Abstract
The Mouse Multiple Tissue Metabolome Database (MMMDB) provides comprehensive and quantitative metabolomic information for multiple tissues from single mice. Manually curated databases that integrate literature-based individual metabolite information have been available so far. However, data sets on the absolute concentration of a single metabolite integrated from multiple resources are often difficult to be used when different metabolomic studies are compared because the relative balance of the multiple metabolite concentrations in the metabolic pathways as a snapshot of a dynamic system is more important than the absolute concentration of a single metabolite. We developed MMMDB by performing non-targeted analyses of cerebra, cerebella, thymus, spleen, lung, liver, kidney, heart, pancreas, testis and plasma using capillary electrophoresis time-of-flight mass spectrometry and detected 428 non-redundant features from which 219 metabolites were successfully identified. Quantified concentrations of the individual metabolites and the corresponding processed raw data; for example, the electropherograms and mass spectra with their annotations, such as isotope and fragment information, are stored in the database. MMMDB is designed to normalize users’ data, which can be submitted online and used to visualize overlaid electropherograms. Thus, MMMDB allows newly measured data to be compared with the other data in the database. MMMDB is available at: http://mmmdb.iab.keio.ac.jp.
INTRODUCTION
Metabolomics, the newest ‘omics’, is defined as the comprehensive identification and quantification of small molecules that provides a holistic view of cellular metabolism. The metabolomic network, downstream of the central dogma, transfers regulatory information from other omics data, such as genomics, transcriptomics and proteomics; thus, the metabolomic profile can be expected to directly reflect cellular phenotype (1). Metabolomic profiling has been used in biological studies in various fields, e.g. microorganism, plant, food, agricultural, pharmaceutical, clinical and medical sciences.
Nuclear magnetic resonance (NMR) (2) and mass spectrometry (MS) (3,4) are the major analytical techniques that are used in metabolomics. The relatively low sensitivity of NMR and spectral overlap limits the number and variety of metabolites that can be observed simultaneously. MS combined with a separation system prior to MS, e.g. gas chromatography (GC)–MS (5), liquid chromatography (LC)–MS (6) and capillary electrophoresis (CE)–MS (7), is currently the leading analytical platform because it provided higher selectivity and sensitivity. Because of the diverse physical and chemical properties of the metabolites, no single analytical method can comprehensively profile data sets, and each method has its own advantages and disadvantages (8). GC–MS is a well-established technology that is capable of profiling only volatile compounds and generally requires an initial derivatization procedure. LC–MS can be used to monitor a wider variety of non-volatile compounds; however, optimization of sample processing and LC column selection that depends on the target analytes is necessary. In contrast, CE–MS can monitor all charged metabolites within two (positive/negative) modes which allows for the simultaneous profiling of many pathways.
Metabolomics has contributed to the accumulation of knowledge about metabolites and their chemical (enzyme) reactions and this information has been stored in various databases. These databases can be classified broadly into two types: (i) databases that contain metabolic pathways based on information from the literature which has been integrated and curated manually and (ii) databases that contain raw or processed data from analytical system like, for example, mass spectrometry, that allow comparisons between stored data and users’ experimental data to be made. Examples of the first type of database include the Kyoto Encyclopedia of Genes and Genomics (KEGG) that contains a large collection of metabolites, enzyme and chemical drug from various species (9), MetaCyc (MetaCyc.org) that includes experimentally verified metabolic pathways and enzyme information (10), MetaCrop (11) containing metabolites from six kinds of crops and Reactome which contains human metabolites (12). These databases visualize metabolic pathways using data from the literature. The Human Metabolome Database (HMDB) contains the endogenous metabolites in human biofluids and their quantified concentrations collected from the literature (13) and the Small Molecular Pathway Database (SMPD), which is fully linked to HMDB, provides manually curated pathways (>350) for the human metabolites (14). There are also several commercial pathway databases that contain integrated knowledge and well-studied pathways, e.g. Cell Signal pathways (www.cellsignal.com), Sigma–Aldrich pathways (http://www.sigmaaldrich.com/life-science/cell-biology/learning-center.html), and Ambion pathways (http://www.ambion.com), and ProteinLounge (http://www.proteinlounge.com/).
