Abstract
Highly compacted DNA nanoparticles, composed of single molecules of plasmid DNA compacted with block copolymers of poly-L-lysine and 10 kDa polyethylene glycol (CK30PEG10k), mediate effective gene delivery to the brain, eyes and lungs in vivo. Nevertheless, we found that CK30PEG10k DNA nanoparticles are immobilized by mucoadhesive interactions in sputum that lines the lung airways of patients with cystic fibrosis (CF), which would presumably preclude the efficient delivery of cargo DNA to the underlying epithelium. We previously found that nanoparticles can rapidly penetrate human mucus secretions if they are densely coated with low MW PEG (2–5 kDa), whereas nanoparticles with 10 kDa PEG coatings were immobilized. We thus sought to reduce mucoadhesion of DNA nanoparticles by producing CK30PEG DNA nanoparticles with low MW PEG coatings. We examined the morphology, colloidal stability, nuclease resistance, diffusion in human sputum and in vivo gene transfer of CK30PEG DNA nanoparticles prepared using various PEG MWs. CK30PEG10k and CK30PEG5k formulations did not aggregate in saline, provided partial protection against DNase I digestion and exhibited the highest gene transfer to lung airways following inhalation in BALB/c mice. However, all DNA nanoparticle formulations were immobilized in freshly expectorated human CF sputum, likely due to inadequate PEG surface coverage.
Keywords: Mucus, Sputum, Cystic Fibrosis, Gene Therapy
1. Introduction
Cystic fibrosis (CF) is the most prevalent lethal genetic disorder among Caucasians, occurring in 1 in 3500 births in the US [1]. CF is caused by a mutation in the cystic fibrosis transmembrane conductance regulator (CFTR) gene, which codes for a chloride ion channel located in the apical membrane of epithelial cells [2]. The consequent imbalanced ion and water transport across the epithelium in CF leads to decreased airway mucus hydration and increased mucus viscoelasticity. In the CF lung, the hyperviscoelastic mucus secretions are less efficiently cleared by mucociliary action and, thus, accumulate in the airways. Mucus accumulation can be severe, leading to airway obstruction, especially during exacerbations triggered by viral infections [3–4]. Reduced mucus clearance causes chronic bacterial infection and inflammation, two major causes of CF-related morbidity and mortality [5–6]. Thus, the lung airways are an important therapeutic target for CF.
The goal of CF lung gene therapy is to restore expression of functional CFTR in the lung epithelium. Despite over two decades of research, development and clinical testing, no patient has yet been cured of CF. A number of synthetic (non-viral) systems have shown promise for CF gene therapy, including the highly compacted rod-shaped DNA nanoparticles composed of block copolymers of poly-L-lysine and polyethylene glycol (CK30PEG10k) and single molecules of plasmid DNA [7–8]. CK30PEG10k DNA nanoparticles traffic to the nucleus in primary airway epithelial cells grown under air-liquid culture within 1 hour [9], and transfect non-dividing cells [8]. These DNA nanoparticles are stable for at least 2 years at 4°C [7], and were non-toxic, non-inflammatory, and non-immunogenic in the human nares [10]. In a Phase I/IIa study in CF subjects, the level of gene transfer to human nasal mucosa mediated by these DNA nanoparticles, as measured by PCR analysis of vector DNA, was comparable to the highest levels observed in an AAV intrasinus trial [10]. Despite these promising results, efficacy has not yet been established in the CF lung airways.
Numerous reports highlight poor sputum penetration as a barrier to successful CF gene therapy [11–13]. Sputum is a porous mesh of biomacromolecules, including mucins, proteins, lipids and DNA [14]. Gene carriers deposited in the airways upon inhalation must penetrate the hyper-viscoelastic sputum layer to reach underlying epithelial cells. However, most synthetic nanoparticles are immobilized in CF sputum [15–17]. We previously found that polymeric particles as large as ~ 200 nm can rapidly penetrate CF sputum if they are densely coated with polyethylene glycol (PEG) with MW between 2–5 kDa [18], whereas nanoparticles coated with 10 kDa PEG were mucoadhesive [19]. Thus, we sought to engineer highly compacted DNA nanoparticles coated with 2 and 5 kDa PEG, and to examine DNA nanoparticle morphology, colloidal stability, nuclease resistance, gene transfer efficiency in mice, and mobility in human CF sputum.
2. Materials and methods
2.1. Materials
All chemicals were purchased from Sigma-Aldrich (St. Louis, MO) and used as received, unless stated otherwise. The trifluoroacetate (TFA) salt of poly-L-lysine of exactly 30 lysine residues in length with an N-terminal cysteine (CK30) was synthesized by Fmoc-mediated solid-phase peptide synthesis using an automated peptide synthesizer (Symphony Quartet, Protein Technologies, Tucson, AZ). CK30 was purified by HPLC and identity confirmed by mass spectrometry. Thiol reactive PEG, methoxy-PEG-maleimide, MW 10, 5, and 2 kDa, was purchased from Rapp Polymere (Tübingen, Germany). Diamine PEG, 2 kDa, was purchased from Nektar Therapeutics (San Carlos, CA). Tissue lysis and luciferase assay reagents were obtained from Promega (Madison, WI). Protein concentration assays were performed using the BCA Protein Assay Kit from Pierce (Rockford, IL). Rhodamine labeled ellipsoidal and rod-shaped CK30PEG10k DNA nanoparticles (provided by Copernicus Therapeutics, Cleveland, OH) were produced by compacting rhodamine labeled DNA with CK30PEG10k associated with either trifluoroacetate or acetate as the counterion, respectively, as previously reported [7–8].
2.2. Formulation of rod-shaped DNA nanoparticles
2.2.1. Preparation of CK30PEG
A block co-polymer of poly-L-lysine and polyethylene glycol, CK30PEG, was prepared as previously described [8], with two exceptions: (1) the MWs of PEG used were 10, 5, and 2 kDa, and (2) TFA counterion was replaced with acetate by size-exclusion chromatography on Sephadex G-25.
2.2.2. Preparation of fluorescently labeled CK30PEG
CK30PEG polymers were fluorescently labeled by conjugating the amine reactive probe Alexa Fluor 488 sulfodichlorophenol (5-SDP) ester (AF488; Invitrogen Corporation, Carlsbad, CA) to the epsilon amines of lysine following manufacturers protocol (reaction stoichiometry: 1 AF488 per CK30PEG). Unreacted AF488 was removed by fractionating the reaction mixture on a Sephadex G25 column (GE Healthcare, Piscataway, NJ) pre-equilibrated with 50 mM ammonium acetate (AA).
2.2.3. Plasmid propagation
The plasmid pd1GL3-RL (7.9 kbp) containing two CMV promoted Luc genes, firefly (GL3) and Renilla (RL), was propagated in Escherichia coli DH5α (Kan/Neo selection). Plasmid DNA was collected and purified using Qiagen EndoFree Plasmid Giga kits (Qiagen Inc., Valencia, CA) per manufacturer’s protocol.
2.2.4. Production of DNA nanoparticles
Rod-shaped DNA nanoparticles were manufactured by the drop-wise addition of 9 volumes of DNA solution (0.222 mg/ml in water) to 1 volume CK30PEG solution at a rate of 1 ml/min while vortexing at room temperature. The final ratio of positively charged primary amines (N) contributed by CK30PEG to negatively charged DNA phosphates (P) was 2:1. After incubating at room temperature for 30 minutes, aggregates were removed by syringe filtration (0.2 µm). To remove free polymers and exchange water for physiologic saline, DNA nanoparticles were diluted 10-fold with 0.9% NaCl and re-concentrated to 0.2 mg/ml using a Vivaspin 6 ultrafiltration device (100,000 MWCO, Vivaproducts, Inc., Littleton, MA) two times. For in vivo experiments, DNA nanoparticles were concentrated to 1 mg/ml and administered the same day. For long-term stability studies, DNA nanoparticles were stored at 4°C.
