Abstract
The cyclic-AMP-dependent protein kinase A (PKA) regulates processes such as cell proliferation and migration following activation of growth factor receptor tyrosine kinases (RTKs), yet the signaling mechanisms that link PKA with growth factor receptors remain largely undefined. Here we report that RTKs can directly modulate the function of the catalytic subunit of PKA (PKA-C) through post-translational modification. In vitro kinase assays revealed that both the epidermal growth factor and platelet derived growth factor receptors (EGFR and PDGFR, respectively) tyrosine phosphorylate PKA-C. Mass spectrometry identified tyrosine 330 (Y330) as a receptor-mediated phosphorylation site and mutation of Y330 to phenylalanine (Y330F) all but abolished the RTK-mediated phosphorylation of PKA-C in vitro. Y330 resides within a conserved region at the C-terminal tail of PKA-C that allosterically regulates enzymatic activity. Therefore, the effect of phosphorylation at Y330 on the activity of PKA-C was investigated. The Km for a peptide substrate was markedly decreased when PKA-C subunits were tyrosine phosphorylated by the receptors as compared to un-phosphorylated controls. Importantly, tyrosine-phosphorylated PKA-C subunits were detected in cells stimulated with EGF, PDGF and FGF2 and in fibroblasts undergoing PDGF-mediated chemotaxis. These results demonstrate a direct, functional interaction between RTKs and PKA-C and identify tyrosine phosphorylation as a novel mechansim for regulating PKA activity.
Keywords: Protein kinase A, Platelet derived growth factor receptor, Epidermal growth factor receptor, Peptide growth factors
Protein kinases are enzymes that transmit signals throughout the intracellular environment. They catalyze the transfer of a phosphate group from ATP to serine, threonine and tyrosine residues within downstream substrates and in this manner alter the biochemical properties of the target proteins. Cyclic-AMP-dependent protein kinase A (PKA) is a member of the large family of AGC protein kinases that also includes PKC, PKB, Rsk and many others. It is one of the most well studied protein kinases to date with a multitude of structural and biochemical studies providing detailed information about how the enzyme functions [Taylor et al., 2008; Taylor et al., 2005]. PKA exists in cells as an inactive holoenzyme consisting of a dimer of two allosteric R (regulatory) subunits, each of which binds to a C (catalytic) subunit. The catalytic subunit (PKA-C) is composed of a conserved catalytic core (aa. 40–300) that is flanked by short amino and carboxy terminal sequences, termed N and C tails. It is now appreciated that the C-tail (residues 301–350) contains particular regions that function as cis-acting regulatory components of PKA catalytic activity [Kannan et al., 2007; Taylor et al., 2008]. An acidic cluster of amino acids (residues 327–336, FDDYEEEEIR), termed the active site tether (AST) is highly dynamic and regulates ATP binding, organization of the active site and the recognition and recruitment of peptide and protein substrates [Batkin et al., 2000; Chestukhin et al., 1996; Kannan et al., 2007; Kennedy et al., 2009; Taylor et al., 2008; Yang et al., 2009]. In cells, the catalytic subunit is assembled as a fully phosphorylated and active enzyme that is kept dormant by its association with an R subunit, in a holoenzyme complex. PKA is typically activated in response to extracellular cues that induce the production of the second messenger, cyclic adenosine monophosphate (cAMP). cAMP binds to the R subunits which induces a dramatic conformational change and the subsequent release of the active C subunits. PKA-C is then free to phosphorylate serine and theronine residues on numerous intracellular substrates that can be found in the cytosol, cytoskeleton, plasma membrane and nucleus. The discrete subcellular compartmentalization of PKA by the A kinase anchoring proteins (AKAPs) provides an important degree of specificity to PKA signaling by helping to match a given extracellular stimulus to a specific subset of cellular targets [Scott, 2003]. PKA-mediated signal transduction controls a myriad of cellular processes including gene transcription, proliferation, differentiation, migration and survival.
Growth factors are one type of extracellular cue known to activate PKA [Bornfeldt and Krebs, 1999; Ciardiello and Tortora, 1998; Fishman et al., 1997]. Soluble peptide growth factors such as epidermal growth factor (EGF) and platelet derived growth factor (PDGF), initiate their effects upon binding to their cognate receptor tyrosine kinases (RTKs), the EGF receptor (EGFR) and PDGF receptor (PDGFR), respectively. Once active, these receptors initiate a host of downstream signaling pathways (eg. AKT/PKB, PKC and Erk) which promote gene transcription, cell proliferation, survival and migration [Hubbard and Miller, 2007]. There are numerous reports that demonstrate a role for PKA in signalling events that occur downstream of the activated EGFR and PDGFRs. A prime example is the demonstration that the RI and C subunits of PKA physically interact with the activated EGFR in a Grb2-dependent manner [Ciardiello and Tortora, 1998]. This interaction was reportedly important for the ability of the EGFR to promote mitogenic signaling events [Ciardiello et al., 1999]. PKA can directly phosphorylate the EGFR and inhibit its tyrosine kinase activity in vitro and cAMP analogs attenuate EGF-induced tyrosine phosphorylation of the EGFR in mammalian fibroblasts [Barbier et al., 1999]. However, PKA’s effect on the EGFR may be cell type specific as PKA was shown to stimulate tyrosine phosphorylation of the EGFR resulting in enhanced kinase activity in PC12 and A431 cells [Piiper et al., 2003]. In response to stimulation of cells with PDGF, PKA is activated and translocated from the cell membrane [deBlaquiere et al., 1994; Graves et al., 1996], and it can either promote or inhibit cellular proliferation and migration depending upon the cell type studied [Bornfeldt and Krebs, 1999; Bornfeldt et al., 1995; deBlaquiere et al., 1994; Deming et al., 2008; Graves et al., 1993; Graves et al., 1996; Howe et al., 2005; Jalvy et al., 2007; O’Connor and Mercurio, 2001; Stork and Schmitt, 2002]. While these connections have been known for some time, the precise manner in which growth factor receptors and PKA activity intersect is poorly understood. The results reported here demonstrate that tyrosyl phosphorylation of PKA regulates its activity and identify a molecular mechanism for crosstalk between growth factor receptor tyrosine kinases and PKA signaling networks.