Examples of the second type of database include the Golm Metablome Database (GMD@CSB.DB) and FiehnLib that contain GC/MS spectra (15), METLIN that provides the mass spectra of metabolites and drugs (16), MassBank that contains mass spectra from various types of MS (17) and HMDB that contains NMR data and the mass spectra of LC–MS and GC/MS along with data comparison and search functions.
Metabolic systems intricately vary in their response to environmental change; these variations are controlled by enzyme regulation. Thus, the simultaneously observed concentrations of metabolites in many pathways are indicators that provide insights to a more holistic understanding of their biological significance. This is in contrast to the integrated profiles available in the literature. Therefore, we established MMMDB, a database that contains a large collection of metabolites in multiple tissues from single mice that were obtained using capillary electrophoresis time-of-flight MS (CE-TOFMS) (18,19).
DATABASE DESCRIPTION
Database content
CE-TOFMS was used to analyze 10 tissues, cerebra, cerebella, thymus, spleen, lung, liver, kidney, heart, pancreas and testis, and also plasma in a non-targeted manner (<1000 Da) so that all possible metabolite peaks were profiled. Wild-type mice have been backcrossed to C57BL/6J for 10 or more generations, and were subsequently bred for 2 years in specific pathogen-free animal housing facilities at Yamagata University Medical School (20). Plastic cage with sawdust on the floor were used with keeping the day lights on 12-h light/dark cycle, maintaining the temperature between 23°C and 24°C. Diet (Oriental MF, Oriental Yeast Co., Tokyo, Japan) and water were available to the mice at all times. Two of 8-week-old male mice were sacrificed between 10:00 am and 12:00 pm. Duplicated data from two mice were included in the database.
Raw data were analyzed using MasterHands software (21), which detects all possible peaks, eliminates noise (e.g. spike noise) and interprets redundant features (e.g. isotopic and adduct noise) commonly observed in ESI–MS data (22). Migration times of CE–MS electropherograms were normalized by dynamic programming-based correlation methods to generate an aligned data matrix (23). We detected 428 kinds of peaks without redundant features [on an average, 351 ± 54 peaks (SD) on each sample] and identified 219 metabolites (192 ± 20).
For details of the measurement methods, the conditions used for CE-TOFMS, and the data processing procedure see Methods in Supplementary Data. Quantified concentration for each peak were identified with matched standard compounds and recorded in the database. To eliminate systematic bias, the peak areas of the 209 unidentified peaks were normalized using the same internal standards and this data was also included in the database. An overview of the profiled data and the results of the multivariate analyses are depicted in Supplementary Figures S1 and S2 using visualization software (24).
Reproducibility of migration times in CE–MS data, i.e. the peak location along the electropherogram axis, is lower than the retention/elution times in LC–MS and GC/MS data. This makes data comparison between different runs difficult. To help address this problem, the migration times of internal standards simultaneously measured in each data were registered for each metabolite entry. Both raw and background-subtracted mass spectrum with their interpreted annotations, such as isotope, fragment, adduct ions, were also registered. Identified metabolites were linked to KEGG (9), HMDB (13), ChEBI (25) and PubChem (http://pubchem.ncbi.nlm.nih.gov/).
Web interface
A screenshot of MMMDB is shown in Figure 1. All quantified data sets can be downloaded as separate csv files for each tissue or plasma. In addition to the search and browse metabolites functions, users can upload their own data and compare it on the website with the data that is stored in the database. The distribution of the quantified data in each tissue can be visualized as a bar graph and the chemical structure is also visualized. The web interface can be customized interactively on the website.
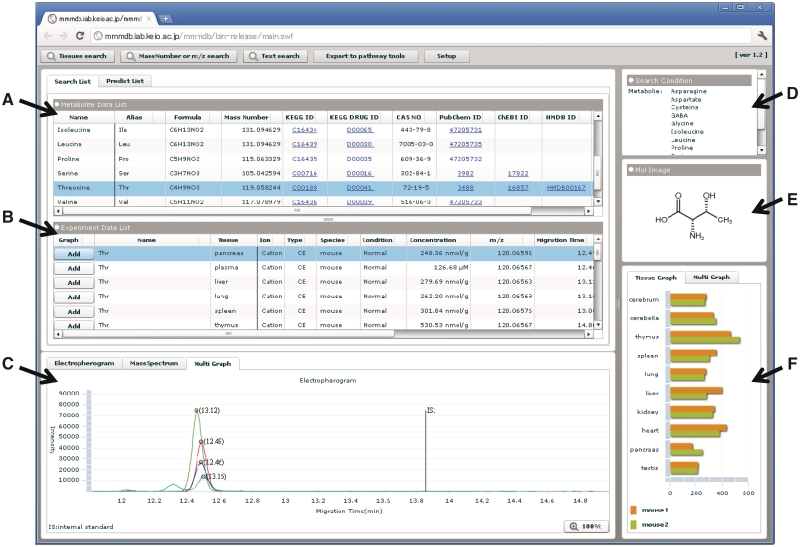
Figure 1.
