Abstract
The polyketide antibiotic frenolicin B harbors a biosynthetically intriguing benzoisochromanequinone core, and has been shown to exhibit promising antiparasitic activity against Eimeria tenella. To facilitate further exploration of its chemistry and biology, we constructed a biosynthetic route to frenolicin B in the heterologous host Streptomyces coelicolor CH999, despite the absence of key enzymes in the identified frenolicin gene cluster. Together with our understanding of the underlying polyketide biosynthetic pathway, this heterologous production system was exploited to produce analogs modified at the C15 position. Both the natural product and these analogs inhibited the growth of Toxoplasma gondii in a manner that reveals sensitivity to the length of the C15 substituent. The ability to construct a functional biosynthetic pathway, despite a lack of genetic information, illustrates the feasibility of a modular approach to engineering medicinally relevant polyketide products.
Introduction
Type II polyketide synthases (PKSs) catalyze the biosynthesis of the carbon skeletons of numerous polyfunctional aromatic natural products from the actinomycetes, including the clinically important antibiotic oxytetracycline and the anticancer agent doxorubicin 1,2. Recent analysis has demonstrated considerable functional modularity within these multienzyme assemblies, which in turn can be exploited to alter priming units and change polyketide backbone length 3,4. At the same time, the substrate flexibility of late-stage tailoring enzymes in these natural product pathways can also be exploited to create designer molecules 4,5. Here we have harnessed each of these modular elements to reconstruct a chimeric pathway to the antiparasitic antibiotic frenolicin B (1b) as well as targeted analogs in a heterologous host. Biological evaluation of our analogs against Toxoplasma gondii has revealed the strong sensitivity of this family of antibiotics to the identity of the C-15 substituent, thereby highlighting a new direction for future antiparasitic drug design.
The benzoisochromanequinone (BIQ) antibiotic frenolicin B, produced by the actinomycete Streptomyces roseofulvus, has been extensively investigated as an anticoccidial agent owing to its potent activity against Eimeria tenella 6–9. Nonetheless, low fermentation titers 10 and challenging synthetic procedures 11,12 appear to have limited further development of this promising anti-infective lead substance. We sought to address both of these problems by reconstructing a biosynthetic pathway to frenolicin B in the heterologous host Streptomyces coelicolor CH999, from which the entire actinorhodin biosynthetic gene cluster has been deleted 13.
Materials and Methods
General
S. coelicolor CH999/pBOOST* 14, which lacks the complete actinorhodin (act) gene cluster, was used as the host for production of polyketides. The plasmid pBOOST* has been shown to cointegrate with vectors containing the SCP2* origin of replication, thereby resulting higher copy numbers and correspondingly improved antibiotic titers14. The transformation of shuttle vectors bearing BIQ pathway genes into S. coelicolor CH999/pBOOST* were performed following standard proceedure15. Deuterated solvents for NMR experiments were purchased from Cambridge Isotope Laboratories, Inc. (Andover, MA) and all other solvents were purchased from Fisher Scientific (Pittsburgh, PA) at the highest available grade. 1H and 13C NMR spectra of purified polyketide products were recorded on Varian Inova 500 or 600 MHz instrument in CD3OD or DMSO-d6. The 1H NMR spectra were referenced to the solvent peak at 3.31 ppm for CD3OD or 2.50 ppm for DMSO-d6. The 13C NMR spectra were referenced to solvent at 49.0 ppm for CD3OD or 39.52 for DMSO-d6. Single-bond 1H -13C, multiple-bond 1H - 1H and 1H -13C connectivity was determined by HSQC, COSY and HMBC respectively on Varian Inova-600 NMR instrument. Mass spectra were obtained by electrospray ionization (ESI) at the Vincent Coates Foundation Mass Spectrometry Laboratory at Stanford University.
Construction of Shuttle Vectors
All cloning steps were performed in either E. coli XL1-Blue (Stratagene, La Jolla, CA), or E. coli DH5α (MCLab, South San Francisco, CA). Plasmids used for cloning included pUC18 (New England Biolabs, Ipswich, MA) and pET28b (Novagen, now EMD Bioscience, Madison, WI). Expression vectors were constructed using standard molecular biology techniques. See the supporting information for a detailed description of the primers and construction.