2.3. Physicochemical characterization of DNA nanoparticles
Rod-shaped DNA nanoparticles were tested for quality assurance by transmission electron microscopy, turbidity, and sedimentation, as described previously [7]. Size and ξ-potential (surface charge) were measured by dynamic light scattering and laser Doppler anemometry, respectively, using a Zetasizer Nano ZS90 (Malvern Instruments, Southborough, MA). Samples were diluted to 0.1 mg/ml in 10 mM NaCl pH 7.1 (viscosity 0.8894 cP and Refractive index 1.33 @ 25°C). Size measurements were performed at 25°C at a scattering angle of 90°. The zeta-potential values were calculated using the Smoluchowski equation.
To determine whether DNA nanoparticles remain stable in presence of CF sputum, dual labeled CK30PEG DNA nanoparticles were formulated by labeling DNA with Cy3 (Mirus Bio LLC, Madison, WI) followed by compaction with AF 488-labeled CK30PEG polymers. Dual labeled DNA nanoparticles were incubated in CF sputum for 30 min and the co-localization of the polymer and DNA was monitored via an inverted epifluorescence microscope (Marianas, Zeiss, Thornwood, NY) equipped with dual Cascade II 512 EMCCD cameras (Photometrics, Tucson, AZ).
2.4. Protection against DNase I
Ten microliters containing 0.5 µg of DNA were incubated with 1µl recombinant DNase I (0.5 U; RNase free; Roche Diagnostics GmbH, Mannheim, Germany) at 37°C for varying durations. After DNase treatment, DNase was inactivated by addition of 5 µl 0.5 M EDTA followed by 15 minute incubation at room temperature. To release DNA for gel electrophoresis, DNA nanoparticles were incubated with 1 µl of 0.25% Trypsin at 37 °C for 10 min. Gel electrophoresis was carried out on a 1% agarose gel containing 50 µg/ml ethidium bromide.
2.5. Polystyrene nanoparticle preparation and characterization
Fluorescent carboxyl-modified polystyrene nanoparticles 200 nm in size (Molecular Probes, Eugene, OR) were covalently conjugated with 2 kDa diamine PEG, as described previously [20]. Size and ζ-potential were determined using a Zetasizer Nano ZS90 (Malvern Instruments, Southborough, MA).
2.6. CF sputum collection
Sputum spontaneously expectorated from male and female CF patients ages 21 – 44, was collected at the Johns Hopkins Adult Cystic Fibrosis Program. The procedures conformed to ethical standards and the sputum collection was performed under informed consent on a protocol approved by the Johns Hopkins Medicine Institutional Review Board. Two to three samples were acquired from the weekly CF outpatient clinic, placed on ice upon collection and during transport, pooled together to minimize patient-to-patient variation, and studied the same day. The total number of individual samples used for the present studies was 12.
2.7. Multiple particle tracking in CF sputum
Nanoparticles were added to CF sputum (3% v/v) and equilibrated for 30 min at 37 °C prior to microscopy. The dynamics of nanoparticles were quantified using multiple particle tracking (MPT) [18, 20]. Briefly, 20 s movies at 67 ms temporal resolution were acquired via dual Cascade II 512 EMCCD cameras on an inverted epifluorescence microscope (3-I Marianas, Zeiss; Thornwood, NY) with 100X/1.4 NA objective. Movies were analyzed with Metamorph software (Universal Imaging, Glendale, WI) to extract x, y positional data over time. Time-averaged mean square displacement (MSD) and effective diffusivity (Deff) for individual DNA nanoparticles were calculated as a function of time scale (τ) [21–23]. CF sputum was assumed to be locally isotropic but not necessarily homogeneous; thus, 2D diffusivity is equal to 3D diffusivity [21]. Bulk transport properties were calculated by geometric ensemble-averaging of individual transport rates. The tracking resolution was 10 nm, determined by tracking the displacement of particles immobilized with a strong adhesive [24]. Particle transport mechanism (immobile, hindered, and diffusive) was classified as discussed previously [23]. The fraction of gene carriers expected to penetrate a 10 µm CF sputum layer was calculated as previously reported [18, 25].
2.8. In vivo airway gene transfer
All procedures performed with mice were approved by the Johns Hopkins University Animal Care and Use Committee. BALB/c mice (female, 6–8 weeks old) were anesthetized by intraperitoneal (IP) injection of tribromoethanol (Avertin) at a dose of 250 mg/kg. Pulmonary administration was performed by oropharyngeal aspiration, as previously described [26]. Briefly, anesthetized mice were suspended at a 45° angle by their incisors on a plexiglass stand. The tongue was gently pulled aside and maintained in full extension by forceps. DNA nanoparticles or naked DNA in saline (1 mg/ml) were pipetted (50 µl) at the base of the tongue and tongue restraint was continued until 2 deep breaths were completed. Control mice were treated with saline. To examine airway gene transfer, we measured the luciferase activity of lung tissue homogenates. Mice were euthanized 24 hrs post pulmonary administration. Hearts were perfused with 3 ml PBS and lung tissues were harvested and homogenized in 1ml of reporter lysis buffer (Promega, Madison, WI) using the TissueLyser LT (Qiagen, Valencia, CA). The homogenates were subjected to a freeze-thaw cycle and supernatants were obtained by centrifugation. Luciferase activity in the supernatant was measured using a standard luciferase assay kit (Promega, Madison, WI) and a 20/20n luminometer (Turner Biosystems, Sunnyvale, CA). Each sample was read in duplicate. Data are shown as relative light unit (RLU)/mg protein.
2.9. PEG conformation comparison
The ratio [Γ/SA] of total unconstrained PEG surface area coverage [Γ] to total particle surface area [SA] indicates whether the PEG coating is in a mushroom or brush conformation, [Γ/SA] < 1 or [Γ/SA] ≥ 1, respectively [27]. To determine the conformation of PEG as a function of MW, the surface area occupied by an unconstrained PEG chain was calculated by random-walk statistics and given by a sphere of diameter (ξ)
| (Eqn 1) |
where mb is the molecular weight of the PEG chain [28]. From the sphere diameter, the area occupied at the surface is π(ξ/2)2. Thus, the surface area occupied by unconstrained PEG of MWs 10, 5 and 2 kDa is 45, 23 and 9 nm2, respectively. The total unconstrained PEG surface area coverage [Γ] was then calculated by multiplying the area occupied at the surface per PEG chain by the total number of PEG chains per DNA nanoparticle, estimated by calculating the number of positively charged DNA binding domains (CK30) bound per single plasmid DNA. Kim et al. recently reported that poly(L-lysine)-b-poly(ethylene glycol) binding to DNA is complete at a nitrogen to phosphate ratio (N:P) of 1.5 [29]; our calculations for [Γ] are based on N:P ratios in the range from 1 to 2. Next, the surface area [SA] on a single DNA nanoparticle was calculated from the TEM size data and assuming cylindrical geometry. It is important to note that PEG moieties are not stained by uranyl acetate, thus only the poly-L-lysine/DNA core contribute to the size as measured by TEM [8].
2.10. Statistical Analysis
Statistical analysis of data was performed by one-way analysis of variance (ANOVA) followed by Tukey HSD test using SPSS 18.0 software (SPSS Inc., Chicago, IL). Differences were considered to be statistically significant at a level of P < 0.05.
