Abstract
The genetic lesions identified in chronic lymphocytic leukemia (CLL) do not entirely recapitulate the disease pathogenesis and the development of serious complications, such as chemorefractoriness. While investigating the coding genome of fludarabine-refractory CLL, we observed that mutations of SF3B1, encoding a splicing factor and representing a critical component of the cell spliceosome, were recurrent in 10 of 59 (17%) fludarabine-refractory cases, with a frequency significantly greater than that observed in a consecutive CLL cohort sampled at diagnosis (17/301, 5%; P = .002). Mutations were somatically acquired, were generally represented by missense nucleotide changes, clustered in selected HEAT repeats of the SF3B1 protein, recurrently targeted 3 hotspots (codons 662, 666, and 700), and were predictive of a poor prognosis. In fludarabine-refractory CLL, SF3B1 mutations and TP53 disruption distributed in a mutually exclusive fashion (P = .046). The identification of SF3B1 mutations points to splicing regulation as a novel pathogenetic mechanism of potential clinical relevance in CLL.
Introduction
The clinical course of chronic lymphocytic leukemia (CLL) ranges from a very indolent disorder with a normal lifespan for the patient to a rapidly progressive disease that leads to death. Occasionally, CLL undergoes a transformation to Richter syndrome (RS).1–3 The variable clinical course of CLL is driven, at least in part, by the disease's immunogenetic and molecular heterogeneity.4
Despite recent advances, the genetic lesions identified to date do not fully recapitulate the molecular pathogenesis of CLL and do not entirely explain the development of severe complications, such as chemorefractoriness, which still represent unmet clinical needs.5 In approximately 40% of cases, refractoriness to fludarabine is attributable to the disruption of TP53, but in a sizeable fraction of patients, the molecular basis of this aggressive phenotype remains unclear.6
Recently, 2 independent studies of the CLL coding genome investigated at disease presentation have revealed a restricted number of mutated genes, including NOTCH1.7,8 These studies have provided a proof of concept that, similar to other malignancies, genome-wide mutational analysis might identify novel lesions of biologic and clinical relevance in CLL. On these grounds, we have embarked on the investigation of the coding genome of fludarabine-refractory CLL to identify genetic lesions associated with chemorefractoriness. The initial phases of this analysis have revealed recurrent mutations of SF3B1, a critical component of the cell spliceosome, pointing to the potential involvement of splicing regulation in CLL pathogenesis and chemorefractoriness.
Methods
Patients
The study population comprised 3 cohorts representative of different disease phases: (1) fludarabine-refractory CLL (n = 59), including cases (n = 11) subjected to whole-exome sequencing (supplemental Table 1, available on the Blood Web site; see the Supplemental Materials link at the top of the online article); (2) a consecutive series of newly diagnosed and previously untreated patients with CLL (n = 301; supplemental Table 2); and (3) clonally related RS (n = 33; all diffuse large B-cell lymphomas; supplemental Table 3). Diagnosis of CLL and of fludarabine-refractoriness were based on International Workshop on CLL–National Cancer Institute (IWCLL-NCI) criteria2; RS was based on histologic criteria.1,9
Peripheral blood tumor samples were obtained as follows: (1) for patients with fludarabine-refractory CLL, immediately before starting treatment to which the patient did not respond because of stable/progressive disease; and (2) for newly diagnosed and previously untreated CLL, at disease presentation. All studies on RS were performed on RS diagnostic biopsies. Germline DNA samples from the same patients were obtained from saliva or from purified granulocytes and confirmed to be tumor-free by PCR of tumor-specific IGHV-D-J rearrangements. Patients provided informed consent in accordance with local institutional review board requirements and the Declaration of Helsinki. The study was approved by the ethical committee of the Ospedale Maggiore della Carità di Novara associated with the Amedeo Avogadro University of Eastern Piedmont (protocol code 59/CE; study number CE 8/11).
Mutation analysis of SF3B1
Mutation analysis of SF3B1 (exons 1-25, including splice sites; RefSeq NM_012433.2) was performed on PCR amplimers obtained from genomic DNA by a combination of Sanger sequencing (ABI PRISM 3100 Genetic Analyzer; Applied Biosystems) and targeted next generation sequencing (Genome Sequencer Junior, 454 Life Sciences; Roche; mean coverage approximately 200×). Additional details of sequencing strategies are in supplemental Methods.