Production, Isolation, and Characterization of Polyketide Products
Each transformed strain was grown on R5 agar plates containing 50 mg/l thiostrepton and 100 mg/l of apramycin at 30°C for 7 days 15, after which metabolites were extracted with either 100% EtOAc or EtOAc/MeOH/Acetic Acid (89:10:1). This crude material was purified by a variety of methods including preparative HPLC and NaHCO3 extraction. Structural determination was performed using a variety of 2D heteronuclear NMR experiments and LC/MS. See the SI for full details.
Toxoplasma gondii Growth Inhibition
Tachyzoites of T. gondii strain RHΔhxgprt YFP 16 were harvested from human foreskin fibroblast (HFF) monolayers grown in 25 cm2 T-flasks. These parasites were then inoculated into a 96-well culture plate containing a confluent layer of HFF cells with ~8×103 tachyzoites/0.32 cm2 well). Parasites were pre-incubated with HFF cells for 6h at 37 °C, 5% CO2. After pre-incubation, the media was exchanged for fresh DMEM complete ((Dulbecco’s modified eagle medium) supplemented with 1% PS (penicillin-streptomycin) and 10 % fetal bovine serum) containing a given concentration of antiparasitic agent. Cultures were then incubated for 72 hours at 37 °C, in 5% CO2. Parasite growth inhibition was monitored in a SpectraMax Gemini EM fluorescent plate reader (Sunnyvale, CA) at 510 nm excitation and 540 nm emission. ED50s were determined using GraphPad Prism (La Jolla, CA).
Cytotoxicity Assay
HFF cell cytotoxicity was determined using a MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxylmethoxylphenyl)-2H-tetrazolium) assay (CellTiter 96 Aqueous One Solution Assay; Promega). HFF cells (2 × 103 cells/well) were grown in a 96 well plate until confluence (~48 hours) in phenol red free DMEM complete at 37 °C, 5% CO2. Varying concentrations of antiparasitic agent were then added to each well and incubated for 48 hours at which point the medium was replaced with 100ul phenol red free DMEM and 20 ul MTS. After an additional 1.5 hrs of incubation at 37 °C, and the absorbance at 490nm was calculated and the ED50 was determined using GraphPad Prism (La Jolla, CA).
Results and Discussion
Determining the Tailoring Enzymes Necessary for BIQ Core Formation
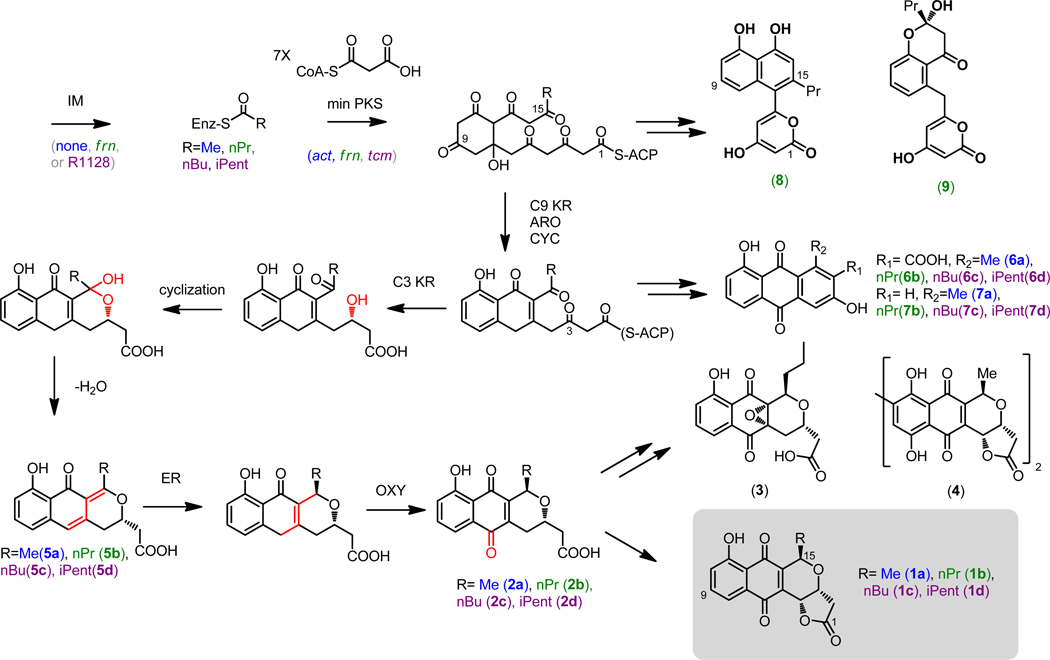
