The overall relationships between sequence identities of the pairs and the iRMSD between both interfaces are shown in Figure 2B. Below 30% sequence identity, 172 interologs were identified with iRMSD below 4 Å providing a number of interesting cases with significant sequence variation but relatively similar structural binding mode. In an attempt to further define the probability to find two protein complexes in the same binding arrangements at a given percent identity, we analysed the proportion of interfaces pairs with a given iRMSD when their binding partners belong to the same CHAIN <70 group. The graph in Supplementary Figure S5A obtained following the methods described in Supplementary Method 1 shows that between 20% and 30% sequence identity, 58% of the interfaces have a iRMSD below 4 Å, while this value drops to 27% below 20% sequence identity. We also tried to check whether iRMSD between orthologous interfaces distributed differently from paralogous interfaces (Supplementary Figure S5B), using a classification performed empirically based on the organism to which each interface belongs and the function of the complex as described in the PDB entry (Supplementary Method 2 for details). While above 50% sequence identity, no significant differences could be noticed between the othologous and the paralogous groups, between 30% and 50% identity the distribution of iRMSD for the ortholog and the paralog groups exhibit significant differences (P = 1.85e-08 from non-parametrical Wilcoxon rank sum test with median iRMSD of 1.7 or 2.5 Å between orthologs and paralogs, respectively). Below 30%, the differences between both distributions were also significant with a P-value of 1.83e-05 and a median iRMSD value of 2.8 and 3.9 Å for orthologs and paralogs, respectively.
COUPLING SEQUENCE ALIGNMENTS AND INTERFACES STRUCTURES
Structural interologs provide explicit snapshots of how evolution effectively reshaped interfaces between related protein families. However, structural data are sparse with respect to the wealth of information available in sequences. To complement the structural information we have derived multiple sequence alignments for every non-redundant interface of the ‘reference complexes’. Our aim was to retrieve as many sequences as possible, exhibiting the highest diversity while restrained to the most likely orthologous sequences. In that way, we can more confidently assume, for every sequence in the alignment, that the selection pressure acted on similar positions to maintain a binding interface and that the evolutionary trace is not blurred by too remotely related sequences. Orthology detection is a difficult task tackled by a number of methods and database (45,46). Our ambition was to provide generic pre-computed interolog alignments with reasonable accuracy in the species retrieved and offer external users the possibility to increase the number of species by re-computing these alignments with alternative parameters and database through the InterEvolAlign web service. A general flow chart explaining the different processes is provided in Supplementary Figure S6.
Pre-computed alignments were processed using fully sequenced genomes to limit the inclusion of too many paralogs or spurious sequences. We used the database of entirely sequenced genomes provided in the OMA database, so-called ‘Entire genomes (OMA)’ (47). For every pair of sequences whose chains interact in the ‘reference complexes’ of the InterEvol database, two iterations of Psi-blast over the ‘Entire genomes (OMA)’ database were performed. To improve alignment coverage, matches selected after an iteration (e-value set at 10e-4) are extended to their full-length sequence (extension can be restricted as an option in InterEvolAlign), realigned with Muscle program (48) and alignment is resized to match the structural limits of the chains. Homologous sequences are kept in subsequent steps only if they respect two conditions, first they should share at least 35% sequence identity with any of the accepted matches and second they should align with the query with a coverage above 50%. After every Psi-blast iteration, the hit with lowest e-value is selected for each species and the corresponding sequence is kept in all subsequent iterations. Doing so, only one sequence per species is retrieved in the final alignment. The sequence profile calculated as an input for the next Psi-blast iteration only contains these best hits for each species aligned together. Once the alignments of both interacting partners are computed, sequences belonging to species common in both alignments are selected as interologs. Redundancy filter is applied to remove pairs of interologs only if the two sequences in the pair share more than 95% sequence identity with their respective homologs. An additional step was added to remove obvious non-orthologous sequences from pre-computed alignments (Supplementary Figure S7). For every sequence of the alignment (one per species), a single reciprocal blast was run on the database to count how often the other sequences of the alignment were indeed ranked as first hit. The ratio between the number of correct reciprocal best hits and the total number of species of the alignment retrieved was calculated and sequences exhibiting significantly lower ratios (less than two standard deviations below the mean ratio) were discarded in an iterative manner. Above a ratio of 0.7, sequences are not discarded anymore thus preventing that all the sequences be excluded while iterating. All the standard values for the parameters mentioned above can be tuned in the InterEvolAlign webtool to create tailored alignments.