MATERIALS AND METHODS
Antibodies and Other Reagents
Primary antibodies were obtained commercially from Santa Cruz Biotechnologies (PKA-Cα, cat # SC904; GFP cat # SC9996), Upstate Biotechnology (phospho-tyrosine, 4G10) and Molecular Probes (GFP, cat # A1112). This antibody is also known to immune-react with YFP), Cell Signaling Technologies (phospho-PKA substrate, cat # 9624L). Horseradish peroxidase-conjugated secondary antibodies were from Jackson Immunolaboratories. Platelet-derived growth factor type BB (PDGF-BB) was from Upstate Biotechnology, Epidermal growth factor (EGF) was from Peprotech, Fibroblast growth factor 2 (FGF2) was from Sigma. Protein G beads were from Calbiochem. Recombinant untagged PKA-Cα was obtained from New England Biolabs and purified active GST-PDGFR and GST-EGFR were from Cell Signaling Technologies. Fibronectin was from BD Biosciences. The site directed mutagenesis kit was from Stratagene. Protease arrest was from G Biosciences and sodium orthovanadate was from Sigma.
Cell Culture and Transfection
COS7 and REF52 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) plus 10% fetal bovine serum. NIH 3T3 fibroblasts were grown in DMEM plus 10% bovine calf serum. COS 7 cells were transfected with FuGENE HD reagent (Roche Applied Science) following the manufacturer’s instructions.
DNA Constructs
The plasmid encoding the PKA-Cα-YFP was a gift from M. Zaccolo (University of Padua) and has been previously described [Zaccolo and Pozzan, 2002]. The plasmid encoding the PKA substrate GFP227RRRRSII [Yang et al., 1999] was obtained from Kevan Shokat (UCSF). The mouse (His)6-tagged PKA-C-α construct in pET15b was a gift from Susan Taylor (UCSD). The Y330F PKA-Cα mutant was generated via site directed mutagenesis using forward 5′CTTTGACGACTTTGAGGAGGAAGAG3′ and reverse 5′CTCTTCCTCCTCAAAGTCGTCAAAG3′ primers and mouse PKA-Cα cDNA in pET15b as a template.
Protein Purification
Recombinant GFP227RRRRSII was produced in E. coli. strain JM109 as previously described [Yang et al., 1999]. Recombinant (His)6-tagged PKA-C subunits were produced in E. coli. strain BL21 by inducing protein expression with 0.2mM IPTG at 18°C overnight. (His)6-tagged proteins were purified with His-Select Beads (Sigma) following the manufacturer’s protocol. The purity of the recombinant proteins was checked by SDS-PAGE and Coomassie staining.
Phosphorylation of PKA-Cα by PDGFR and EGFR
For in vitro kinase assays with the receptors and PKA-C subunits, 200 ng of receptor was incubated with 500 ng of C subunit in kinase reaction buffer (20 mM Tris-Cl, pH 7.4, 100 mM NaCl, 200 μM ATP, 15 mM MgCl2) at 30°C for 1 hour.
PKA Kinase Assay
For in-vitro PKA activity assays from whole cell extracts, cells were serum starved overnight, stimulated with growth factor for the various times indicated and then washed with ice cold PBS two times. Cells were harvested in 200μl PKA Activity Buffer (50mM Tris pH 7.5, 0.5mM EDTA, 50mM β-glycerolphosphate, 1mM NaF, 0.5mM EGTA, Protease Inhibitor Cocktail (Pierce)) and allowed to incubate on ice for 10 minutes. Cells were then lysed by sonication using a Virsonic 100 Ultrasonic Cell Disruptor (VirTis) by pulsing for 10 seconds on setting 7 twice. Protein concentration was assayed using a standard BCA protocol (Pierce). For each reaction, 100 ng of PKA substrate (GFP227RRRRSII) pre-bound to 25 μl of His-Select HF Nickel Affinity Gel (Sigma) in PKA Activity Buffer was mixed with 10 μg of whole cell extract in 75 μl PKA Activity Buffer containing kinase reaction mix (15mM MgCl2, 200μM ATP, and 0.5 mM DTT, final concentrations). The samples were incubated at 32°C for 20 minutes, after which the reaction was stopped by the addition of 5 X Laemmli sample buffer. Samples were then subjected to SDS-Page and immunoblot analysis using anti-phospho-PKA substrate and anti-GFP antibodies (to ensure equal loading of substrate). Densitometry was performed using Image J analysis software. The relative amount of growth factor-induced PKA kinase activity was calculated by normalizing to the unstimulated control. The GFP227RRRRSII substrate was highly specific for PKA in this assay as incubation with protein kinase inhibitor peptide (5–24, BioMol) completely ablated phosphorylation (Supplemental Fig. 1).