Before embarking on the reconstruction of the frenolicin pathway, we first attempted to complete the biosynthesis of the simplest BIQ, deoxykalafungin 2a (Scheme 1)17, which does not require an initiation module and is an intermediate in the actinorhodin (4) pathway 18. Previous studies have shown that introduction of the plasmid pRM5 (which encodes act KS/CLF, ACP, C-9 KR, ARO, and CYC genes; for details, see Scheme 1) into CH999 leads to the production of 3,8-dihydroxy-1-methylanthraquinone-2-carboxylic acid (DMAC) (6a) and aloesaponarin II (7a), as summarized in Table 1 13,14. The next step in BIQ production is stereoselective reduction of the C-3 ketone into an alcohol capable of cyclizing the third ring of the scaffold (Scheme 1 in red). To accomplish this, we cloned the C-3 ketoreductase gene, encoded by actVI-ORF1, onto pRM5, yielding the plasmid pJF6 (Table 1). As predicted, S. coelicolor CH999/pJF6/pBOOST* yielded 5a after cyclization and loss of water (Figure S16) 5,19.
Scheme 1. A Biosynthetic Pathway to the Benzoisochromanequinones.
The presumed biosynthetic route to 1a-d is shown along with isolable intermediates and shunt products. Abbreviations: IM (initiation module = ketosynthase III, acyl carrier protein, acyl-ACP thioesterase); min PKS (minimal polyketide synthase = ketosynthase/chain length factor (KS/CLF), acyl carrier protein (ACP), malonyl-CoA:ACP transacylase (MAT)); C9 KR = C-9 ketoreductase encoded by actIII; ARO = 1st ring aromatase encoded by actVII; CYC = 2nd ring cyclase encoded by actIV; C3 KR = C-3 ketoreductase encoded by actVI-ORF1; ER = enoyl reductase encoded by actVI-ORF2; OXY = oxygenase, encoded by an unidentified enzyme in the S. coelicolor genome. The natural product frenolicin A, (3) produced by S. roseofulvus, and the natural product actinorhodin, (4) produced by S. coelicolor, are also shown forming from their corresponding carboxylic acids, 2. For details, see text.
Table 1. Plasmid Constructs and Resulting Polyketide Products.
| Plasmida | KS-CLF | ACP | Initiation | Other relevant PKS genes |
Tailoringc | Polyketide Productsd |
|---|---|---|---|---|---|---|
| module | ||||||
| pRM5b | act | act | act KR, act ARO, act CYC | actVB | 6a, 7a | |
| pJF6 | act | act | act KR, act ARO, act CYC | actVI-ORF1, actVB | 5a, 6a, 7a | |
| pCR66 | act | act | act KR, act ARO, act CYC | actVI-ORF1, actVI-ORF2, actVB | 2a, 5a, 6a, 7a | |
| pCR67 | act | act | act KR, act ARO, act CYC | actVI-ORF1, actVI-ORF2, actVI-ORF4, actVB | 2a, 5a, 6a, 7a | |
| pJF35 | frn | frn | frn | act KR, act ARO, act CYC | actVI-ORF1, actVI-ORF2, actVB | 2b, 6b, 8, 9 |
| pYT127b | tcm | R1128 | R1128 | act KR, act ARO, act CYC | actVB | 6c, 6d, 7c, 7d |
| pJF7 | tcm | R1128 | R1128 | act KR, act ARO, act CYC | actVI-ORF1, actVB | 5c, 5d 6c, 6d, 7c, 7d |
| pJF9 | tcm | R1128 | R1128 | act KR, act ARO, act CYC | actVI-ORF1, actVI-ORF2, actVB | 2c, 2d, 5c, 5d, 6c, 6d, 7c, 7d |
Each pRM5 derived shuttle vector was introduced into S. coelicolor CH999/pBOOST* via transformation.