FISH karyotype; mutation analysis of IGHV, TP53, and NOTCH1; copy number analysis; and gene expression profile analysis
FISH analysis was performed with the use of probes LSI13 and LSID13S319, CEP12, LSIp53, and LSIATM (Abbott).3 IGHV sequences were aligned to ImMunoGeneTics directory and considered mutated if their identity to corresponding germline genes was < 98%.3 TP53 and NOTCH1 mutations were analyzed by Sanger sequencing.3,7 Genome-wide DNA profiles were obtained with the Affymetrix Genome-Wide Human SNP Array 6.0. Gene expression profile analysis was performed with the use of Affymetrix HG-U133Plus2 arrays. Further details are reported in supplemental Methods.
Statistical analysis
Overall survival was measured from date of diagnosis to date of death (event) or last follow-up (censoring). Treatment-free survival was measured from date of diagnosis to date of progression to symptomatic disease requiring treatment according to IWCLL-NCI guidelines (event),2 death, or last follow-up (censoring). Further details are reported in supplemental Methods.
Results and discussion
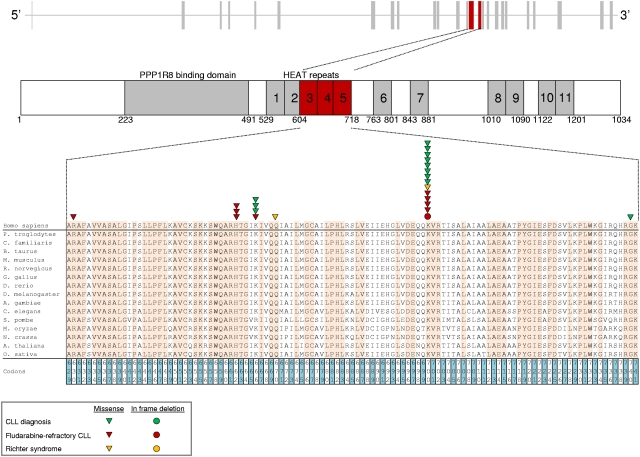
After the initial observation of recurrent SF3B1 mutations in 3 of 11 fludarabine-refractory CLL analyzed by whole-exome sequencing, we performed targeted resequencing of the SF3B1 coding sequence and splice sites in 48 additional cases of progressive and fludarabine-refractory CLL (total number of cases analyzed: 59; supplemental Table 1). SF3B1 was altered in 10 of 59 (17%) fludarabine-refractory CLL by missense mutations (n = 9) or in-frame deletions (n = 1) clustering in the HEAT3, HEAT4, and HEAT5 repeats of the SF3B1 protein (Figures 1 and 2A; supplemental Table 4). Two sites that are highly conserved interspecies (codon 662 and codon 700) were recurrently mutated in 3 and 5 cases, respectively (Figure 1). SF3B1 mutations were monoallelic and were predicted to be functionally significant according to the PolyPhen-2 algorithm (supplemental Table 4).10 These data document that mutations of SF3B1, a splicing factor that is a critical component of the spliceosome, recurrently associate with fludarabine-refractory CLL.
Figure 1.
SF3B1 mutations in CLL and RS. Schematic diagram of the human SF3B1 gene (top) and protein (bottom) with its functional domains (PPP1R8 binding domain and HEAT repeats), and multiple alignment of the HEAT3, HEAT4, and HEAT5 amino acid sequences of the human SF3B1 protein with orthologous SF3B1 proteins (n = 15). Amino acids conserved among species are highlighted. Color-coded shapes indicate the position of the mutations found in CLL at diagnosis, in fludarabine-refractory CLL, and in RS.
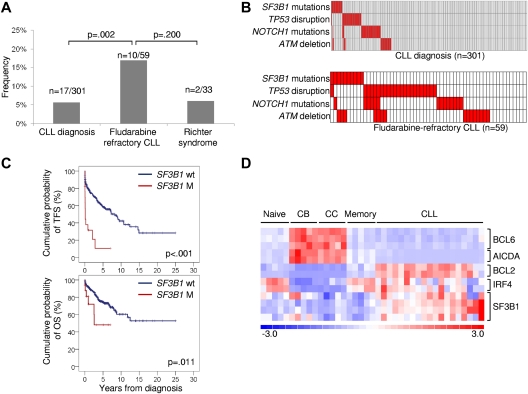
Figure 2.