3. Results
3.1. Physicochemical characterization of rod-shaped DNA nanoparticles
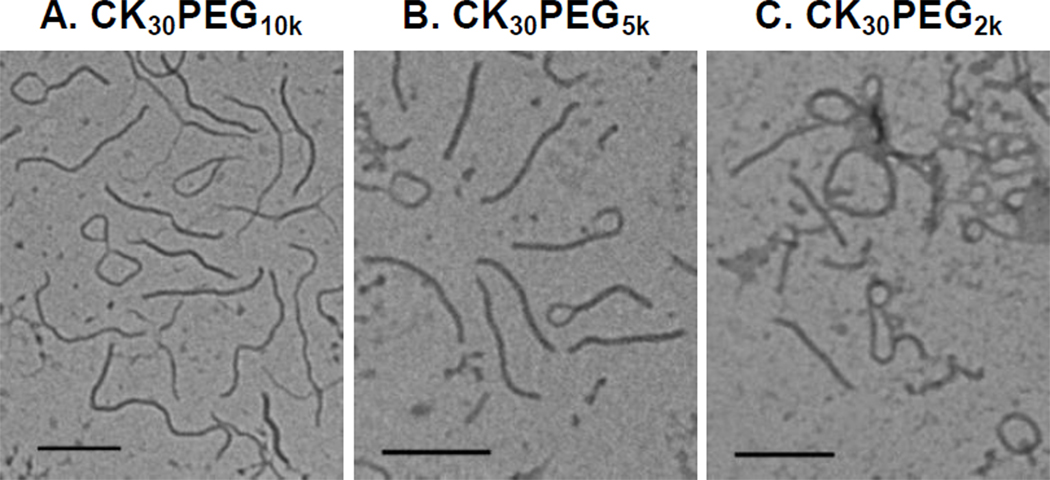
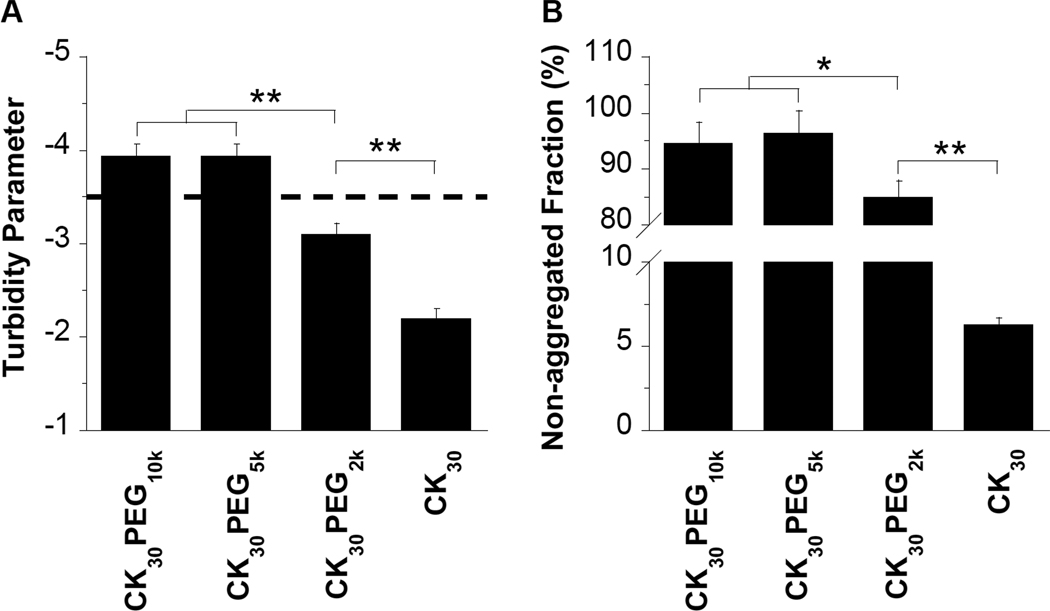
We prepared nanoparticles composed of DNA compacted with CK30 peptide conjugated to PEG of varying MWs (10 kDa, 5 kDa, or 2 kDa). Following purification by syringe filtration (0.2 µm), the yield for CK30PEG10k and CK30PEG5k DNA nanoparticles were both > 95 percent, whereas the yield for CK30PEG2k DNA nanoparticles was ~ 85 percent. Following ultrafiltration, the final formulation yield for CK30PEG10k and CK30PEG5k DNA nanoparticles were both ~ 70 percent, whereas the yield for CK30PEG2k DNA nanoparticles was only ~ 20 percent. Morphological examination via transmission electron microscopy (TEM) revealed that all formulations produced flexible rod like nano-structures, similar to previous reports for CK30PEG10k (Figure 1) [7]. However, a significant fraction of CK30PEG2k DNA nanoparticles remained in small aggregated clusters. There was a statistically significant correlation between PEG MW and DNA nanoparticle length/width; as PEG MW increased, nanoparticle length increased while width decreased. The average dimensions were ~ 220 × 15 nm, ~ 300 × 13 nm and ~ 350 × 11 nm for CK30PEG2k, CK30PEG5k and CK30PEG10k DNA nanoparticles, respectively (Table 1). All formulations had near neutral surface charge (as measured by ζ-potential), whereas DNA compacted with CK30 (i.e. no PEG) exhibited a highly positive surface charge (Table 1). We monitored the colloidal stability in saline after storage at 4°C for 6 months using turbidity and sedimentation assays, dynamic light scattering, and laser Doppler anemometry. CK30PEG10k and CK30PEG5k DNA nanoparticles both exhibited acceptable turbidity parameters indicative of negligible aggregation (Figure 2A), which is in good agreement with results from a sedimentation assay that indicated < 6% of DNA compacted with CK30PEG10k or CK30PEG5k sedimented (Figure 2B). In comparison, CK30PEG2k DNA nanoparticles exhibited a significantly higher turbidity parameter, indicative of particle aggregation, as supported by sedimentation assay results where ~ 15% of DNA compacted with CK30PEG2k sedimented. The size, particle size distribution (PDI), and ξ-potential of CK30PEG2k, CK30PEG5k and CK30PEG10k DNA nanoparticles did not change significantly after storage at 4°C for 6 months (Supplementary Table S1). However, the distribution of particles sizes for CK30PEG2k DNA nanoparticles was significantly higher than that for CK30PEG5k or CK30PEG10k DNA nanoparticles, before and after storage at 4°C.
Figure 1.
Transmission electron microscopy of compacted DNA nanoparticles formulated with (A) CK30PEG10k (B) CK30PEG5k and (C) CK30PEG2k. Scale bar represents 200 nm.
Table 1.
Particle Size and ζ-potentials (surface charge) for various DNA nanoparticles
| Formulation | Length, nm a | Width, nm a | ζ-potential, mV b |
|---|---|---|---|
| CK30PEG10k | 350 ± 7 | 11 ± 1 | −1.5 ± 3.6 |
| CK30PEG5k | 300 ± 11* | 13 ± 0.2 | −1.4 ± 0.6 |
| CK30PEG2k | 220 ± 8* | 15 ± 0.8* | −3.0 ± 6.0 |
| CK30 | n/a | n/a | 25 ± 2.0 |
DNA nanoparticle length and width as measured from TEM images, using ImageJ software package. Data represents the average of 3 independent experiments +/− the standard deviation. * denotes statistical significance (P < 0.05) as compared to CK30PEG10k.