In-Gel Digestion, Mass Spectrometry and Data Analysis
Cubed coomassie-stained gel bands of the catalytic subunit of murine PKA alpha that had previously been subjected to an in vitro kinase reaction with or without PDGFR were rinsed with water; destained with 50% acetonitrile (MeCN), 50% ammonium bicarbonate; dehydrated with 100% MeCN; and subjected to in-gel digestion with 6 ng/μl sequencing grade modified trypsin (Promega,) in 50mM ammonium bicarbonate for 16 hours at 37oC. Peptides were extracted once with 50% MeCN, 2.5% formic acid (FA); and once with 100% MeCN. Dried peptides were resuspended in 2.5% MeCN, 2.5% FA and loaded using a Micro AS autosampler (Thermo Electron) and a Surveyor MS Pump Plus (Thermo Electron) onto a nano-electrospray microcapillary column packed with 12cm of reverse phase MagicC18 material (5 μm, 200Å, Michrom Bioresources, Inc.). Elution was performed with a 5–35% MeCN (0.15 % FA) gradient over 45 minutes, after a 15 minute isocratic loading at 2.5% MeCN, 0.15% FA. Solvent A was 2.5% MeCN, 0.15% FA and Solvent B was 99.85% MeCN, 0.15% FA. Mass spectra were acquired in a LTQ XL linear ion trap mass spectrometer (Termo Electron). Throughout the entire run a precursor survey (MS1) scan was followed by seven data-dependent MS/MS scans on the most abundant ions (dynamic exclusion repeat count 1 and duration 180s) as well as three MS/MS scans on the following m/z values 1267.4, 876.4 and 1027.4 (each +/− 1.5). These values correspond to the average molecular mass of putative phosphotyrosyl-tryptic peptides harboring pY236 or pY248 (triply-charged, oxidized metionine); pY307 (doubly-charged); and pY331 (doubly-charged). Mass spectral data were searched against a murine PKA-C alpha protein database using Turbo SEQUEST (Thermo Electron, Version 27, Revision 12) requiring no enzyme specificity, a 2 Da precursor mass tolerance, and a 1Da fragment ion mass tolerance. Cysteine residues were required to have a static increase in 71.0 Da for acrylamide (C3H5ON) adduction. Differential modification of 16.0 Da on methionine and 80.0 Da on serine, threonine and tyrosine was permitted.
Generation of phosphorylation site specific pY330 PKA-C polyclonal antibody
A polyclonal antibody specific for phosphorylated Y330 (pY330) on PKA-C was generated by Yenzym Antibodies (San Franciso, CA). Briefly, a peptide corresponding to phosphorylated mouse PKA-Cα at Y330 (FDDpYEEEIR) was synthesized, conjugated to KLH and used to immunize rabbits following a standard immunization protocol. Crude sera were collected and tested for immunoreactivity by ELISA. Sera were then pooled and affinity purified using the immunizing phosphor-peptide followed by absorption with an unphosphorylated peptide matrix to remove cross-reactive antibodies.
Kinetic analysis of PKA-C subunits
Kinetic analysis of control or pre-phosphorylated C-subunits were performed by radioisotope [γ-32P] ATP labeling using the peptide TQAKRKKSLAMA as a substrate as previously described [Dostmann et al., 1999]. The Vmax and Km values were derived using non-linear regression analysis in GraphPad Prism software. A sigmoidal dose response (variable slope) was generated.
Assessment of tyrosine phosphorylated PKA-C from whole cell extracts, cell bodies and pseudopods
Proteins were isolated from cell bodies and pseudopods of NIH 3T3 fibroblasts responding to a chemotactic gradient of PDGF as previously described (Howe et al., 2005). Briefly, serum starved cells were replated for 2 hours on fibronectin-coated, 3 μm-pore polycarbonate membranes in Costar Transwell inserts. PDGF (10 ng/mL) was added to the lower chamber for 1 hour. To harvest pseudopodia, inserts were washed in PBS, cell bodies (CB) were removed from the upper surface, and the pseudopodia (Pd) on the underside were scraped into lysis buffer. Alternatively, Pd were removed, and CB on the upper surface were harvested. Whole cell extracts were prepared from COS 7 cells by rinsing twice with ice cold phosphate buffered saline following by harvesting in modified RIPA buffer (50 mM Tris-Cl pH 7.5, 150 mM NaCl, 1% Nonidet, 0.5% NaDOC, 5mM EDTA, protease arrest cocktail and 1 mM sodium orthovanadate). REF 52 cells were harvested in RIPA buffer (50 mM Tris-Cl pH 7.5, 150 mM NaCl, 0.1% sodium dodecyl sulfate, 1% Nonidet, 0.5% NaDOC, 5mM EDTA, protease arrest cocktail and 1 mM sodium orthovanadate). All lysates were clarified by centrifugation and protein content quantified by bicinchoninic acid assay (Pierce). Antibodies directed against PKA-C and GFP/YFP, were used to immunoprecipite endogenous PKA-C and the PKA-C-YFP fusion protein, respectively. Briefly, cell extracts were incubated with PKA-C or GFP antibody for 1 hour at 4°C. Protein G beads were then added for an additional thirty minutes after which the immunoprecipitates were washed three times with lysis buffer. Immunoprecipitates and protein extracts were subjected to SDS-PAGE and immunoblot analysis using the antibodies indicated in the figure legends.
Protein Data Base
The accession number for crystal structure of the α catalytic subunit of PKA used in this study is PBD:3FJQ.