Prevalence, mutual relationship with other genetic lesions, and clinical impact of SF3B1 mutations in CLL. (A) Prevalence of SF3B1 mutations in CLL at diagnosis, in fludarabine-refractory CLL, and in RS; numbers on top indicate the actual number of mutated samples over the total number analyzed. (B) Mutual relationship of SF3B1 mutations with other genetic lesions in CLL at diagnosis and in fludarabine-refractory CLL. In the heat map, rows correspond to identical genes, and columns represent individual patients color-coded based on the gene status (white: wild type; red: mutations of SF3B1, mutations of NOTCH1, mutations and/or deletion of TP53, deletion of ATM). (C) Kaplan-Meier estimates of treatment-free survival (TFS) and overall survival (OS) from diagnosis in the consecutive series of newly diagnosed and previously untreated CLL (n = 301). SF3B1 wild-type (SF3B1 wt) are represented by the blue line. SF3B1 mutated cases (SF3B1 M) are represented by the red line. (D) Gene expression levels of BCL6, AICDA, BCL2, IRF4, and SF3B1 in normal B-cell subpopulations (Naive; centroblasts, CB; centrocytes, CC; memory) and CLL samples. Relative levels of gene expression are depicted with a color scale: red represents the greatest level of expression and blue represents the lowest level.
The biologic characteristics of fludarabine-refractory CLL harboring SF3B1 mutations are summarized in supplemental Table 1. Mutations occurred irrespective of the IGHV mutation status, CD38 expression, and ZAP70 expression. At the time of fludarabine-refractoriness, SF3B1 mutations were enriched in cases harboring a normal FISH karyotype (P = .008; supplemental Table 1). In addition, SF3B1 mutations distributed in a mutually exclusive fashion compared with TP53 disruption tested by deletion and/or mutation (mutual information I = 0.0609; P = .046; Figure 2B). By combining SF3B1 mutations with other genetic lesions enriched in chemorefractory cases (TP53 disruption, NOTCH1 mutations, ATM deletion),7,11–13 fludarabine-refractory CLL appeared to be characterized by multiple molecular alterations that, to some extent, are mutually exclusive (Figure 2B).
To investigate whether SF3B1 mutations are restricted to chemorefractory cases, we then compared the prevalence of mutations observed at the time of fludarabine-refractoriness to the prevalence of mutations observed in other disease phases. In a consecutive series evaluated at CLL diagnosis, SF3B1 mutations were rare (17/301; 5%; Figure 2A; supplemental Table 4) and occurred irrespective of other molecular and immunogenetic features (supplemental Table 2; supplemental Table 5; supplemental Figure 1). Remarkably, 5 of 17 (29%) CLL mutated at diagnosis were primary fludarabine-refractory patients. In these 5 cases, TP53 disruption and NOTCH1 mutations occurred in 1 case each. None of the 12 remaining cases harbored TP53 disruption or NOTCH1 mutations. By univariate analysis, SF3B1 mutations showed a crude association with short treatment-free survival (P < .001) and overall survival (P = .011; Figure 2C). By multivariate analysis, the increased risk of death predicted by SF3B1 mutations was independent (hazard ratio 3.02; 95% confidence interval 1.24-7.35; P = .015) of confounding clinical and biologic variables (supplemental Table 6). Confirmation within the frame of prospective clinical trials will be helpful to fully assess the generalization of SF3B1 mutations as a CLL prognostic marker.
In CLL investigated at diagnosis, the hotspot distribution and molecular spectrum of SF3B1 mutations, as well as their mutual relationship with other genetic lesions, were similar to those observed in fludarabine-refractory CLL (Figures 1 and 2B; supplemental Table 4). SF3B1 mutations were only found in 2 of 33 (6%) clonally related RS (Figures 1 and 2A; supplemental Table 4). Across the different disease phases investigated, mutations were confirmed to be somatically acquired in all cases (n = 18) for which germline DNA was available (supplemental Table 4). Among the 3 SF3B1 mutated cases for which serial samples were analyzed, SF3B1 mutations were acquired in 2 cases. One fludarabine-refractory CLL (case 7915 in supplemental Table 4) acquired the c.2044A > G p.K666E mutation at the time of refractoriness, and one RS (case 7509 in supplemental Table 4) acquired the c.2146A > G p.K700E mutation at the time of transformation. In the remaining case (case 8343 in supplemental Table 4), the SF3B1 mutation was present in all disease phases.