Measured at pH 7.1. Data represents the average of 3 independent experiments +/− the standard deviation.
Figure 2.
Colloidal stability of rod-shaped DNA nanoparticles after storage at 4°C for 6 months. (A) Turbidity parameters, an indicator of colloidal stability, for various DNA nanoparticle formulations. A turbidity parameter less than −3.5 (above the dashed line) indicates DNA nanoparticles that are compacted and non-aggregated, whereas values greater than −3.5 (below the dashed line) indicate significant aggregation of DNA nanoparticles. (B) Non-aggregated fraction of compacted DNA nanoparticles as determined by sedimentation. Data represents the average of 3 independent experiments +/− the standard deviation. * denotes statistical significance (* P < 0.05, ** P < 0.01).
3.2. Protection against DNase I
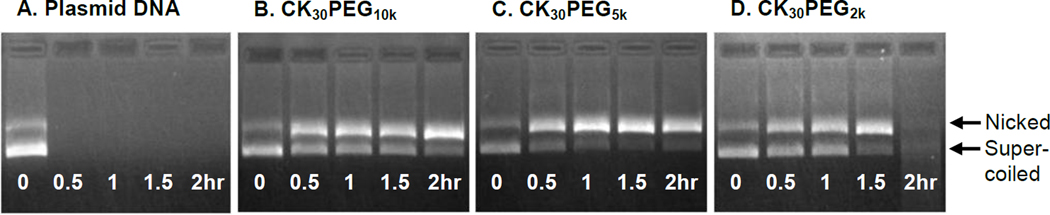
To assess the ability of nanoparticles, composed using PEG of various MWs, to protect DNA against DNase digestion, we incubated free DNA and rod-shaped DNA nanoparticles with DNase I for various durations before gel electrophoresis. Free DNA was completely degraded within 30 minutes of incubation with DNase I (Figure 3). DNA compacted with CK30PEG10k or CK30PEG5k was partially protected from DNase I digestion for at least 2 hrs, as shown by the increasing ratio of nicked vs. supercoiled DNA. In contrast, DNA compacted with CK30PEG2k was completely degraded by 2 hrs.
Figure 3.
Stability against DNase I digestion over time of (A) free DNA, or DNA compacted with (B) CK30PEG10k, (C) CK30PEG5k, or (D) CK30PEG2k. Upper and lower bands represent nicked circular and supercoiled conformations of DNA.
3.3. Gene transfer in mice
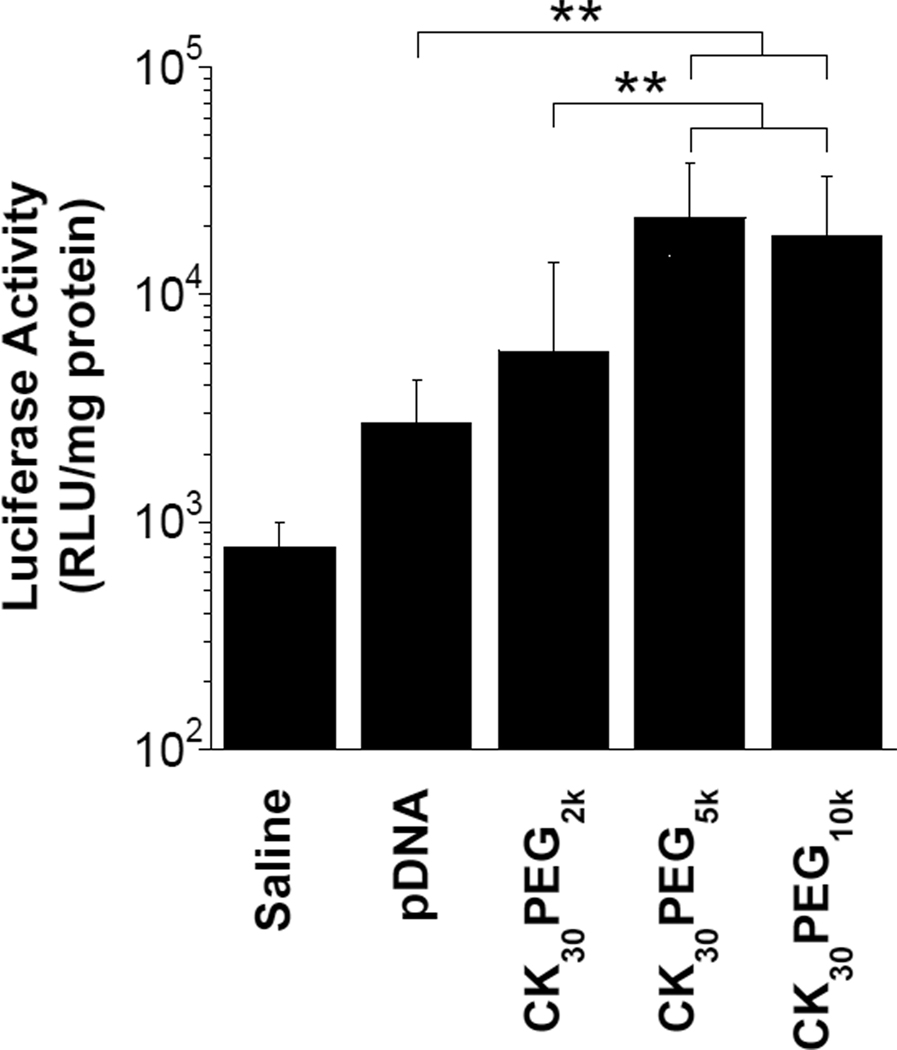
To determine in vivo gene transfer efficiency, we administered various rod-shaped DNA nanoparticles composed of compacted pd1GL3-RL plasmids encoding luciferase via oropharyngeal aspiration to the lungs of BALB/c mice. We first tested CK30PEG10k DNA nanoparticles at a dose of 50 µg DNA per mouse to determine the duration of luciferase gene expression. We observed maximum luciferase enzyme activity at 24 hrs post administration, with declining activities at later time points likely due to silencing of the CMV promoter (data not shown). Mice receiving either CK30PEG10k or CK30PEG5k DNA nanoparticles exhibited the highest overall expression of luciferase (Figure 4), significantly higher (P < 0.01) than both naked DNA and CK30PEG2k DNA nanoparticles. The reduced gene transfer by CK30PEG2k DNA nanoparticles may be a result of their increased susceptibility to nuclease attack and/or their tendency to aggregate, which may hinder efficient cellular uptake and/or trafficking to the nucleus.
Figure 4.
In vivo gene expression following pulmonary administration. BALB/c mice (n = 8) received 50 µg compacted or naked pd1GL3-RL DNA in saline by oropharyngeal aspiration. Control mice (n=6) received saline alone. Luciferase activity in lung homogenates was measured 24 hrs after pulmonary administration. Data represents the average +/− the standard deviation. * denotes statistical significance (** P < 0.01).