RESULTS
RTKs phosphorylate PKA-C in vitro
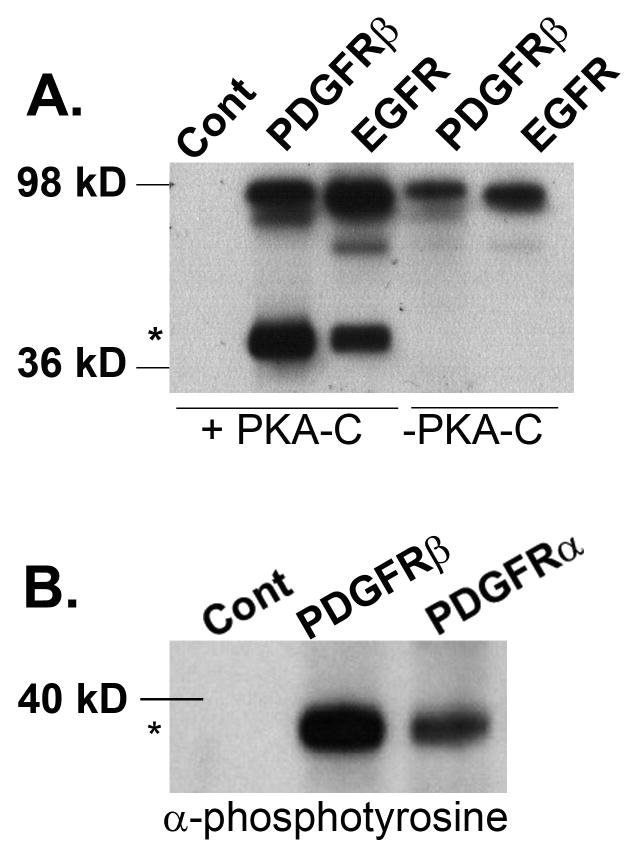
Our recent work established an important role for PKA in regulating cytoskeletal events and membrane dynamics during PDGF- and EGF-mediated chemotaxis [Deming et al., 2008; Howe et al., 2005]. However, the mechanism/s by which the PDGFR and EGFR communicate to PKA was not defined. Given that the C subunit of PKA was shown previously to physicially interact with the EGFR, we wondered whether the catalytic subunit might be also be phosphorylated directly by RTKs. To investigate this possibility, we performed in vitro kinase assays with purified PKA-C α subunits and the active cytoplasmic domains of PDGFRβ and EGFR and monitored tyrosine phosphorylation via western blot using an anti-phosphotyrosine antibody. As shown in Figure 1 A, the C subunit of PKA (molecular weight 38 kD) was tyrosine phosphorylated by both the PDGFRβ and EGFR. There was no phosphotyrosine signal at 38kD in the samples containing RTK alone. The α PDGFR like the PDGFRβ, also phosphorylated PKA-C albeit to a lesser extent (Fig. 1B). Taken together, these results demonstrate that both the EGFR and PDGFR are able to phosphorylate PKA-C in vitro.
Figure 1.
(A) Buffer (Cont), active GST-tagged- PDGFRβ (PDGFRβ) or EGFR (EGFR) were subjected to a cold kinase assay in the presence (+PKA-C) or absence (−PKA-C) of purified PKA-Cα. The samples were then processed for immunoblot analysis using an antibody that recognizes pan phosphotyrosine. * denotes the position of PKA-C (mw 38 kD). The upper band migrating near the 98 kD marker corresponds to the molecular weights of the active, tyrosine-phosphorylated, bacterially purified, GST-receptors. (B) Recombinant PKA-Cα was incubated with buffer only (Cont) or bacterially purified, GST-tagged PDGFRβ or PDGFR α under cold kinase assay conditions and samples were processed as described in (A).
PKA-C subunit is tyrosine phosphorylated in growth factor stimulated cells
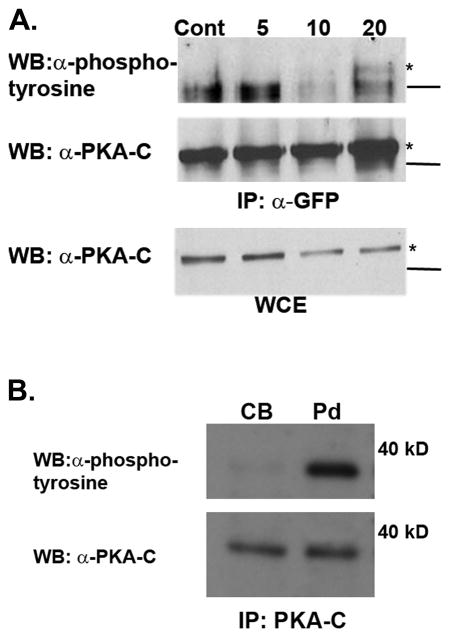
In order to determine whether this tyrosine phosphorylation event on PKA-C occurs in cells undergoing growth factor stimulation, the phosphorylation status of PKA-C following treatment of cells with EGF was investigated. Exogenously expressed YFP-PKA-C was immunoprecipitated from unstimulated and EGF-stimulated COS7 cells, known to express the EGFR, and then assessed for total phosphotyrosine via western blot using a pan anti-phosphotyrosine antibody. While there was no detectable tyrosine phosphorylation on YFP-PKA-C in unstimulated cells, tyrosine phosphorylation was slightly detectable at 10 minutes and increased by 20 minutes following stimulation with EGF (Fig 2A). The previously demonstrated role for PKA in growth factor-induced chemotaxis [Howe et al., 2005] and PDGF-induced membrane ruffling in fibroblasts [Deming et al., 2008], led us next to investigate whether PKA-C may be tyrosine phosphorylated in cells undergoing PDGF-mediated chemotaxis. To address this possibility, endogenous PKA-C subunits were isolated from cell body and pseudopod extracts prepared from NIH 3T3 fibroblasts migrating toward a gradient of PDGF. Although the amount of total PKA-C was similar in the cell body and pseudopod fractions, tyrosine- phosphorylated PKA-C was relegated to the pseudopod (Fig. 2B). This together with our previous finding that PKA activity (but not total PKA-C protein levels) is markedly enriched within pseudopods suggest that phosphorylation of PKA-C may regulate PKA function during cellular migration. These results provide evidence that a pool of PKA-C becomes phosphorylated on a tyrosine residue/s in cells stimulated with EGF and PDGF.