An MMMDB screenshot. (A) List of metabolites that fulfilled the search condition. (B) List of the registered entries of the metabolites selected from panel (A). (C) Overlaid electropherograms displayed by the Multi Graph option. The other options, Electropherogram and Mass Spectrum, will display a single electropherogram or the mass spectrum, respectively. (D) The search conditions used. (E) The molecular structure of the displayed metabolite. (F) Tissue graph showing the quantified concentrations of the selected metabolites in each tissue.
Similar to the search in HMDB and MassBank, users can use m/z or molecular weight as a query to search the metabolites. The search results list the possible metabolites along with redundant features, such as isotope and fragment, which may help identify the interesting peaks.
For users to be able to compare their CE–MS data with the stored data, user can upload a data file containing the electropherogram, mass spectrum and migration times of internal standards in a structured csv format. In MMMDB, the migration times of the electropherogram are corrected using a polynomial equation derived from the migration times of multiple internal standards (26), and the overlaid electropherograms are visualized. Based on the difference between m/z and migration times, the searched results are evaluated and assigned scores which indicate the matched possibility. For non-CE–MS users, the electropherogram can be omitted from the upload data, and comparison are then made based only on mass spectrometry data. These functions help identify the peaks in the users’ data sets. To use the data visualization tools in the pathway mode, a part of the data sets, for example, all the metabolites in a tissue, can be exported and used as the input data file for the Pathway Projector (27) and for Vanted (28).
In addition to the data sets, web interfaces provide a data browsing navigator, a tutorial movie and document, and an example of users’ data that can be compared on-line. A tool for formatting users’ data is also provided.
Database implementation
The sever programs are designed as a versatile independent to operating system (OS). Client Adobe Flex 3 was used for the framework server program which was implemented on a Java platform [standard edition 6.0 (1.6.0_26)]. Although users are required to install the Adobe Flash Player, the interactive operations without reloading that are used, for example, to visualize overlaid electropherograms, are enabled and the user interface does not depend on the browser programs. We used Tomcat 6.0 for the web server and PostgreSQL 9.0.4 for the database. The server programs were implemented on CentOS 5.6 with 4 GB memory.
DISCUSSION
The principal feature of MMMDB is the comprehensive large collection of the absolute concentrations of metabolites in various pathways in multiple tissues from single mice using a single measurement platform. In contrast to changes in a single metabolite, profiles of multiple metabolites in many pathways are important for metabolomics research. Metabolic pathway databases containing a large amount of related information are currently available; KEGG (9), HumanCyc for human specific metabolic pathways (29), BioCarta Collections visualizing well-curated pathways (http://www.biocarta.com/genes/allPathways.asp) and Ingenuity Pathways Analysis (IPA) (http://www.ingenuity.com) providing both pathway and structured literature information are examples of these. MouseCyc is a mouse specific metabolic database and a subset of MetaCyc, provides various build-in pathway visualization tools and a pathway characteristics comparison function for mouse and human (30).
Excellent tools to visualize the quantified concentrations of multiple metabolites in metabolic pathways are also available. Pathway Projector (27) has distributed various graphs of KEGG pathways using Google Maps technologies and KEGG Atlas (31). This is useful to explore the relation of multiple metabolites from large metabolic pathways using various search functions. Vanted (28) provides an editable pathway using a template that can be downloaded from several databases, such as KEGG (9) and MetaCrop (11), with several statistical analyses tools, such as, for example, a self-organizing map (SOM) that can be used to depict the metabolite relations by reducing the metabolic complexity. With these visualization tools, the profiled data sets in MMMDB provide both an overview and the characteristic relations in complicated metabolic systems.