The automatic protocol described above was found rather robust to the inclusion of spurious sequences in the alignments. For instance, a typical difficult case is that of the complex between Nas6 proteasome chaperone and Rpt3 domain, an AAA ATPase part of the 19S proteasome regulatory particle (PDB code: 2dzn). Rpt3 is closely related to the 5 AAA ATPase subunits forming the base of the proteasome (sharing more than 45% sequence identity with Rpt1-6 paralogs). Nas6 belongs to the widespread ankyrin superfamily very likely to retrieve homologs unrelated to Nas6 function. In the pre-computed alignment, more than 25 sequences were retrieved from yeast to human with a mean sequence identity of 34% to the query. Expert analysis of the alignment showed that no false positive was retrieved although Nas6 has no orthologs in insects. The reciprocal blast hit procedure cleaned up spurious homologs initially detected from insect genomes. From the InterEvolAlign webtool, it is also possible to reproduce the search with an additional iteration over the NCBI reference database which provide up to 69 sequences of aligned orthologs without any insect sequence. The InterEvolAlign interface is also useful to explore alternative thresholds and retrieve more homologous sequences. For instance, the complex between RecO and RecR was solved in Deinococcus Radiodurans in which RecO sequence diverged significantly (pdb code: 2v1c). Only three interologs could be retrieved using the standard thresholds in the pre-computed alignments. However, decreasing the minimal identity threshold to 25% instead of 35% helped retrieving up to 300 sequences of both partners aligned. Detailed inspection showed that they were all orthologs of RecO and RecR, underscoring the interest of tuning the parameters to improve alignment completion. Overall, the possibility to use either pre-computed alignments or the InterEvolAlign webtool alleviates much of the tedious efforts of aligning the most likely orthologs for a pair of interologs.
Overall, for more than 1200 interfaces of heteromers the pre-computed multiple sequence alignment contained more than 10 sequences with an average size of 47 sequences (Table 1 and Figure 3A). A global estimation for the rates of evolution for the positions involved at those interfaces could be obtained by using the Rate4Site algorithm (49). Mapping the estimated rates on the structure of the chains, we defined the ‘conserved’ positions as those exhibiting rates of evolution comprised in the first third of the rates computed over the whole sequence. As previously observed in other datasets (50,51), we confirmed that overall interfaces gather relatively conserved positions (Figure 3B) but that the conservation signal remains not specific enough to define unambiguously interaction patches since a given interface contains less than a third of the positions conserved over the whole surface (Figure 3C). Following that track the InterEvol database provides additional means to explore coevolution events for these slowly evolving positions as shown below.
Figure 3.
(A) Histogram reporting the number of sequences retrieved in the interologs multiple sequences alignments which were pre-calculated in the InterEvol database for every heteromeric binary interaction. (B) Evolutionary rates for every position of a chain were computed with the Rate4Site algorithm (49) for the heterodimer interologs alignments containing at least 10 sequences. These evolutionary rates were binned into nine classes of conservation and positions were considered ‘conserved’ when they belong to the three most conserved classes. The histogram reports the number of heterodimer interfaces containing a given percentage of ‘conserved’ positions. (C) The ‘conserved’ positions were identified as in (B). For every binary heterodimer, the ratio between the number of conserved positions at the interface and the total number of conserved positions exposed at the surface of the chain was calculated. For every range of ratio, the histogram represents the number of binary heteromers obtained.