Figure 2.
(A) Cos7 cells were transfected with a plasmid encoding PKA-Cα fused to YFP. Twenty four hours later cells were serum starved overnight and then treated with vehicle control (Cont) or EGF (100 ng/mL) for 5, 10 or 20 minutes after which they were harvested in mRIPA buffer. The PKA-Cα-YFP fusion protein was immunoprecipitated, then subjected to SDS-Page and immunoblot analysis using antibodies directed against pan-phosphotyrosine (upper panel) and the catalytic subunit of PKA-Cα (middle panel). The lower panel illustrates the amount of PKA-C-α–YFP in the whole cell extract and is equivalent to 1/20th the input. The lines to the right represent the position of a 60 kD molecular weight marker and the asterisk (*) denotes the PKA-Cα-YFP fusion protein. The experiment was repeated three times and yielded similar results. (B) Cell bodies (Cb) and pseudopods (Pd) were isolated from NIH3T3 cells as described in Materials and Methods. PKA-Cα was immunoprecipitated from each of these fractions, separated by SDS-PAGE, and immunoblotted with antibodies against pan-phosphotyrosine and the C subunit (PKA-C). The numbers to the right indicate the position of a 40 kD molecular weight marker. The result shown is representative of two separate experiments.
RTKs phosphorylate PKA-C on tyrosine 330
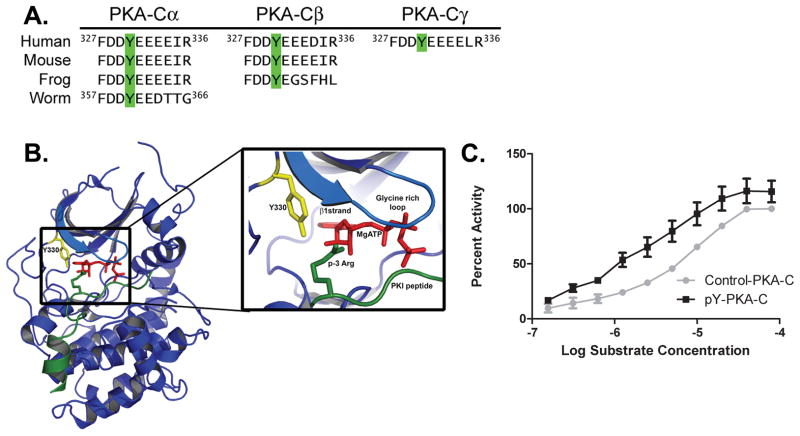
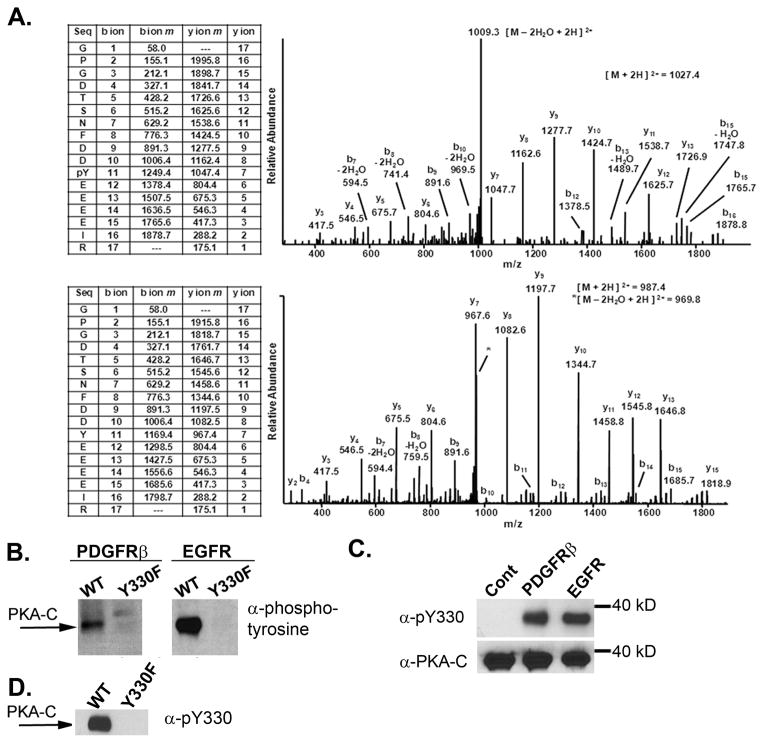
We next wished to map which tyrosine/s on PKA-C were phosphorylated by the PDGFR and EGFR. To do this, we first subjected PKA-C to an in vitro kinase reaction with or without the PDGFR. The reaction was terminated by adding SDS sample buffer and heating to 95oC for 5 minutes. Following SDS-PAGE PKA-C was cut from the gel and subjected to in gel tryptic digestion. Tryptic peptides were then subjected to liquid chromatography tandem mass spectrometry (LC-MS/MS) in a linear ion trap mass spectrometer as described in the Methods. In addition to randomly choosing peptides for fragmentation analysis we also performed targeted MS/MS analysis on masses corresponding to phosphotyrosyl-tryptic peptides that would contain tyrosines on the surface of the enzyme based on its crystal structure [Fig. 4B]. We identified one phosphotyrosyl peptide in the PDGFR-treated sample and none in mock-treated sample. The upper panel of Fig. 3A shows the MS/MS spectrum of the identified phosphotyrosyl peptide harboring phosphorylated tyrosine 330. The lower panel is an MS/MS spectrum of the same peptide in its unphosphorylated state which was identified in both samples. It is clear that both phosphorylated and unphosphorylated peptides show strikingly similar collision-induced dissociation fragmentation patterns. Similar results were obtained when PKA-C was phosphorylated by the EGFR (not shown). Importantly, when Y330 was mutated to a non-phosphorylatable phenylalanine residue (Y330F) the ability of the PDGFRβ and EGFR to tyrosine phosphorylate PKA-C α in vitro was essentially ablated (Fig. 3B). These data suggest that Y330 is the major (if not only) tyrosine phosphorylated by PDGFRβ and EGFR in vitro. We next generated a peptide antibody targeted against phosphorylated Y330 (pY330). The antibody specifically recognized wild type PKA-C α subunits phosphorylated by PDGFR and EGFR in vitro but not control subunits or the Y330F mutant (Fig 3C & D). The antibody was raised against a region in PKA-C that is conserved across all three isoforms (Materials and Methods and Fig 4A) and therefore is predicted to react with phosphorylated PKA-C α, β and γ subunits.