Although the relative expression of SF3B1 in CLL was greater compared with normal B-cell subsets (Figure 2D), extensive investigation by single-nucleotide polymorphism array analysis ruled out focal copy number abnormalities of SF3B1 in this leukemia (n = 0/323). SF3B1 mutations were consistently absent among mature B-cell neoplasms (n = 136) other than CLL (supplemental Table 7). These data document that SF3B1 mutations: (1) are specific for CLL among mature B-cell neoplasms; (2) occur at a low rate at CLL presentation, whereas they are enriched in fludarabine-refractory cases; and (3) play a minor role in RS transformation, corroborating the notion that CLL histologic shift is molecularly distinct from chemorefractory progression without RS transformation.3
Our identification of SF3B1 mutations in CLL, and the recent discovery of SF3B1 mutations in myelodysplasia, points to the involvement of splicing regulation as a novel pathogenetic mechanism in hematologic malignancies.14,15 SF3B1 is a critical component of both major (U2-like) and minor (U12-like) spliceosomes,16–18 which enact the precise excision of introns from pre-mRNA.19–21 The precise biologic role of SF3B1 mutations in CLL is currently elusive and will require dedicated studies. The pathogenicity of SF3B1 mutations in CLL is strongly supported by the clustering of these mutations in evolutionarily conserved hotspots localized within HEAT domains, which are tandemly arranged curlicue-like structures serving as flexible scaffolding on which other components can assemble.22,23 In addition, the observation that SF3B1 regulates the alternative splicing program of genes controlling cell-cycle progression and apoptosis points to a potential contribution of SF3B1 mutations in modulating tumor cell proliferation and survival.20,24,25 In addition to pathogenetic implications, SF3B1 mutations might also provide a therapeutic target for SF3B1 inhibitors,24,25 which are currently under preclinical development as anticancer drugs.
Supplementary Material
Acknowledgments
This study was supported by grants from AIRC, Special Program Molecular Clinical Oncology, 5 x 1000, No. 10007, Milan, Italy (to G.G. and R.F.); Progetto FIRB-Programma “Futuro in Ricerca” 2008 (to D.R.), PRIN 2008 (to G.G.), and PRIN 2009 (to D.R.), MIUR, Rome, Italy; Progetto Giovani Ricercatori 2008 (to D.R.) and Ricerca Sanitaria Finalizzata 2008 (to G.G.), Ministero della Salute, Rome, Italy; Compagnia di San Paolo, Turin, Italy (to R.F.); NIH Grant PO1-CA092625 and a Specialized Center of Research grant from the Leukemia & Lymphoma Society (both to R.D.-F.); and Helmut Horten Foundation and San Salvatore Foundation (to F.B.). S.M. and S. Cresta are being supported by fellowships from Novara-AIL Onlus, Novara.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: D.R., L.P., R.F., R.D.-F., and G.G. designed the study, interpreted data, and wrote the manuscript; A.B., V.S., S.R., M.M., S.M., M.C., S. Cresta, and E.G. performed and interpreted mutational analysis; C.D. performed and interpreted FISH studies; H.K. and R.R. performed and interpreted bioinformatics studies; T.V. and S.D. contributed to molecular data analysis and interpretation; M.F., S. Chiaretti, A.G., I.D.G., and F.F. provided well-characterized clinical samples; V.G. performed immunophenotypic studies; and F.B. and L.P. interpreted SNP array data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Riccardo Dalla-Favera, MD, Institute for Cancer Genetics and the Herbert Irving Comprehensive Cancer Center, Columbia University, St Nicholas Ave, Rm 508, New York, NY 10032; e-mail: rd10@columbia.edu; and Gianluca Gaidano, MD, PhD, Division of Hematology, Department of Clinical and Experimental Medicine, Amedeo Avogadro University of Eastern Piedmont, Via Solaroli 17, 28100 Novara, Italy; e-mail gaidano@med.unipmn.it.