Previously reported constructs (see text).
Function of gene products: actVB flavin reductase; actVI-ORF1 C-3 ketoreductase; actVI-ORF2 enoyl reductase; actVI-ORF4 secondary enoyl reductase.
See SI for compound characterization.
The next step in the pathway is enoyl reduction of 5a followed by oxygenation. Mutants of the actinorhodin producing strain S. coelicolor A3(2), in which the enoyl reductase gene actVI-ORF2 has been inactivated, produce no actinorhodin, and mutants of the secondary enoyl reductase gene actVI-ORF4 produce only 18% of the actinorhodin of the native strain 20. We therefore constructed two derivatives of pJF6, pCR66 and pCR67, harboring actVI-ORF2 and actVI-ORF2/actVI-ORF4, respectively (Table 1). The product profiles of S. coelicolor harboring either plasmid was identical, and included 2a as the next isolable intermediate (Figure S16). This finding has two interesting implications for BIQ biosynthesis. First, overexpression of actVI-ORF2 in a pRM5 derived vector obviates the need for a supplementary enoyl reductase in a heterologous system. Second, and more interestingly, the inferred product of the enoyl reductase reaction (Scheme 1 bottom left) was not observed, nor was there a large buildup of 5a. It therefore appears that, although the act gene cluster harbors dedicated oxygenase genes 21,22, the S. coelicolor genome either has a non-specific oxygenase capable of catalyzing the same reaction, or that quinone formation proceeds spontaneously in air.
Production of Frenolicin B
Having identified the genes required to produce 2a in S. coelicolor CH999/pBOOST*, we turned our attention to frenolicin B and analogs thereof. Whereas the minimal PKS responsible for frenolicin production has been previously defined 23, the initiation module and downstream tailoring enzymes have not yet been characterized. Intriguingly, homologs of the actVI-ORF1 and actVI-ORF2 genes are not known to exist in the frenolicin (frn) gene cluster 24. In fact, constructs harboring all putative biosynthetic genes from the identified frn gene cluster failed to produce frenolicin or any related BIQ antibiotic (data not shown). In the face of insufficient genetic data, we hypothesized that the tailoring enzymes identified in production of 2a could be functionally repurposed for the biosynthesis of longer chain analogs. To accommodate the longer alkyl priming chain, we replaced the octaketide act KS-CLF genes with the mixed octaketide/nonaketide frn KS-CLF genes, and also included the genes thought to encode the initiation module of the frn PKS.
The frn gene cluster harbors homologs of the genes encoding the well-characterized initiation module of the R1128 cluster 25. Specifically, frnI is a homolog of zhuH, frnK is a homolog of zhuC, and frnJ is a homolog of zhuN. These genes encode a homodimeric KS responsible for chain initiation, an acyl-ACP thioesterase (AATE), and a dedicated ACP for the initiation module, respectively. Co-expression of these three frn genes with the genes encoding the frn KS-CLF, the frnN ACP the act C-9 KR, the act ARO and the act CYC, as well as the actVI-ORF1 and actVI-ORF2 genes (pJF35; Table 1) yielded a mixture of butyryl-primed nonaketide derivatives that arise from off-path cyclization reactions (Figure S17). In addition, very small quantities of 2b were also observed. Aside from 6b the two other major shunt products were propyl analogs of the previously identified compounds dehydromutactin, and SEK34 26–28. They form either due to the inability of the act ARO to aromatize the first ring or the inability of the act CYC to form the second 6-membered ring (Scheme 1). Notwithstanding the relative abundance of these shunt products, the ability of actVI-ORF1 and actVI-ORF2 to replace the unidentified components of the frenolicin pathway underscores the potential for rational design of a biosynthetic scheme using a diverse PKS toolbox in the face of incomplete genetic information.