Multivariable analyses of the different tissue profiles allowed their tissue-specific bias to be visualized. Principal component (PC) analysis (Supplementary Figure S1) showed, as expected, that along with the first PC axis, the profiles in liver and kidney were exceptionally different from those in other tissues. Plots along the third PC axis also showed that the differences between kidney and liver profiles were larger than the differences between any of the other tissue profiles. Plots of cerebra and cerebella profiles exist close together in all PC spaces, indicating that these two profiles are similar. Interestingly, compared to the other tissues, the testis profiles were closest to the cerebra and cerebella profiles. Clustering (Supplementary Figure S2) also produced consistent results revealing, for example, the similarities in the brain profiles. Metabolites categorized as amino acids (betaine and taurine), organic acids (malate, lactate, succinate and citrate), those synthesized from arginine (creatine and carnitine), and involved in nucleotide pathways (uridine), tryptophan metabolism (indole-3-acetaldehyde), glutathione metabolism (5-oxoproline) and other pathways (glycolate, 2-hydroxypentanoate, allantoin and acetohydroxamate) were consistently higher than other metabolites in both the averaged tissue profiles and in the plasma profile. The profiles in the other metabolites in plasma are more constant than those in the tissues.
MMMDB has several limitations. The observed profiles include the metabolites in glycolysis, the tricarboxylic acid cycle, the pentose phosphate pathway, the nucleotide pathway, the amino acid pathway and the urea cycle. Because CE–MS can detect only soluble and charged metabolites, molecules with other feature, for example, lipids, volatile metabolites and molecules in secondary metabolism, are not covered in the current data sets. Using LC–MS and GC/MS that are complementary to the CE–MS data would raise the coverage; however, when non-CE–MS data is included, several metabolites that are also monitored by CE-MS make it necessary to include the instrument and the correlation of the overlapped metabolite concentrations need to be evaluated for the data sets to be extended. Another limitation of MMMDB is that several metabolites in the data are not in KEGG and therefore the Pathway Projector and Vanted cannot visualize this data. The development of a data converter for other visualization tools or for the original pathway visualization function is also necessary.
As for many other databases, the development and expansion of MMMDB is on-going. Because, the profiling technique itself does not limit its possible applications (32,33), new data sets of more segmented brain tissue and of various biofluids such as urine and saliva, are being developed. In addition to data from healthy mice (the control), data from mice with various diseases (34) could also be integrated. In contrast to profile data, metabolomic data using stable isotope labeling is also useful for tracing the metabolite flux distribution (35). Ideally, the identification of all unknown peaks would make more comprehensive improve analyses possible. We also aim to replace the currently unknown data with computational estimations of the unknown peaks (36) and their quantification (37).
CONCLUSION
In summary, MMMDB contains the concentrations of a large number of metabolites simultaneously profiled using of CE-TOFMS in a non-targeted manner. The collected profiles are from 10 tissues and the plasma of single mice. Quantified values of each metabolite and the annotated mass spectra and electropherograms are also provided. A functional web interface provides various data search options and an on-line data comparison function between data uploaded by users and stored data from mass spectrum and normalized electropherograms. The database system itself was developed as a versatile system that can contain data sets from any other species or from any tissue. Thus, the next task in the development of MMMDB is to add data from other organisms and tissues, obtained under different conditions using a variety of measurement instruments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1 and 2 and Supplementary Methods.
FUNDING
Funding for open access charge: Yamagata Prefectural Government and Tsuruoka City, Japan.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge Shinobu Abe and Nami Abe of the Institute for Advanced Biosciences, Keio University, Japan, for their help with measurement and data analysis.