3.4. Effect of PEG MW on the diffusion of rod-shaped DNA nanoparticles in CF sputum
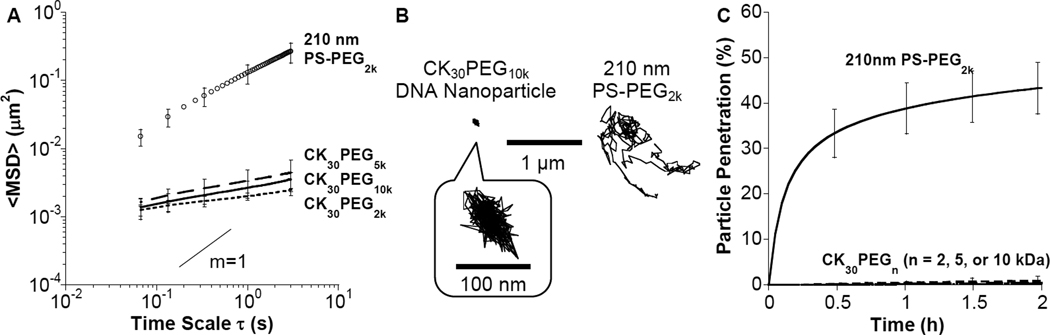
DNA nanoparticles were formulated with CK30PEG covalently conjugated with Alexa Fluor 488 (AF488) to allow measurement of their diffusion rates in freshly expectorated human CF sputum. The fluorophores were conjugated to the ε-amines on the CK30 peptide and, hence, likely localized only to the core and not the surface of compacted DNA nanoparticles. Physicochemical characterization of AF488 labeled DNA nanoparticles, including TEM (Supplementary Figure S1), ξ-potential and sedimentation (data not shown), suggested the fluorophore did not alter nanoparticle physicochemical properties. We used multiple particle tracking (MPT) to measure the diffusion rates of hundreds of individual fluorescent DNA nanoparticles in purulent sputum samples freshly expectorated by CF patients. Co-localization experiments with Cy3 labeled DNA confirmed that DNA nanoparticles remained intact in CF sputum (Supplementary Figure S2). Despite having a roughly 4-fold smaller hydrodynamic diameter, the diffusion of DNA nanoparticles, regardless of PEG MW, was 40-fold slower in CF sputum than 210 nm polystyrene nanoparticles densely coated with 2 kDa diamine PEG (PS-PEG2k), as measured by geometric ensemble mean-squared displacements (<MSD>) at a time scale, τ, of 1 second (Figure 5A). Overall, the effective diffusivity (Deff) of DNA nanoparticles at τ = 1s was reduced by over 5,000-fold as compared to the theoretical diffusivity of particles with the same hydrodynamic diameter (~ 60 – 80 nm) in water (Table 2). By fitting <MSD> to the equation, MSD = 4Doτα, where Do is the time scale-independent diffusion coefficient, τ is the time scale, and α ranges from 0 (completely immobile particles) to 1 (unobstructed Brownian diffusion, such as that of a particle in water), the extent of impediment to particle diffusion can be determined. Average α values were 0.24, 0.25, and 0.17 for CK30PEG10k, CK30PEG5k and CK30PEG2k compacted DNA nanoparticles, respectively, indicative of highly obstructed transport, whereas 210 nm PS-PEG2k nanoparticles had an average α value of 0.71 in the same sputum samples.
Figure 5.
Transport of rod-shaped DNA nanoparticles and PEG coated polystyrene (PS-PEG2k) nanoparticles in fresh, undiluted CF sputum. (A) Ensemble-averaged geometric mean square displacement (<MSD>) of compacted DNA nanoparticles and PS-PEG2k in CF sputum as a function of time scale (τ). The slope of m = 1 corresponds to unobstructed diffusive behavior. Data represents 3 independent experiments, with n ≥120 particles per experiment. Error bars represent standard error of the mean. (B) Representative trajectories of CK30PEG10k DNA nanoparticles and 210 nm PS-PEG2k particles in CF sputum during 20 s movies. (C) The fraction of particles predicted to penetrate a 10 µm think CF sputum layer over time using Fick’s second law and diffusion coefficients obtained from tracking experiments.
Table 2.
Characterization of nanoparticle PEG surface coverage and nanoparticle diffusivity in human CF sputum (Deff) compared to in water (Dw)
| Formulation | Hydrodynamic Diameter a (nm) |
PEG Surface Density b (molecules/100 nm2) |
[Γ/SA] c | PEG Regime d | Dw/Deffe |
|---|---|---|---|---|---|
| CK30PEG10k | 60 | 4.5 ± 0.4 – 9.0 ± 0.8 | 2.1 ± 0.2* – 4.1 ± 0.3* | Brush | 6,000 |
| CK30PEG5k | 56 | 4.3 ± 0.2 – 8.6 ± 0.4 | 0.98 ± 0.05* – 2.0 ± 0.1* | Brush | 5,000 |
| CK30PEG2k | 81 | 5.0 ± 0.1 – 10 ± 0.2 | 0.45 ± 0.01 – 0.9 ± 0.02 | Mushroom | 6,000 |
| PS-PEG2k | 210 | 120 ± 4** | 11 ± 1** | Dense Brush | 30 |
Hydrodynamic diameter measured by dynamic light scattering.
For DNA nanoparticles, PEG surface density calculated assuming stoichiometric ratios of poly-L-lysine ε-amines to DNA phosphate groups (N:P) in the range from 1 to 2. For PS-PEG2k, PEG surface coverage reported previously [19]. ** denotes statistical significance (P < 0.01) as compared to all other groups.
Ratio of total unconstrained PEG surface area coverage [Γ] to total particle surface area [SA]. For DNA nanoparticles, [Γ/SA] calculated assuming N:P ratios in the range from 1 to 2. * denotes statistical significance (P < 0.05) as compared to CK30PEG2k at the same N:P ratio. ** denotes statistical significance (P < 0.01) as compared to all other groups.
PEG regime defined as being mushroom coverage for [Γ/SA] < 1 and brush coverage for [Γ/SA] > 1, as illustrated in Figure 6.
Dw is the theoretical diffusivity of particles in water calculated from the Stokes-Einstein equation and the hydrodynamic diameter measured by dynamic light scattering. Deff is the effective diffusivity in CF sputum measured at a time scale of 1 s. The Dw/Deff ratio indicates by what multiple the average particle movement rate in fresh CF sputum is slower than in pure water.
To further understand the mechanism of particle transport, we assigned the diffusion of DNA nanoparticles to three non-overlapping transport modes; immobile (I), hindered (H), and diffusive (D) (where the rates of movement ~ D > H > I), based on their time scale-dependent effective diffusion coefficients [20]. For all DNA nanoparticle formulations, the large majority of particles (> 99%) were either immobilized or hindered by CF sputum, as indicated by their highly constrained non-Brownian time lapse traces (Figure 5B). Using Fick’s second law of diffusion [18, 25] and the measured Deff of hundreds of DNA nanoparticles, we estimated that <1% of DNA nanoparticles are expected to penetrate a 10 µm thick CF sputum layer within 2 hrs, compared to ~ 40% for PS-PEG2k nanoparticles (Figure 5C).
3.5. Effect of shape on the diffusion of compacted DNA nanoparticles in CF sputum
To determine if rod-shaped CK30PEG DNA nanoparticles were slowed by adhesive interactions or steric obstruction with the sputum mesh, we also measured the mobility of rhodamine labeled ellipsoidal DNA nanoparticles that possessed major diameters (~ 40 nm) much smaller than the average CF sputum 3D mesh spacing (provided by Copernicus Therapeutics, Cleveland, OH) [8]. Physicochemical characterization of rhodamine labeled DNA nanoparticles, including TEM, ξ-potential and sedimentation, suggested the fluorophore did not alter nanoparticle physicochemical properties (data not shown). Ellipsoidal and rod-shaped DNA nanoparticles had similar hydrodynamic diameters (~ 60 nm) and near neutral surface charge (~ −2 mV). The diffusion of DNA nanoparticles, regardless of shape, was 60-fold slower in CF sputum than 210 nm PS-PEG2k nanoparticles as measured by <MSD> at a time scale, τ, of 1 second (Supplementary Figure S3). Overall, the effective diffusivity (Deff) of DNA nanoparticles at τ = 1s was reduced by over 7,000-fold as compared to the theoretical diffusivity of particles with the same hydrodynamic diameter (~ 60 nm) in water. Average α values were 0.11 and 0.14 for ellipsoidal and rod-shaped DNA nanoparticles, respectively, indicative of highly obstructed transport. The large majority of particles (> 99%), regardless of shape, were either immobilized or hindered by CF sputum. Using Fick’s second law of diffusion and the measured Deff of hundreds of DNA nanoparticles, less than 1% of ellipsoidal or rod-shaped DNA nanoparticles are expected to penetrate a 10 µm thick CF sputum layer within 2 hrs.