STRUCTURAL INSIGHTS INTO INTERFACES COEVOLUTION
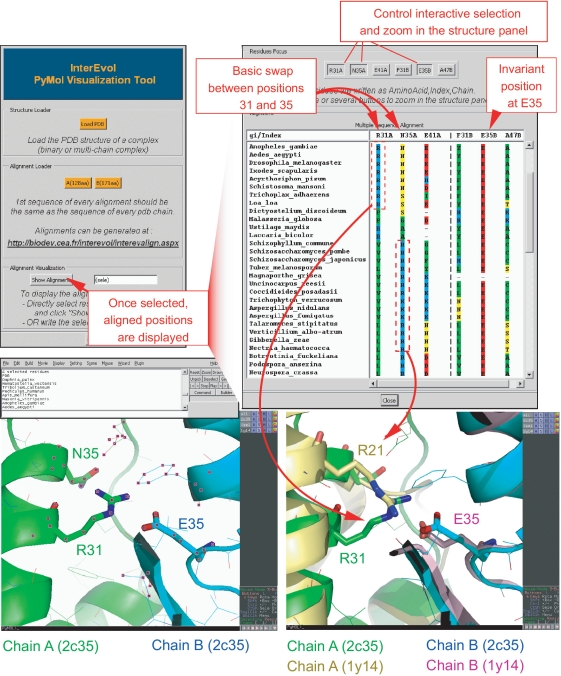
Evolutionary properties of interfaces are particularly difficult to analyse from sequence alignments because alignments of both partners have to be manipulated simultaneously and neighbouring positions at the interface are not necessarily contiguous in the alignments. To overcome this issue, we developed a visualization tool implemented as a plugin into the popular PyMol program. The plugin can read generic alignment fasta files together with a structure but is best optimized to process alignments generated with InterEvolAlign webtool since species names can be displayed. The InterEvol PyMol plugin can be very helpful in revealing compensatory changes at interfaces in a complex such as the conserved Rpb4-Rpb7 interaction for which InterEvol identified 4 structural interologs including one in Homo sapiens (pdb: 2c35) and another from Saccharomyces cerevisiae (pdb: 1y14).
After loading the structure of the interface available in the InterEvol database (chains were renumbered with respect to the original PDB) (pdb: 2c35) and the alignments in the InterEvol PyMol plugin, a user can select a subset of residues in PyMol and generate an alignment panel restricted to this set of residues (Figure 4). Let us focus on the buried salt bridge between R31 (R44 in original PDB) and E35 at the interface between Rpb4 (chain A) and Rpb7 (chain B), respectively. After selecting this pair together with neighbouring positions in the structure comprising N35 and E41 in chain A (corresponding to N48 and E54 in original PDB) and F31 and A47 in chain B, a specific alignment panel restricted to the positions pops up. Although E35 (in Rpb7) has been strictly conserved throughout evolution, its contacting residue R31 (in Rpb4) switches from basic to polar or even hydrophobic residue in fungi genomes. How the deleterious effect of this mutation was buffered in these species is suggested from the alignment of position N35, neighbouring R31, which swapped to basic when position R31 lost its basic character in a correlated manner. The interest of the InterEvol database is precisely to provide the structural solution to that question since the complex from S. cerevisiae (pdb:1y14) was identified as a structural interolog. Structural superposition between both human and yeast complex confirm the swapping behaviour of the basic position to maintain a buried salt-bridge at Rpb4–Rpb7 interface. Hence, analyzing the plasticity at the interface of a complex can be carried out in an interactive manner repeating the selection/alignment display cycle several times.
Figure 4.