Analogs of Frenolicin B
With a route in hand to the bioactive natural product frenolicin B, we turned our attention to the rational design of biosynthetic analogs. Earlier work with Eimeria tenella had shown that extension of the C-15 methyl substituent of 1a to a propyl substituent (1b) led to improved antiparasitic activity.6 We therefore reasoned that analogs 1c and 1d could be useful to investigate the steric effect of the C-15 substituent on biological activity. Because chemical synthesis of such analogs is not straightforward, we replaced the genes encoding the frn initiation module and ACP with homologs from the R1128 cluster, which incorporates bulkier primer units into the polyketide backbone 29. At the same time, to compensate for the anticipated increase in overall chain length of the polyketide skeleton, we also replaced the frn KS-CLF genes with homologs from the decaketide tetracenomycin (tcm) PKS 30. In combination with the act KR, act ARO, and act CYC, this hybrid bimodular PKS gene cluster yielded a mixture of compounds including 6c, 6d, 7c and 7d (pYT127; Table 1 and Figure S18) 23.
Further inclusion of the actVI-ORF1 and actVI-ORF2 genes yielded S. coelicolor CH999/pJF9, which produced a spectrum of products including the desired 2c and 2d (Table 1 and Figure S18). Although this strain produced a wide range of uncharacterized polyketide products, the ratio of the desired products 2c and 2d to the shunt products 6c and 6d was considerably higher in this recombinant strain than in S. coelicolor CH999/pJF35/pBOOST*. The lack of large quantities of butyl- and isopropyl-primed analogs of 8 and 9 indicate that act ARO and CYC interface with the tcm min PKS better than with the frn PKS. In addition, this suggests that substrate incompatibility arising from increased alkyl priming unit length is unlikely to be the cause of the large amounts of 8 and 9 observed in S. coelicolor CH999/pJF35/pBOOST*. These rationally designed analogs highlight the versatility of our approach to polyketide construction, and enabled evaluation of their antiparasitic activities.
Biological Evaluation against Toxoplasma gondii
Previous studies with E. tenella showed that the lactone forms of the BIQs 1a and 1b were more potent than the free acid forms (2a and 2b) 6. We therefore converted our isolated analogs 2a-d into their corresponding lactones 1a-d by previously reported methods 31. The structures of the novel compounds 1c and 1d were confirmed by NMR and mass spectrometric analysis (Tables S3, S5 and S6). The compounds were assayed against Toxoplasma gondii, the causative agent of toxoplasmosis, along with the known antiparasitic agent pyrimethamine as a reference.32 A slight modification of a previously reported yellow fluorescence protein reporter assay was used for this purpose 16 (Figures S1 and S2). As summarized in Table 2, within the C1-C4 range, the alkyl substituent has little effect on antiparasitic activity against T. gondii, but the longer branched substituent leads to a notable decrease in activity. Thus, the benefit of the longer substituent against E. tenella does not hold in our assays, and could be due to improved pharmacokinetics or differential species specificity. To assess their relative tolerability by mammalian cells, we also measured the cytotoxicity of 1a-d against HFF-1 cells using a MTS-based assay (Figure S3). The therapeutic indices of 1a-d, reported in Table 2, appear to be comparable.
Table 2. Bioactivity of compounds 1a–d and pyrimethamine.
Each compound was tested at least in triplicate. The therapeutic index is calculated by dividing the cytotoxic ED50 by the EC50 against T. gondii. See Figures S1 and S2 for growth inhibition curves.
| Compound | T. gondii ED50 (nM) | HFF1 cytotoxicity ED50 (nM) |
Therapeutic Index |
|---|---|---|---|
| 1a | 230 ± 15 | 5700 ± 800 | 25 |
| 1b | 260 ± 15 | 2400 ± 300 | 9 |
| 1c | 420 ± 30 | - | - |
| 1d | 1200 ± 60 | 8000 ± 2000 | 7 |
| Pyrimethamine | 820 ± 60 | >50000 | >60 |
Conclusion
In summary, notwithstanding incomplete genetic data for the natural product biosynthetic pathway, we have successfully engineered a chimeric, heterologous system for the production of frenolicin B. Furthermore, we have exploited this heterologous system to produce novel frenolicin analogs that are difficult to access by alternative methods. Our results showcase the utility of a modular approach to the creation of medicinally relevant polyketide products of type II PKSs. Last but not least, we have shown that the new compounds have antiparasitic activity against T. gondii in vitro. Further modification of the biosynthetic system presented here could yield additional improvements in BIQ productivity as well as additional analogs of frenolicin B.