4. Discussion
Hyperviscoelastic sputum coating the airways of CF patients has been cited as a critical barrier to successful CF gene therapy [13]. This is highlighted by our finding that the only polymeric system currently being tested clinically in CF patients, the CK30PEG10k DNA nanoparticles, are trapped in CF sputum, thereby precluding the carriers from efficiently reaching the airway epithelium. We sought to create DNA nanoparticles with low MW PEG coatings by substituting 10 kDa PEG in CK30PEG10k with 5 kDa and 2 kDa PEG. We were able to formulate CK30PEG5k DNA nanoparticles that were stable at 4°C for at least 6 months, resistant against DNase attack and transfected the lungs of mice as well as CK30PEG10k DNA nanoparticles. However, despite having low MW PEG coatings, CK30PEG5k and CK30PEG2k DNA nanoparticles were trapped in CF sputum to the same extent as CK30PEG10k DNA nanoparticles.
DNA nanoparticles may be trapped in CF sputum via steric and/or adhesive interactions. While the length of the shortest CK30PEG2k DNA nanoparticles (~ 220 nm) is greater than the average CF sputum 3D mesh spacing of ~ 140 ± 50 nm [18], well-PEGylated PS-PEG2k nanoparticles 210 nm in diameter exhibited markedly higher diffusion rates in the same sputum samples, presumably due to the large range of pore sizes found in CF sputum (60 – 300 nm) [18]. Thus, it is likely that CK30PEG2k DNA nanoparticles are trapped predominantly by adhesive interactions rather than by steric obstruction. Given their greater lengths, CK30PEG5k and CK30PEG10k DNA nanoparticles may be trapped in CF sputum by a combination of steric and adhesive interactions. If steric interactions were responsible for trapping rod-shaped DNA nanoparticles, then ellipsoidal-shaped DNA nanoparticles, with a major diameter (~ 40 nm) much less than the average CF sputum 3D mesh spacing [8], would be expected to have greater mobility. However, ellipsoidal-shaped CK30PEG10k DNA nanoparticles were trapped in CF sputum to the same extent as rod-shaped CK30PEG10k DNA nanoparticles. Furthermore, we recently discovered that N-acetylcysteine (NAC), a mucolytic that increases the average CF sputum 3D mesh spacing [30], enhanced the penetration of rod-shaped CK30PEG10k DNA nanoparticles through CF sputum [unpublished data]. Nevertheless, a large fraction of CK30PEG10k DNA nanoparticles remained hindered in CF sputum treated with NAC. These results suggest that adhesive interactions with sputum constituents are most likely the primary factor responsible for the limited mobility of these DNA nanoparticles.
That compacted DNA nanoparticles are likely immobilized by adhesive interactions with sputum constituents suggests that insufficient PEG surface coverage exists on the CK30PEG DNA nanoparticles. Insufficient PEG coverage may allow sputum components to adhere to the poly-L-lysine/DNA core via hydrogen bonding, van der Waals forces, polymer chain interpenetration, hydrophobic forces, and/or electrostatic interactions [31]. We estimated the PEG surface density on rod-shaped DNA nanoparticles in this work to be ~ 4 – 10 molecules/100 nm2 (Table 2), which is roughly 12 – 30-fold less than that estimated to be on sputum-penetrating 210 nm PS-PEG2K nanoparticles. Due to a decrease in surface area by a factor of ~ 2 for ellipsoidal versus rod-shaped DNA nanoparticles, the PEG surface density on ellipsoidal-shaped DNA nanoparticles is expected to be ~ 8 – 20 molecules/100 nm2. Previously, Wang et al. reported that as little as a 40% reduction in PEG coverage led to a 700-fold decrease in the average diffusivity of 200 nm PS-PEG2K nanoparticles in cervicovaginal mucus [19]. These results suggest that inadequate PEG coatings achieved on DNA nanoparticles, regardless of PEG MW or particle shape, are responsible for particle trapping via mucoadhesion.
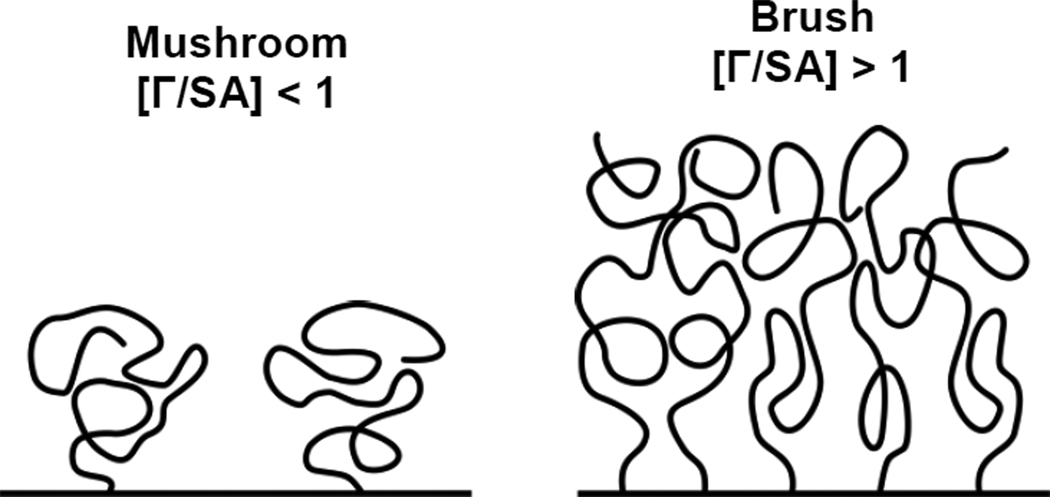
We sought to gain improved mechanistic insight into how PEG density may alter the conformation of PEG on the particle surface and, consequently, colloidal stability and mucoaffinity. If the average distance between neighboring PEG chains on a nanoparticle surface is greater than the Flory radius (RF ~ aN3/5, where N is the degree of polymerization and a is the effective monomer length) of the PEG chain, neighboring PEG chains do not overlap and are in the so-called “mushroom” conformational regime (as illustrated in Figure 6) [32]. In this regime [Γ/SA] < 1, where Γ is the total unconstrained PEG surface area coverage and [SA] is the total particle surface area. However, when the grafting density or PEG MW increases to an extent such that neighboring PEG chains overlap ([Γ/SA] > 1), the PEG chains stretch away from the surface, forming a “brush” layer, due to excluded-volume effects [32]. The least stable CK30PEG2k DNA nanoparticles displayed PEG coatings in the mushroom conformation, as indicated by [Γ/SA] ≈ 0.45 - 0.9 (Table 2). Increasing the PEG MW to 5 kDa significantly (P < 0.05) improved the PEG coating to the brush regime ([Γ/SA] ≈ 0.98 - 2.0), providing improved yield, colloidal stability, DNase resistance and in vivo transfection efficiency. DNA nanoparticles formulated with CK30PEG10k also displayed PEG coatings in the brush regime ([Γ/SA] ≈ 2.1 - 4.1). For comparison, muco-inert 200 nm PS-PEG2K nanoparticles ([Γ/SA] ≈ 11) and mucoadhesive PS-PEG2K nanoparticles ([Γ/SA] ≈ 7) both have PEG coatings in the very dense PEG brush regime [19]. These results indicate that while a brush conformation is adequate for colloidal stability and protection against DNase, a very dense PEG brush coating, in excess of [Γ/SA] ≈ 7 for latex particles, may be required to resist mucoadhesion. It is noted that the precise [Γ/SA] required to resist mucoadhesion will likely depend on the material(s) forming the core of the particle.