Figure 4.
(A) PKA is evolutionarily conserved. The amino acid sequence in the conserved AST region of PKA-C is shown Human, Mouse, Frog and Worm. The following NCBI sequence identifiers were used: for PKA-Cα: Human: NP_002721, Mouse: NP_032880, Frog: AAI69356, Worm: CAB41352; for PKA-Cβ: Human: NP_002722, Mouse: NP_035230.1, Frog: NP_001080696 and for PKA-C γ: Human: AAH39888. (B) Structure of the active catalytic subunit of PKA-α, PDB: 3FJQ. Tyrosine 330 is highlighted in yellow. Note the close proximity of Y330 to the β1 strand and the glycine rich loop (marine), and the Arginine that lies in the p-3 position of the PKI peptide inhibitor (green). MgATP is colored in red. (C) Kinetic analysis of PKA-C subunits. Control or tyrosine-phosphorylated (pY-PKA-C) PKA-Cα subunits were subjected to in vitro kinase assays as described in Methods. The percent activity was calculated as the reaction velocity (μM/min/μg) normalized to the maximum velocity of the control and then plotted as a function of the log of the substrate concentration (μM). The graph represents the average of four independent experiments and error bars indicate the standard error ±S.E.
Figure 3.
(A) Low energy MS/MS spectra identifying tryptic peptides with Y331 in a phosphorylated state (upper panel) or unophosphorylated state (lower panel). Sequest Xcorr scores were 3.5 and 5.7 respectively. See Methods for details. (B) Mutation of Y330 to Y330F ablates PDGFR- and EGFR-induced tyrosine phosphorylation. His-tagged wildtype or Y330F PKA-C-α subunits were incubated together with GST-PDGFRβ or GST-EGFR and subjected to a cold in vitro kinase assay. Samples were then analyzed by SDS-Page and immunoblot analysis using an antibody directed against pan-phosphotyrosine. (C) Phospho-specific antibody that recognizes Y330. In vitro kinase assays were performed with buffer alone (Cont), PDGFR, or EGFR and recombinant PKA-C. Immunoblot analysis was performed using the Y330 phosphospecific antibody as described in Materials and Methods. Membranes were stripped and re-probed with an antibody that recognizes total PKA-C. (D) In vitro kinase assays were performed with or EGFR and His(6)WT or His(6) Y330F PKA-Cα subunits. Immunoblot analysis was performed using a Y330 phosphospecific antibody as described in Materials and Methods.
Tyrosine phosphorylation of PKA-C enhances its kinetic activity
Y330 lies within a region of the C terminal tail of PKA-C that is highly evolutionarily conserved and Y330 is found in the α, β and γ isoforms of PKA-C (Fig 4A). The C tail region, wherein Y330 resides is malleable and in the closed (i.e. ATP- and substrate-bound) conformation of the enzyme, lies in close proximity to the ATP binding pocket, glycine rich loop, and p-3 subsite of the substrate (Fig. 4B). Therefore, we reasoned that phosphorylation at this residue might have effects on the activity of the enzyme. To address this possibility, the kinetic activity of unphosphorylated C subunit was compared to that of C-subunit that had been pre-phosphorylated with the EGFR. As demonstrated in Figure 4C, tyrosine phosphorylation of PKA-C altered the Km for a known peptide substrate. Specifically, phosphorylation on PKA-C reduced the Km by 3 fold, resulting in a significant overall increase in the catalytic efficiency of the enzyme. The Vmax/Km was 0.980 for control and 3.982 for pY-PKA-C subunits, respectively (Table 1). Similar results were obtained when PKA-C was pre-phosphorylated with PDGFRβ (data not shown). Although tyrosine phosphorylation enhanced PKA kinase activity, the ability of the protein kinase inhibitor (PKI) peptide (residues 5–24 of full length PKI) to inhibit catalytic activity was not altered (data not shown). Thus, it appears that phosphorylation of PKA-C at Y330 by RTKs enhances the overall activity of the enzyme in vitro, in particular by increasing the affinity for substrate binding.
Table 1. Kinetic Summary of PKA-C subunits.