Supplementary Material
Acknowledgements
This research was supported by a grant from the National Institutes of Health to C.K. (R01 CA 77248). T. gondii strain RHΔhxgprt YFP was a gift from Dr. Matthew Bogyo. We thank Dr. Sandeep Ravindran for his help culturing T. gondii.
References
- 1.Khosla C, Ridley CP. Encyclopedia of Microbiology. San Diego, CA: Academic Press; 2009. pp. 472–481. [Google Scholar]
- 2.O’Hagan D. The Polyketide Metabolites. Chichester, UK: Ellis Horwood; 1991. [Google Scholar]
- 3.Das A, Khosla C. Biosynthesis of aromatic polyketides in bacteria. Acc. Chem. Res. 2009;42:631–639. doi: 10.1021/ar8002249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hertweck C. The biosynthetic logic of polyketide diversity. Angew. Chem., Int. Ed. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 5.Hopwood D, Taguchi T, Ebizuka Y, Ichinose K. A new mode of stereochemical control revealed by analysis of the biosynthesis of dihydrogranaticin in Streptomyces violaceoruber Tu 22. J. Am. Chem. Soc. 2001;123:11376–11380. doi: 10.1021/ja015981+. [DOI] [PubMed] [Google Scholar]
- 6.Omura S, Tsuzuki K, Iwai Y. Anticoccidal activity of frenolicin B and its derivatives. J. Antibiot. 1985;38:1447–1448. doi: 10.7164/antibiotics.38.1447. [DOI] [PubMed] [Google Scholar]
- 7.Armer RE, et al. Anticoccidial activity of novel semi-synthetic analogues of deoxyfrenolicin and Frenolicin B (Part I) Heterocycl. Commun. 1998;4:309–315. [Google Scholar]
- 8.Armer RE, et al. Anticoccidial activity of novel semi-synthetic analogues of deoxyfrenolicin and Frenolicin B (Part II) Heterocycl. Commun. 1998;4:345–350. [Google Scholar]
- 9.Armer RE, et al. Carbocyclic frenolicin analogues: novel anticoccidial agents. Bioorg. Med. Chem. Lett. 1998;8:139–142. doi: 10.1016/s0960-894x(97)10200-1. [DOI] [PubMed] [Google Scholar]
- 10.Omura S, Iwai Y, Awaya J, Oiwa R. Compound, frenolicin B which is useful as an antibiotic. #4199514. US Patent. 1980
- 11.Masquelina T, Hengartnerb U, Streith J. Naphthopyranquinone antibiotics: novel enantioselective syntheses of frenolicin B and some of its stereoisomers. Helv. Chim. Acta. 1997;80:43–58. [Google Scholar]
- 12.Brimble MA, Narin MR, Prabaharan H. Synthetic strategies towards pyranonaphthoquinone antibiotics. Tetrahedron. 2000;56:1937–1992. [Google Scholar]
- 13.McDaniel R, Ebert-Khosla S, Hopwood D, Khosla C. Engineered biosynthesis of novel polyketides. Science. 1993;262:1546–1550. doi: 10.1126/science.8248802. [DOI] [PubMed] [Google Scholar]
- 14.Hu Z, Hopwood DA, Hutchinson CR. Enhanced heterologous polyketide production in Streptomyces by exploiting plasmid co-integration. J. Ind. Microbiol. Biotechnol. 2003;30:516–522. doi: 10.1007/s10295-003-0064-y. [DOI] [PubMed] [Google Scholar]
- 15.Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA. Practical Streptomyces Genetics. Norwich, UK: The John Innes Foundation; 2000. pp. 472–481. [Google Scholar]
- 16.Gubbels MJ, Li C, Striepen B. High-throughput growth assay for Toxoplasma gondii using yellow fluorescent protein. Antimicrob. Agents Chemother. 2003;47:309–316. doi: 10.1128/AAC.47.1.309-316.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoekesema H, Krueger WC, Kalifungin II. Chemical Transformations and the Absolute Configuration. J. Antibiot. 1976;29:704–709. doi: 10.7164/antibiotics.29.704. [DOI] [PubMed] [Google Scholar]
- 18.Hopwood DA. Genetic contributions to understanding polyketide synthases. Chem. Rev. 1997;97:2465–2497. doi: 10.1021/cr960034i. [DOI] [PubMed] [Google Scholar]