Screen capture to illustrate the interest of the InterEvol PyMol plugin for diving into the structures of complex interfaces interactively with their evolutionary properties. The example focuses on the Rpb4-Rpb7 complex crystallized in both H. sapiens (pdb code:2c35) and S. cerevisiae (pdb code:1y14). The interface coordinates files were downloaded from InterEvol database in which the chains have been renumbered to match the positions in the alignment [R31, N35 and E41 in Rpb4 (2c35) stand for R44, N48 and E54 in the original PDB]. After selecting a small set of positions neighbouring at the interface it is possible to display a restricted view of the multiple sequence alignment for the 2c35 complex. Specific position can then be selected from the alignment panel and focused on in the structural panel allowing a cross talk between both dimensions. A compensatory switch between two positions of Rpb4 to maintain a salt–bridge interaction with an invariant acidic residue of Rpb7 is presented. The pre-computed alignment reveals this coordinated switch between positions 31 and 35. Retrieving the superimposed structures of the 1y14 complex reveals how structural micro-environments can adapt to allow for the plasticity of the interface in response to potentially deleterious sequence changes.
CONCLUSION AND OUTLOOK
The development of the InterEvol database reveals that the number of structures of hetero and homo-oligomers has sufficiently increased over the recent years to provide a representative set of structural interologs. Together with the multiple sequences alignments of the interologs computed in the database, a wealth of data is now available to challenge our understanding of protein complex coevolution. We believe the InterEvol database will interest a large audience because it can be accessed with a variety of purposes. Bioinformaticians will find a rich set of data to run statistics not only at the structural level but also at the sequence and evolutionary levels. Structural biologists solving a new structure of complex, may submit their sequences to discover remotely related interfaces. Also they can capture important constraints at the interface of their complex first by generating the alignments of both binding partners and diving into the evolutionary history of their interface, position by position, using the interactive InterEvol PyMol plugin. As an extension InterEvol can also be used to discover templates for comparative modelling. Other natural clients of the InterEvol database will be evolutionary biologists who wish to improve their phylogenetic models of interface evolution by examining the ready-to-use alignments pairs for coevolution analyses. As suggested from the example of the Rpb4–Rpb7 complex remarkable cases of interface plasticity and molecular epistasis might be revealed in the future by bringing together structures and sequences information.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures S1–7 and Supplementary Methods 1–2.
FUNDING
Funding for open access charge: Commissariat à l'Energie Atomique (CEA) and ANR HPGenVar (2009–2013).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Arnaud Martel for his help in the development of the web interface and the three referees for their insightful remarks.
REFERENCES
- 1.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. The BioGRID interaction database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Dam TJ, Snel B. Protein complex evolution does not involve extensive network rewiring. PLoS Comput. Biol. 2008;4:e1000132. doi: 10.1371/journal.pcbi.1000132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pazos F, Valencia A. Protein co-evolution, co-adaptation and interactions. EMBO J. 2008;27:2648–2655. doi: 10.1038/emboj.2008.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Levy ED, Pereira-Leal JB. Evolution and dynamics of protein interactions and networks. Curr. Opin. Struct. Biol. 2008;18:349–357. doi: 10.1016/j.sbi.2008.03.003. [DOI] [PubMed] [Google Scholar]
- 6.Walhout AJ, Sordella R, Lu X, Hartley JL, Temple GF, Brasch MA, Thierry-Mieg N, Vidal M. Protein interaction mapping in C. elegans using proteins involved in vulval development. Science. 2000;287:116–122. doi: 10.1126/science.287.5450.116. [DOI] [PubMed] [Google Scholar]