Effect of phosphorylation at Y330 of PKA- C on Km and Vmax values. Non-linear regression analysis was performed on the data represented in Figure 4C to obtain the average Km and Vmax for control or phosphorylated (pY-PKA-C) PKA-Cα subunits. Each value represents the average result (±S.E.) of four independent experiments. A paired students T test demonstrated a significant difference in the Km **p<0.005 and the Vmax/Km, *<0.05 between the control and pY-PKA-C subunits.
| Km (μM) | Vmax(μM/min/mg) | Vmax/Km | |
|---|---|---|---|
| Control PKA-C | 7.97 ± .19 | 7.8±1.6 | 0.980 ± .218 |
| pY-PKA-C | **2.61 ± .59 | 9.1± 2.0 | *3.982 ± 1.343 |
Growth factors induce phosphorylation of Y330 in cells
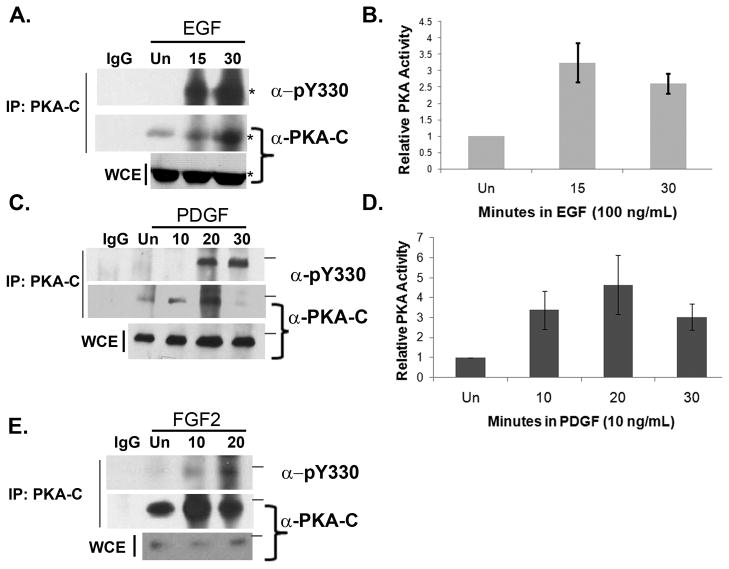
We next used the pY330 antibody to monitor the phosphorylation status of PKA-C upon stimulation of cells with growth factors. Consistent with the observed increased in total phosphotyrosine on PKA described in Fig. 2A, stimulation of COS 7 cells with EGF induced the phosphorylation of Y330 at 15 minutes, which remained high for at least 30 minutes (Fig 5A). Assessment of EGF-induced PKA activity in COS 7 cells revealed increases in PKA activity at time points coincident with the phosphorylation of Y330 (Fig. 5B). In REF 52 fibroblasts, which are known to express the PDGFR, little to no basal phosphorylation of Y330 was observed in unstimulated cells or those treated with PDGF for 10 minutes (Fig. 5C). However, phosphorylation of Y330 was evident 20 and 30 minutes following stimulation with PDGF (Fig. 5C). Interestingly, like for EGF, peak PKA activity following PDGF treatment was coincident with the timing of phosphorylation on Y330 (Fig 5C & 5D). It should be noted that treatment of REF 52 cells with PDGF appeared to slightly increase the total amount of PKA-C in the whole cell extract used for immunoprecipitation (Fig. 5C), which could have contributed to the rise in activity. We also wished to determine whether phosphorylation of PKA-C on Y330 might occur in response to growth factors other than PDGF and EGF. In addition to the PDGFR, fibroblasts express the fibroblast growth factor receptor tyrosine kinase (FGFR). In NIH 3T3 fibroblasts, FGFR is activated by fibroblast growth factor 2 (FGF2) and FGF2 activates PKA in these cells [Pursiheimo et al., 2000]. Therefore, the ability of FGF-2 to induce phosphorylation of Y330 on PKA-C was investigated in NIH 3T3 cells. Indeed, while there was little phosphorylation of Y330 in unstimulated NIH 3T3 cells, FGF2 increased the levels of phosphorylation at 10 and 20 minutes following treatment (Fig 5E). Taken together, our results demonstrate that PKA-C subunits are phosphorylated on Y330 in cells following stimulation with three different growth factors and suggest that phosphorylation of PKA-C on Y330 may be a general intermediate event of RTK signaling.
Figure 5.
(A) Cos7 cells were transfected with a plasmid encoding PKA-Cα fused to YFP. Twenty four hours later cells were serum starved overnight and then treated with vehicle control (Un) or EGF (100 ng/mL) for 15 or 30 minutes after which they were harvested in RIPA buffer. The PKA-Cα-YFP fusion protein was immunoprecipitated, then subjected to SDS-Page and immunoblot analysis using antibodies directed against pY330 (upper panel) and the catalytic subunit of PKA-Cα (middle panel). The lower panel illustrates the amount of PKA-C-α–YFP in the whole cell extract. The results depicted are representative of three independent experiments. The asterisk (*) denotes the PKA-Cα-YFP fusion protein. (B) Cos7 cells were serum starved overnight and then treated with vehicle control (Un) or 100 ng/mL EGF for 15 or 30 minutes. Cells were harvested in PKA assay buffer and processed for PKA kinase activity as described in Materials and Methods. The graph depicts the relative amount of PKA activity induced upon stimulation with EGF (100 ng/ml) normalized to the unstimulated control. The data represents the average of three independent experiments and the error bars indicate the standard error (± S.E.). (C). REF 52 fibroblasts were serum starved overnight and then treated with vehicle control (Un) or PDGF (20 ng/mL) for the times indicated and endogenous PKA-C subunits were immunoprecipitated as described in Materials and Methods. Immunoprecipitates and whole cell extracts (WCE) were subjected to SDS-Page and immunoblot analysis using the pY330 antibody. The membrane was stripped and re-blotted with an antibody directed against PKA-C. The line denotes the position of the 40 kD molecular weight marker. The results are representative of four independent experiments (D) REF 52 fibroblasts were serum starved overnight and then treated with vehicle control (Un) or PDGF (10 ng/mL) for the times indicated. PKA kinase activity was assessed from whole cell extracts as described in Materials and Methods. The graph depicts the relative amount of PKA activity induced upon stimulation with PDGF normalized to the unstimulated control. The data shown represent the average of three independent experiments and the error bars indicate the standard error (±S.E.). (E). NIH 3T3 cells were serum starved overnight and then treated with vehicle control (Un) or FGF2 (25 ng/mL) for the times indicated. Endogenous PKA-C immunoprecipiates and whole cell extracts (WCE) were subjected to SDS Page and western blot analysis as described in (C). The line denotes the position of the 40 kD molecular weight marker. The results shown are representative of two independent experiments.