REFERENCES
- 1.Fiehn O. Metabolomics–the link between genotypes and phenotypes. Plant Mol. Biol. 2002;48:155–171. [PubMed] [Google Scholar]
- 2.Reo NV. NMR-based metabolomics. Drug Chem. Toxicol. 2002;25:375–382. doi: 10.1081/dct-120014789. [DOI] [PubMed] [Google Scholar]
- 3.Aharoni A, Ric de Vos CH, Verhoeven HA, Maliepaard CA, Kruppa G, Bino R, Goodenowe DB. Nontargeted metabolome analysis by use of Fourier Transform Ion Cyclotron Mass Spectrometry. Omics. 2002;6:217–234. doi: 10.1089/15362310260256882. [DOI] [PubMed] [Google Scholar]
- 4.Castrillo JI, Hayes A, Mohammed S, Gaskell SJ, Oliver SG. An optimized protocol for metabolome analysis in yeast using direct infusion electrospray mass spectrometry. Phytochemistry. 2003;62:929–937. doi: 10.1016/s0031-9422(02)00713-6. [DOI] [PubMed] [Google Scholar]
- 5.Fiehn O, Kopka J, Trethewey RN, Willmitzer L. Identification of uncommon plant metabolites based on calculation of elemental compositions using gas chromatography and quadrupole mass spectrometry. Anal. Chem. 2000;72:3573–3580. doi: 10.1021/ac991142i. [DOI] [PubMed] [Google Scholar]
- 6.Plumb R, Granger J, Stumpf C, Wilson ID, Evans JA, Lenz EM. Metabonomic analysis of mouse urine by liquid-chromatography-time of flight mass spectrometry (LC-TOFMS): detection of strain, diurnal and gender differences. Analyst. 2003;128:819–823. doi: 10.1039/b304296k. [DOI] [PubMed] [Google Scholar]
- 7.Soga T, Ohashi Y, Ueno Y, Naraoka H, Tomita M, Nishioka T. Quantitative metabolome analysis using capillary electrophoresis mass spectrometry. J. Proteome Res. 2003;2:488–494. doi: 10.1021/pr034020m. [DOI] [PubMed] [Google Scholar]
- 8.Monton MR, Soga T. Metabolome analysis by capillary electrophoresis-mass spectrometry. J. Chromatogr. A. 2007;1168:237–246. doi: 10.1016/j.chroma.2007.02.065. discussion 236. [DOI] [PubMed] [Google Scholar]
- 9.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grafahrend-Belau E, Weise S, Koschutzki D, Scholz U, Junker BH, Schreiber F. MetaCrop: a detailed database of crop plant metabolism. Nucleic Acids Res. 2008;36:D954–D958. doi: 10.1093/nar/gkm835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Frolkis A, Knox C, Lim E, Jewison T, Law V, Hau DD, Liu P, Gautam B, Ly S, Guo AC, et al. SMPDB: the Small Molecule Pathway Database. Nucleic Acids Res. 2010;38:D480–D487. doi: 10.1093/nar/gkp1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kopka J, Schauer N, Krueger S, Birkemeyer C, Usadel B, Bergmuller E, Dörmann P, Weckwerth W, Gibon Y, Stitt M, et al. GMD@CSB.DB: the Golm Metabolome Database. Bioinformatics. 2005;21:1635–1638. doi: 10.1093/bioinformatics/bti236. [DOI] [PubMed] [Google Scholar]
- 16.Smith CA, O'Maille G, Want EJ, Qin C, Trauger SA, Brandon TR, Custodio DE, Abagyan R, Siuzdak G. METLIN: a metabolite mass spectral database. Ther. Drug Monit. 2005;27:747–751. doi: 10.1097/01.ftd.0000179845.53213.39. [DOI] [PubMed] [Google Scholar]
- 17.Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, et al. MassBank: a public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010;45:703–714. doi: 10.1002/jms.1777. [DOI] [PubMed] [Google Scholar]
- 18.Soga T, Baran R, Suematsu M, Ueno Y, Ikeda S, Sakurakawa T, Kakazu Y, Ishikawa T, Robert M, Nishioka T, et al. Differential metabolomics reveals ophthalmic acid as an oxidative stress biomarker indicating hepatic glutathione consumption. J. Biol. Chem. 2006;281:16768–16776. doi: 10.1074/jbc.M601876200. [DOI] [PubMed] [Google Scholar]
- 19.Soga T, Igarashi K, Ito C, Mizobuchi K, Zimmermann HP, Tomita M. Metabolomic Profiling of Anionic Metabolites by Capillary Electrophoresis Mass Spectrometry. Anal. Chem. 2009;81:6165–6174. doi: 10.1021/ac900675k. [DOI] [PubMed] [Google Scholar]
- 20.Sato H, Shiiya A, Kimata M, Maebara K, Tamba M, Sakakura Y, Makino N, Sugiyama F, Yagami K, Moriguchi T, et al. Redox imbalance in cystine/glutamate transporter-deficient mice. J. Biol. Chem. 2005;280:37423–37429. doi: 10.1074/jbc.M506439200. [DOI] [PubMed] [Google Scholar]