- 7.Nooren IM, Thornton JM. Structural characterisation and functional significance of transient protein-protein interactions. J. Mol. Biol. 2003;325:991–1018. doi: 10.1016/s0022-2836(02)01281-0. [DOI] [PubMed] [Google Scholar]
- 8.Janin J, Bahadur RP, Chakrabarti P. Protein-protein interaction and quaternary structure. Quart. Rev. Biophys. 2008;41:133–180. doi: 10.1017/S0033583508004708. [DOI] [PubMed] [Google Scholar]
- 9.Kinjo AR, Nakamura H. Geometric similarities of protein-protein interfaces at atomic resolution are only observed within homologous families: an exhaustive structural classification study. J. Mol. Biol. 2010;399:526–540. doi: 10.1016/j.jmb.2010.04.028. [DOI] [PubMed] [Google Scholar]
- 10.Gong S, Yoon G, Jang I, Bolser D, Dafas P, Schroeder M, Choi H, Cho Y, Han K, Lee S, et al. PSIbase: a database of protein structural interactome map (PSIMAP) Bioinformatics. 2005;21:2541–2543. doi: 10.1093/bioinformatics/bti366. [DOI] [PubMed] [Google Scholar]
- 11.Davis FP, Sali A. PIBASE: a comprehensive database of structurally defined protein interfaces. Bioinformatics. 2005;21:1901–1907. doi: 10.1093/bioinformatics/bti277. [DOI] [PubMed] [Google Scholar]
- 12.Ogmen U, Keskin O, Aytuna AS, Nussinov R, Gursoy A. PRISM: protein interactions by structural matching. Nucleic Acids Res. 2005;33:W331–W336. doi: 10.1093/nar/gki585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levy ED, Pereira-Leal JB, Chothia C, Teichmann SA. 3D complex: a structural classification of protein complexes. PLoS Comput. Biol. 2006;2:1395–1406. doi: 10.1371/journal.pcbi.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Winter C, Henschel A, Kim WK, Schroeder M. SCOPPI: a structural classification of protein - protein interfaces. Nucleic Acids Res. 2006;34:D310–D314. doi: 10.1093/nar/gkj099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tuncbag N, Gursoy A, Guney E, Nussinov R, Keskin O. Architectures and functional coverage of protein-protein interfaces. J. Mol. Biol. 2008;381:785–802. doi: 10.1016/j.jmb.2008.04.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Teyra J, Paszkowski-Rogacz M, Anders G, Pisabarro MT. SCOWLP classification: structural comparison and analysis of protein binding regions. BMC Bioinformatics. 2008;9:9. doi: 10.1186/1471-2105-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunther S, von Eichborn J, May P, Preissner R. JAIL: a structure-based interface library for macromolecules. Nucleic Acids Res. 2009;37:D338–D341. doi: 10.1093/nar/gkn599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lo YS, Chen YC, Yang JM. 3D-interologs: an evolution database of physical protein- protein interactions across multiple genomes. BMC Genomics. 2010;11:S3–S7. doi: 10.1186/1471-2164-11-S3-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shoemaker BA, Zhang D, Thangudu RR, Tyagi M, Fong JH, Marchler-Bauer A, Bryant SH, Madej T, Panchenko AR. Inferred biomolecular interaction server–a web server to analyze and predict protein interacting partners and binding sites. Nucleic Acids Res. 2010;38:D518–D524. doi: 10.1093/nar/gkp842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein A, Ceol A, Aloy P. 3did: identification and classification of domain-based interactions of known three-dimensional structure. Nucleic Acids Res. 2011;39:D718–D723. doi: 10.1093/nar/gkq962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xu QF, Dunbrack RL. The protein common interface database (ProtCID)-a comprehensive database of interactions of homologous proteins in multiple crystal forms. Nucleic Acids Res. 2011;39:D761–D770. doi: 10.1093/nar/gkq1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andreeva A, Howorth D, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. SCOP database in 2004: refinements integrate structure and sequence family data. Nucleic Acids Res. 2004;32:D226–D229. doi: 10.1093/nar/gkh039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cuff AL, Sillitoe I, Lewis T, Clegg AB, Rentzsch R, Furnham N, Pellegrini-Calace M, Jones D, Thornton J, Orengo CA. Extending CATH: increasing coverage of the protein structure universe and linking structure with function. Nucleic Acids Res. 2011;39:D420–D426. doi: 10.1093/nar/gkq1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Levy ED, Boeri Erba E, Robinson CV, Teichmann SA. Assembly reflects evolution of protein complexes. Nature. 2008;453:1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Teyra J, Pisabarro MT. Characterization of interfacial solvent in protein complexes and contribution of wet spots to the interface description. Proteins. 2007;67:1087–1095. doi: 10.1002/prot.21394. [DOI] [PubMed] [Google Scholar]