- 19.Ichinose K, et al. Proof that the actVI genetic region of Streptomyces coelicolor A3(2) is involved in stereospecific pyran ring formation in the biosynthesis of actinorhodin. Bioorg. Med. Chem. Lett. 1999;9:395–400. doi: 10.1016/s0960-894x(99)00011-6. [DOI] [PubMed] [Google Scholar]
- 20.Taguchi T, et al. Chemical characterisation of disruptants of the Streptomyces coelicolor A3 (2) actVI genes involved in actinorhodin biosynthesis. J. Antibiot. 2000;53:144. doi: 10.7164/antibiotics.53.144. [DOI] [PubMed] [Google Scholar]
- 21.Okamoto S, Taguchi T, Ochi K, Ichinose K. Biosynthesis of actinorhodin and related antibiotics: discovery of alternative routes for quinone formation encoded in the act gene cluster. Chemistry & biology. 2009;16:226–236. doi: 10.1016/j.chembiol.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 22.Kendrew SG, Hopwood DA, Marsh NG. Identification of a monooxygenase from Streptomyces coelicolor A3(2) involved in biosynthesis of actinorhodin: purification and characterization of the recombinant enzyme. J. Bacteriol. 1997;179:4305–4310. doi: 10.1128/jb.179.13.4305-4310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tang Y, Lee TS, Khosla C, Lee HY. Exploring the biosynthetic potential of bimodular aromatic polyketide synthases. Tetrahedron. 2004;60:7659–7671. [Google Scholar]
- 24.Bibb MJ, Sherman DH, Omura S, Hopwood DA. Cloning, sequencing and deduced functions of a cluster of Streptomyces genes probably encoding biosynthesis of the polyketide antibiotic frenolicin. Gene. 1994;142:31–39. doi: 10.1016/0378-1119(94)90351-4. [DOI] [PubMed] [Google Scholar]
- 25.Tang Y, Lee TS, Khosla C. Engineered biosynthesis of regioselectively modified aromatic polyketides using bimodular polyketide synthases. PLoS Biol. 2004;2:227–238. doi: 10.1371/journal.pbio.0020031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McDaniel R, Ebert-Khosla S, Hopwood DA, Khosla C. Engineered biosynthesis of novel polyketides: actVII and actIV genes encode aromatase and cyclase enzymes, respectively. J. Am. Chem. Soc. 1994;116:10855–10859. [Google Scholar]
- 27.Zhang H, et al. Mutactin, a novel polyketide from Streptomyces coelicolor. Structure and biosynthetic relationship to actinorhodin. J. Org. Chem. 1990;55:1682–1684. [Google Scholar]
- 28.Khosla C, et al. Genetic construction and functional analysis of hybrid polyketide synthases containing heterologous acyl carrier proteins. J. Bacteriol. 1993;175:2197–2204. doi: 10.1128/jb.175.8.2197-2204.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Marti T, Hu Z, Pohl NL, Shah aN, Khosla C. Cloning, nucleotide sequence, and heterologous expression of the biosynthetic gene cluster for R1128, a non-steroidal estrogen receptor antagonist. J. Biol. Chem. 2000;275:33443–33448. doi: 10.1074/jbc.M006766200. [DOI] [PubMed] [Google Scholar]
- 30.Bibb MJ, Biró S, Motamedi H, Collins JF, Hutchinson CR. Analysis of the nucleotide sequence of the Streptomyces glaucescens tcmI genes provides key information about the enzymology of polyketide antibiotic biosynthesis. EMBO J. 1989;8:2727–2736. doi: 10.1002/j.1460-2075.1989.tb08414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li T, Ellison RH. Stereoselective total synthesis of racemic kalafungin and nanaomycin A. J. Am. Chem. Soc. 1978;78:6264–6265. [Google Scholar]
- 32.McCabe R, Oster S. Current recommendations and future prospects in the treatment of toxoplasmosis. Drugs. 1989;38:973–987. doi: 10.2165/00003495-198938060-00008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.