Figure 6.
Schematic illustration of PEG regimes. The mushroom regime exists when neighboring PEG chains do not overlap, [Γ/SA] < 1, where Γ is the total unconstrained PEG surface area coverage and [SA] is the total particle surface area [27]. The brush regime exists when neighboring PEG chains overlap, [Γ/SA] > 1, and stretch away from the surface.
Achieving PEG surface densities on DNA nanoparticles at levels comparable to PS-PEG2K particles represents the next step towards developing DNA nanoparticles that can readily penetrate CF sputum. In addition, the affects of high PEG surface density on DNA nanoparticle mediated gene transfer need to be determined, as it is possible that high PEG surface density may alter the cellular uptake, for example through binding of DNA nanoparticles to nucleolin, a cell surface receptor that appears to be involved in CK30PEG DNA nanoparticle uptake and trafficking to the nucleus [9, 33]. However, it is unclear whether additional PEG can be incorporated without markedly altering the stability or morphology of CK30PEG DNA nanoparticles. The presence of too much PEG may inhibit the stability of DNA nanoparticles if the electrostatic interactions between CK30 and DNA are not sufficient to maintain the association of PEG to the particle surface. In addition, high PEG content may shield the electrostatic charge on CK30 and interfere with DNA compaction. In the present study, morphological analysis of DNA nanoparticles via TEM revealed an interesting correlation between PEG conformation and the dimensions of the elongated rod like structures (Figure 1). As PEG conformation transitioned from mushroom to brush, the length of DNA nanoparticles significantly increased while the width decreased (Table 1). This “stretching” phenomenon may be attributed to repulsive forces between neighboring PEG chains on the nanoparticle surface. The hydrophilic and flexible nature of PEG chains is responsible for its extended conformation as a free polymer in solution [34], a maximum entropy state. However, when PEG containing polymers are compacted into nanoparticles, the PEG chains are forced to assume a higher energy conformation with increasing entropic penalty for greater [Γ/SA]. Thus, as PEG MW increases, the corresponding increase in repulsive forces may cause the carriers to elongate, leading to larger surface areas that minimize the entropic penalty from less favorable PEG conformations. This suggests the morphology of DNA nanoparticles may be further elongated as PEG surface density increases, leading to the formation of long carriers that may be trapped by steric obstruction in CF sputum. This work underscores the importance of considering particle morphology in the development of DNA nanoparticles for CF.
In summary, despite having a surface-density of low MW PEG sufficient to provide long-term (6 month) colloidal stability, stability against nuclease attack, and effective gene delivery to mouse airways, the CK30PEG5k DNA nanoparticles developed were unable to diffuse through human CF sputum. Insufficient PEG coverage on the DNA nanoparticles, as compared to mucus-penetrating PS-PEG2K nanoparticles, suggests that inadequate PEG surface density is the critical limiting factor for the development of sputum penetrating gene carriers. To achieve PEG surface densities comparable to PS-PEG2K nanoparticles, the amount of PEG on DNA nanoparticles may need to be increased perhaps as much as 30-fold or more. In addition to treating CF lung airway disease, the development of mucus penetrating DNA nanoparticles may find use for the local delivery of therapeutic DNA at various mucosal sites.
Supplementary Material
Acknowledgements
Funding was provided by the National Institutes of Health (NIH R01EB003558 and P01HL51811) and a postdoctoral fellowship from the Croucher Foundation to S.K.L. We thank Professor Alexander Klibanov (Department of Chemistry, the Massachusetts Institute of Technology) for providing the plasmid pd1GL3-RL. We also thank Meghan Ramsay and Sharon Watts at the Johns Hopkins Adult Cystic Fibrosis Center for cystic fibrosis sputum collection. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Biomedical Imaging and Bioengineering, the National Heart, Lung, and Blood Institute, or the National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Pringle IA, Hyde SC, Gill DR. Non-viral vectors in cystic fibrosis gene therapy: recent developments and future prospects. Expert opinion on Biological Therapy. 2009;9:991–1003. doi: 10.1517/14712590903055029. [DOI] [PubMed] [Google Scholar]
- 2.Welsh MJ, Smith AE. Molecular mechanisms of CFTR chloride channel dysfunction in cystic fibrosis. Cell. 1993;73:1251–1254. doi: 10.1016/0092-8674(93)90353-r. [DOI] [PubMed] [Google Scholar]
- 3.Donaldson SH, Bennett WD, Zeman KL, Knowles MR, Tarran R, Boucher RC. Mucus Clearance and Lung Function in Cystic Fibrosis with Hypertonic Saline. New Engl. J. Med. 2006;354:241–250. doi: 10.1056/NEJMoa043891. [DOI] [PubMed] [Google Scholar]
- 4.Tarran R, Button B, Picher M, Paradiso AM, Ribeiro CM, Lazarowski ER, Zhang L, Collins PL, Pickles RJ, Fredberg JJ, Boucher RC. Normal and Cystic Fibrosis Airway Surface Liquid Homeostasis. J. Biol. Chem. 2005;280:35751–35759. doi: 10.1074/jbc.M505832200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsui H, Verghese MW, Kesimer M, Schwab UE, Randell SH, Sheehan JK, Grubb BR, Boucher RC. Reduced three-dimensional motility in dehydrated airway mucus prevents neutrophil capture and killing bacteria on airway epithelial surfaces. J. Immunol. 2005;175:1090–1099. doi: 10.4049/jimmunol.175.2.1090. [DOI] [PubMed] [Google Scholar]
- 6.Matsui H, Wagner VE, Hill DB, Schwab UE, Rogers TD, Button B, Taylor Ii RM, Superfine R, Rubinstein M, Iglewski BH, Boucher RC. A physical linkage between cystic fibrosis airway surface dehydration and Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. U. S. A. 2006;103:18131–18136. doi: 10.1073/pnas.0606428103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziady AG, Gedeon CR, Miller T, Quan W, Payne JM, Hyatt SL, Fink TL, Muhammad O, Oette S, Kowalczyk T, Pasumarthy MK, Moen RC, Cooper MJ, Davis PB. Transfection of airway epithelium by stable PEGylated poly-L-lysine DNA nanoparticles in vivo. Mol. Ther. 2003;8:936–947. doi: 10.1016/j.ymthe.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 8.Liu G, Li D, Pasumarthy MK, Kowalczyk TH, Gedeon CR, Hyatt SL, Payne JM, Miller TJ, Brunovskis P, Fink TL, Muhammad O, Moen RC, Hanson RW, Cooper MJ. Nanoparticles of compacted DNA transfect postmitotic cells. J. Biol. Chem. 2003;278:32578–32586. doi: 10.1074/jbc.M305776200. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Kube DM, Cooper MJ, Davis PB. Cell Surface Nucleolin Serves as Receptor for DNA Nanoparticles Composed of Pegylated Polylysine and DNA. Mol Ther. 2007;16:333–342. doi: 10.1038/sj.mt.6300365. [DOI] [PubMed] [Google Scholar]