- 27.Tuncbag N, Gursoy A, Keskin O. Identification of computational hot spots in protein interfaces: combining solvent accessibility and inter-residue potentials improves the accuracy. Bioinformatics. 2009;25:1513–1520. doi: 10.1093/bioinformatics/btp240. [DOI] [PubMed] [Google Scholar]
- 28.Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. J. Mol. Biol. 2003;332:989–998. doi: 10.1016/j.jmb.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 29.Eames M, Kortemme T. Structural mapping of protein interactions reveals differences in evolutionary pressures correlated to mRNA level and protein abundance. Structure. 2007;15:1442–1451. doi: 10.1016/j.str.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franzosa EA, Xia Y. Structural determinants of protein evolution are context-sensitive at the residue level. Mol. Biol. Evol. 2009;26:2387–2395. doi: 10.1093/molbev/msp146. [DOI] [PubMed] [Google Scholar]
- 31.Rahat O, Yitzhaky A, Schreiber G. Cluster conservation as a novel tool for studying protein-protein interactions evolution. Proteins. 2008;71:621–630. doi: 10.1002/prot.21749. [DOI] [PubMed] [Google Scholar]
- 32.Mintseris J, Weng Z. Structure, function, and evolution of transient and obligate protein-protein interactions. Proc. Natl Acad. Sci. USA. 2005;102:10930–10935. doi: 10.1073/pnas.0502667102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weigt M, White RA, Szurmant H, Hoch JA, Hwa T. Identification of direct residue contacts in protein-protein interaction by message passing. Proc. Natl Acad. Sci. USA. 2009;106:67–72. doi: 10.1073/pnas.0805923106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madaoui H, Guerois R. Coevolution at protein complex interfaces can be detected by the complementarity trace with important impact for predictive docking. Proc. Natl Acad. Sci. USA. 2008;105:7708–7713. doi: 10.1073/pnas.0707032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317:1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Levin KB, Dym O, Albeck S, Magdassi S, Keeble AH, Kleanthous C, Tawfik DS. Following evolutionary paths to protein-protein interactions with high affinity and selectivity. Nat. Struct. Mol. Biol. 2009;16:1049–1055. doi: 10.1038/nsmb.1670. [DOI] [PubMed] [Google Scholar]
- 37.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 38.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 40.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics. 2010;26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 42.Kawabata T. MATRAS: A program for protein 3D structure comparison. Nucleic Acids Res. 2003;31:3367–3369. doi: 10.1093/nar/gkg581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mendez R, Leplae R, De Maria L, Wodak SJ. Assessment of blind predictions of protein-protein interactions: current status of docking methods. Proteins. 2003;52:51–67. doi: 10.1002/prot.10393. [DOI] [PubMed] [Google Scholar]
- 44.Zhu H, Domingues FS, Sommer I, Lengauer T. NOXclass: prediction of protein-protein interaction types. BMC Bioinformatics. 2006;7:27. doi: 10.1186/1471-2105-7-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gabaldon T, Dessimoz C, Huxley-Jones J, Vilella AJ, Sonnhammer EL, Lewis S. Joining forces in the quest for orthologs. Genome Biol. 2009;10:403. doi: 10.1186/gb-2009-10-9-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Altenhoff AM, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput. Biol. 2009;5:e1000262. doi: 10.1371/journal.pcbi.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altenhoff AM, Schneider A, Gonnet GH, Dessimoz C. OMA 2011: orthology inference among 1000 complete genomes. Nucleic Acids Res. 2011;39:D289–D294. doi: 10.1093/nar/gkq1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18(Suppl. 1):S71–S77. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- 50.Von Eichborn J, Gunther S, Preissner R. Structural features and evolution of protein-protein interactions. Genome Inform. 2010;22:1–10. [PubMed] [Google Scholar]
- 51.Choi YS, Yang JS, Choi Y, Ryu SH, Kim S. Evolutionary conservation in multiple faces of protein interaction. Proteins. 2009;77:14–25. doi: 10.1002/prot.22410. [DOI] [PubMed] [Google Scholar]