References
- 1.Müller-Hermelink HK, Montserrat E, Catovsky D, Campo E, Harris NL, Stein H. Chronic lymphocytic leukemia/small lymphocytic lymphoma. In: Swerdlow SH, Campo E, Harris NL, et al., editors. World Health Organization Classification of Tumours, Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC; 2008. pp. 180–182. [Google Scholar]
- 2.Hallek M, Cheson BD, Catovsky D, et al. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute—Working Group 1996 guidelines. Blood. 2008;111(12):5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossi D, Spina V, Deambrogi C, et al. The genetics of Richter syndrome reveals disease heterogeneity and predicts survival after transformation. Blood. 2011;117(12):3391–3401. doi: 10.1182/blood-2010-09-302174. [DOI] [PubMed] [Google Scholar]
- 4.Chiorazzi N, Rai KR, Ferrarini M. Chronic lymphocytic leukemia. N Engl J Med. 2005;352(8):804–815. doi: 10.1056/NEJMra041720. [DOI] [PubMed] [Google Scholar]
- 5.Kay NE, O'Brien SM, Pettitt AR, Stilgenbauer S. The role of prognostic factors in assessing ‘high-risk’ subgroups of patients with chronic lymphocytic leukemia. Leukemia. 2007;21(9):1885–1891. doi: 10.1038/sj.leu.2404802. [DOI] [PubMed] [Google Scholar]
- 6.Stilgenbauer S, Zenz T. Understanding and managing ultra high-risk chronic lymphocytic leukemia. Hematology Am Soc Hematol Educ Program. 2010;2010:481–488. doi: 10.1182/asheducation-2010.1.481. [DOI] [PubMed] [Google Scholar]
- 7.Fabbri G, Rasi S, Rossi D, et al. Analysis of the chronic lymphocytic leukemia coding genome: role of NOTCH1 mutational activation. J Exp Med. 2011;208(7):1389–1401. doi: 10.1084/jem.20110921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puente XS, Pinyol M, Quesada V, et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature. 2011;475(7354):101–105. doi: 10.1038/nature10113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stein H, Warnke RA, Chan WC, Jaffe ES. Diffuse large B-cell lymphoma. In: Swerdlow SH, Campo E, Harris NL, et al., editors. World Health Organization Classification of Tumours, Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues. Lyon, France: IARC; 2008. pp. 233–237. [Google Scholar]
- 10.Adzhubei IA, Schmidt S, Peshkin L, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Döhner H, Stilgenbauer S, Benner A, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–1916. doi: 10.1056/NEJM200012283432602. [DOI] [PubMed] [Google Scholar]
- 12.Rossi D, Cerri M, Deambrogi C, et al. The prognostic value of TP53 mutations in chronic lymphocytic leukemia is independent of Del17p13: implications for overall survival and chemorefractoriness. Clin Cancer Res. 2009;15(3):995–1004. doi: 10.1158/1078-0432.CCR-08-1630. [DOI] [PubMed] [Google Scholar]
- 13.Zenz T, Häbe S, Denzel T, et al. Detailed analysis of p53 pathway defects in fludarabine-refractory chronic lymphocytic leukemia (CLL): dissecting the contribution of 17p deletion, TP53 mutation, p53-p21 dysfunction, and miR34a in a prospective clinical trial. Blood. 2009;114(13):2589–2597. doi: 10.1182/blood-2009-05-224071. [DOI] [PubMed] [Google Scholar]
- 14.Visconte V, Makishima H, Jankowska A, et al. SF3B1, a splicing factor is frequently mutated in refractory anemia with ring sideroblasts [published online ahead of print September 2, 2011]. Leukemia. doi: 10.1038/leu.2011.232. doi: 10.1038/leu.2011.232. [DOI] [PubMed] [Google Scholar]
- 15.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;365(15):1384–1395. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 16.Luke MM, Della Seta F, Di Como CJ, Sugimoto H, Kobayashi R, Arndt KT. The SAP, a new family of proteins, associate and function positively with the SIT4 phosphatase. Mol Cell Biol. 1996;16(6):2744–2755. doi: 10.1128/mcb.16.6.2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang C, Chua K, Seghezzi W, Lees E, Gozani O, Reed R. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 1998;12(10):1409–1414. doi: 10.1101/gad.12.10.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das BK, Xia L, Palandjian L, Gozani O, Chyung Y, Reed R. Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol Cell Biol. 1999;19(10):6796–6802. doi: 10.1128/mcb.19.10.6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wahl MC, Will CL, Lührmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 20.David CJ, Manley JL. Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 2010;24(21):2343–2364. doi: 10.1101/gad.1973010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ward AJ, Cooper TA. The pathobiology of splicing. J Pathol. 2010;220(2):152–163. doi: 10.1002/path.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andrade MA, Bork P. HEAT repeats in the Huntington's disease protein. Nat Genet. 1995;11(2):115–116. doi: 10.1038/ng1095-115. [DOI] [PubMed] [Google Scholar]
- 23.Andrade MA, Perez-Iratxeta C, Ponting CP. Protein repeats: structures, functions, and evolution. J Struct Biol. 2001;134(2-3):117–131. doi: 10.1006/jsbi.2001.4392. [DOI] [PubMed] [Google Scholar]
- 24.Kaida D, Motoyoshi H, Tashiro E, et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat Chem Biol. 2007;3(9):576–583. doi: 10.1038/nchembio.2007.18. [DOI] [PubMed] [Google Scholar]
- 25.Corrionero A, Miñana B, Valcárcel J. Reduced fidelity of branch point recognition and alternative splicing induced by the antitumor drug spliceostatin A. Genes Dev. 2011;25(5):445–459. doi: 10.1101/gad.2014311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.