- 10.Konstan MW, Davis PB, Wagener JS, Hilliard KA, Stern RC, Milgram LJ, Kowalczyk TH, Hyatt SL, Fink TL, Gedeon CR, Oette SM, Payne JM, Muhammad O, Ziady AG, Moen RC, Cooper MJ. Compacted DNA nanoparticles administered to the nasal mucosa of cystic fibrosis subjects are safe and demonstrate partial to complete cystic fibrosis transmembrane regulator reconstitution. Hum. Gene Ther. 2004;15:1255–1269. doi: 10.1089/hum.2004.15.1255. [DOI] [PubMed] [Google Scholar]
- 11.Garcia-Contreras L, Hickey AJ. Aerosol treatment of cystic fibrosis. Crit. Rev. Ther. Drug Carrier Syst. 2003;20:317–356. doi: 10.1615/critrevtherdrugcarriersyst.v20.i5.10. [DOI] [PubMed] [Google Scholar]
- 12.Laube BL. The expanding role of aerosols in systemic drug delivery gene therapy, and vaccination. Respiratory care. 2005;50:1161–1176. [PubMed] [Google Scholar]
- 13.Montier T, Delépine P, Pichon C, Férec C, Porteous DJ, Midoux P. Non-viral vectors in cystic fibrosis gene therapy: progress and challenges. Trends in Biotechnology. 2004;22:586–592. doi: 10.1016/j.tibtech.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Sanders N, Rudolph C, Braeckmans K, De Smedt SC, Demeester J. Extracellular barriers in respiratory gene therapy. Adv. Drug Del. Rev. 2009;61:115–127. doi: 10.1016/j.addr.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanders N, De Smedt SC, Van Rompaey E, Simoens P, De Baets F, Demeester J. Cystic Fibrosis Sputum. A Barrier to the Transport of Nanospheres. Am. J. Respir. Crit. Care Med. 2000;162:1905–1911. doi: 10.1164/ajrccm.162.5.9909009. [DOI] [PubMed] [Google Scholar]
- 16.Braeckmans K, Peeters L, Sanders NN, De Smedt SC, Demeester J. Three-Dimensional Fluorescence Recovery after Photobleaching with the Confocal Scanning Laser Microscope. Biophys. J. 2003;85:2240–2252. doi: 10.1016/s0006-3495(03)74649-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dawson M, Wirtz D, Hanes J. Enhanced Viscoelasticity of Human Cystic Fibrotic Sputum Correlates with Increasing Microheterogeneity in Particle Transport. J. Biol. Chem. 2003;278:50393–50401. doi: 10.1074/jbc.M309026200. [DOI] [PubMed] [Google Scholar]
- 18.Suk JS, Lai SK, Wang Y-Y, Ensign LM, Zeitlin PL, Boyle MP, Hanes J. The penetration of fresh undiluted sputum expectorated by cystic fibrosis patients by non-adhesive polymer nanoparticles. Biomaterials. 2009;30:2591–2597. doi: 10.1016/j.biomaterials.2008.12.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang YY, Lai SK, Suk JS, Pace A, Cone R, Hanes J. Addressing the PEG mucoadhesivity paradox to engineer nanoparticles that "slip" through the human mucus barrier. Angew. Chem. Int. Ed. Engl. 2008;47:9726–9729. doi: 10.1002/anie.200803526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lai SK, O'Hanlon DE, Harrold S, Man ST, Wang Y-Y, Cone R, Hanes J. Rapid transport of large polymeric nanoparticles in fresh undiluted human mucus. Proc. Natl. Acad. Sci. U. S. A. 2007;104:1482–1487. doi: 10.1073/pnas.0608611104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suh J, Dawson M, Hanes J. Real-time multiple-particle tracking: applications to drug and gene delivery. Adv. Drug Del. Rev. 2005;57:63–78. doi: 10.1016/j.addr.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 22.Suh J, Wirtz D, Hanes J. Efficient active transport of gene nanocarriers to the cell nucleus. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3878–3882. doi: 10.1073/pnas.0636277100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suk JS, Suh J, Lai SK, Hanes J. Quantifying the intracellular transport of viral and nonviral gene vectors in primary neurons. Exp Biol Med. 2007;232:461–469. [PubMed] [Google Scholar]
- 24.Apgar J, Tseng Y, Fedorov E, Herwig MB, Almo SC, Wirtz D. Multiple-particle tracking measurements of heterogeneities in solutions of actin filaments and actin bundles. Biophys J. 2000;79:1095–1106. doi: 10.1016/S0006-3495(00)76363-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang BC, Dawson M, Lai SK, Wang Y-Y, Suk JS, Yang M, Zeitlin P, Boyle MP, Fu J, Hanes J. Biodegradable polymer nanoparticles that rapidly penetrate the human mucus barrier. Proc. Natl. Acad. Sci. U. S. A. 2009;106:19268–19273. doi: 10.1073/pnas.0905998106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rao GV, Tinkle S, Weissman DN, Antonini JM, Kashon ML, Salmen R, Battelli LA, Willard PA, Hoover MD, Hubbs AF. Efficacy of a technique for exposing the mouse lung to particles aspirated from the pharynx. J Toxicol Environ Health A. 2003;66:1441–1452. doi: 10.1080/15287390306417. [DOI] [PubMed] [Google Scholar]
- 27.Auguste DT, Furman K, Wong A, Fuller J, Armes SP, Deming TJ, Langer R. Triggered release of siRNA from poly(ethylene glycol)-protected, pH-dependent liposomes. J. Controlled Release. 2008;130:266–274. doi: 10.1016/j.jconrel.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brandrup J, Immergut EH. Polymer Handbook. 3rd ed. New York: Wiley and Sons; 1989. [Google Scholar]
- 29.Kim W, Yamasaki Y, Jang W-D, Kataoka K. Thermodynamics of DNA Condensation Induced by Poly(ethylene glycol)-block-polylysine through Polyion Complex Micelle Formation. Biomacromolecules. 2010;11:1180–1186. doi: 10.1021/bm901305p. [DOI] [PubMed] [Google Scholar]
- 30.Suk JS, Lai SK, Boylan NJ, Dawson MR, Boyle MP, Hanes J. Rapid transport of muco-inert nanoparticles in cystic fibrosis sputum treated with N-acetyl cysteine. Nanomedicine (Lond) 2011;6:365–375. doi: 10.2217/nnm.10.123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lai SK, Wang Y-Y, Hanes J. Mucus-penetrating nanoparticles for drug and gene delivery to mucosal tissues. Adv. Drug Del. Rev. 2009;61:158–171. doi: 10.1016/j.addr.2008.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeRouchey J, Walker GF, Wagner E, Rädler JO. Decorated Rods: A "Bottom-Up" Self-Assembly of Monomolecular DNA Complexes. J. Phys. Chem. B. 2006;110:4548–4554. doi: 10.1021/jp053760a. [DOI] [PubMed] [Google Scholar]
- 33.Chen X, Shank S, Davis PB, Ziady AG. Nucleolin-Mediated Cellular Trafficking of DNA Nanoparticle Is Lipid Raft and Microtubule Dependent and Can Be Modulated by Glucocorticoid. Mol Ther. 2011;19:93–102. doi: 10.1038/mt.2010.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Owens DE, III, Peppas NA. Opsonization, biodistribution, and pharmacokinetics of polymeric nanoparticles. Int. J. Pharm. 2006;307:93–102. doi: 10.1016/j.ijpharm.2005.10.010. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.