- 21.Sugimoto M, Wong DT, Hirayama A, Soga T, Tomita M. Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics. 2010;6:78–95. doi: 10.1007/s11306-009-0178-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brown M, Dunn WB, Dobson P, Patel Y, Winder CL, Francis-McIntyre S, Begley P, Carroll K, Broadhurst D, Tseng A, et al. Mass spectrometry tools and metabolite-specific databases for molecular identification in metabolomics. Analyst. 2009;134:1322–1332. doi: 10.1039/b901179j. [DOI] [PubMed] [Google Scholar]
- 23.Baran R, Kochi H, Saito N, Suematsu M, Soga T, Nishioka T, Robert M, Tomita M. MathDAMP: a package for differential analysis of metabolite profiles. BMC Bioinformatics. 2006;7:530. doi: 10.1186/1471-2105-7-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34:374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 25.Degtyarenko K, Hastings J, de Matos P, Ennis M. ChEBI: an open bioinformatics and cheminformatics resource. Curr. Protoc. Bioinformatics. 2009 doi: 10.1002/0471250953.bi1409s26. Chapter 14, Unit 14.19. [DOI] [PubMed] [Google Scholar]
- 26.Reijeng JC, Martens JH, Giuliani A, Chiari M. Pherogram normalization in capillary electrophoresis and micellar electrokinetic chromatography analyses in cases of sample matrix-induced migration time shifts. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2002;770:45–51. doi: 10.1016/s0378-4347(01)00527-8. [DOI] [PubMed] [Google Scholar]
- 27.Kono N, Arakawa K, Ogawa R, Kido N, Oshita K, Ikegami K, Tamaki S, Tomita M. Pathway projector: web-based zoomable pathway browser using KEGG atlas and Google Maps API. PLoS One. 2009;4:e7710. doi: 10.1371/journal.pone.0007710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Junker BH, Klukas C, Schreiber F. VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics. 2006;7:109. doi: 10.1186/1471-2105-7-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Romero P, Wagg J, Green ML, Kaiser D, Krummenacker M, Karp PD. Computational prediction of human metabolic pathways from the complete human genome. Genome Biol. 2004;6:R2. doi: 10.1186/gb-2004-6-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evsikov AV, Dolan ME, Genrich MP, Patek E, Bult CJ. MouseCyc: a curated biochemical pathways database for the laboratory mouse. Genome Biol. 2009;10:R84. doi: 10.1186/gb-2009-10-8-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Okuda S, Yamada T, Hamajima M, Itoh M, Katayama T, Bork P, Goto S, Kanehisa M. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008;36:W423–W426. doi: 10.1093/nar/gkn282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baena B, Cifuentes A, Barbas C. Analysis of carboxylic acids in biological fluids by capillary electrophoresis. Electrophoresis. 2005;26:2622–2636. doi: 10.1002/elps.200410329. [DOI] [PubMed] [Google Scholar]
- 33.Ramautar R, Mayboroda OA, Somsen GW, de Jong GJ. CE-MS for metabolomics: developments and applications in the period 2008–2010. Electrophoresis. 2011;32:52–65. doi: 10.1002/elps.201000378. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Perez I, Whitfield P, Bartlett A, Angulo S, Legido-Quigley C, Hanna-Brown M, Barbas C. Metabolic fingerprinting of Schistosoma mansoni infection in mice urine with capillary electrophoresis. Electrophoresis. 2008;29:3201–3206. doi: 10.1002/elps.200800031. [DOI] [PubMed] [Google Scholar]
- 35.Fan TW, Lane AN, Higashi RM, Yan J. Stable isotope resolved metabolomics of lung cancer in a SCID mouse model. Metabolomics. 2011;7:257–269. doi: 10.1007/s11306-010-0249-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sugimoto M, Hirayama A, Robert M, Abe S, Soga T, Tomita M. Prediction of metabolite identity from accurate mass, migration time prediction and isotopic pattern information in CE-TOFMS data. Electrophoresis. 2010;31:2311–2318. doi: 10.1002/elps.200900584. [DOI] [PubMed] [Google Scholar]
- 37.Chalcraft KR, Lee R, Mills C, Britz-McKibbin P. Virtual quantification of metabolites by capillary electrophoresis-electrospray ionization-mass spectrometry: predicting ionization efficiency without chemical standards. Anal. Chem. 2009;81:2506–2515. doi: 10.1021/ac802272u. [DOI] [PubMed] [Google Scholar]