DISCUSSION
The results presented in this report describe an unprecedented role for tyrosine phosphorylation of the C subunit of PKA during growth factor signaling and build upon the existing role of the C tail as a critical regulatory region. The C tail allosterically regulates the enzyme on two levels; it directly regulates the catalytic core and functions to facilitate interactions with cellular components that modulate catalytic activity [Kannan et al., 2007]. Our data show that RTKs directly phosphorylate PKA-C on a tyrosine residue/s and identified Y330 within the C tail as the major phosphorylation site in vitro. We demonstrate that phosphorylation of PKA-C by RTKs functionally modifies the enzymatic activity of PKA in vitro and lastly establish that this phosphorylation event occurs in response to stimulation of cells with several different growth factors.
Although the functional relevance of the AST of PKA-C has been extensively studied with regard to how it allosterically regulates the catalytic core, post-translational modification of this regulatory region has never been reported. In mammalian cells, post-translational modification of PKA-C is reported to occur through autophosphorylation on serine residues (S10, S110, and S338) and phosphorylation in the activation loop on T197 by PDK1 or a PDK1-like enzyme [Cheng et al., 1998; Yonemoto et al., 1993]. Large scale phosphoproteomic screens performed in mouse brain and cell lines expressing oncogenic tyrosine kinases have also observed phosphorylation on tyrosine 69, however, the kinase/s responsible for this phosphorylation event and the functional relevance have not yet been determined [Ballif et al., 2008; Guo et al., 2008; Rikova et al., 2007]. It is formally possible that other tyrosine residues in addition to Y330 are phosphorylated by RTKs at a lower abundance. However, given that there was no detectable tyrosine phosphorylation on sites other than Y330 via mass spectrometry and that mutation of Y330 to Y330F ablated phosphorylation, it is likely that Y330 is the major, if not only, residue phosphorylated by the PDGFR and EGFR.
The kinetic studies described here demonstrate that tyrosine phosphorylation of the catalytic subunit enhances the enzymatic activity of PKA. Y330 specifically appears to be involved in the hydrophobic packing of the backbone of the β1 strand that flanks the glycine rich loop (Fig. 4B) [Yang et al., 2009]. During the catalytic cycle, Y330 moves into close proximity to the p-3 subsite of the substrate, a site known to confer specificity for PKA [Kemp et al., 1977; Taylor et al., 1990; Walsh et al., 1992]. A previous report by Batkin et al. demonstrated that mutation of Y330 to a panel of amino acids resulted a marked increase in the Km and decrease in catalytic efficacy of PKA-C [Batkin et al., 2000]. Interestingly, of the mutants tested, the phenylalanine mutant (Y330F) exhibited the smallest although yet significant difference in Km as compared to control PKA-C. As conversion of tyrosine to phenylalanine retains the phenyl ring but loses the phenolic hydroxyl these results suggested that both the phenyl ring and phenolic hydroxyl of Y330 contribute to the Km of substrate. This is thought to occur through packing or Van der Waals interactions between the phenyl ring and residues in the conserved core and hydrogen bonding between the phenolic hydroxyl and the basic amino acid at the p-3 position, respectively. The decreased Km observed with the tyrosine phosphorylated PKA-C subunit are consistent with this idea that Y330 is involved in creating preferential affinity of kinase for substrate as the addition of a phosphate to the hydroxyl group would be predicted to strengthen a hydrogen bonding interaction with the basic amino acid at p-3 (Fig 4B). The timing of Y330 phosphorylation in EGF and PDGF-treated cells was coincident with growth factor-induced PKA activity (Fig. 5A&5B, 5C&D), and the tyrosine phosphorylated C subunit was restricted to the pseudopod of chemotaxing fibroblasts, a structure previously reported to have highly localized PKA activity [Howe et al., 2005]. However, whether phosphorylation of Y330 confers specificity for PKA to a particular subset of substrates or is required for growth factor-induced PKA kinase activity remains to be determined.
A bioinformatics study revealed that the C-tail of AGC kinases is highly conserved, suggesting a global mode of regulation for this family of kinases [Kannan et al., 2007]. In addition to directly contributing to the catalytic efficacy of the enzyme, the C tail of some AGC kinases contains protein interaction motifs that recruit cellular regulatory factors. For example, the PXXP motif in PKCβII was shown to bind to two chaperone proteins (Hsp90 and cdc37) and contribute to protein stability [Gould et al., 2009]. Moreover, a recent study identified a conserved FD(X)1–2Y/D motif within the tail that is essential for docking of the activating kinase, PDK1 [Romano et al., 2009]. Interestingly, in PKA, this motif encompasses Y330. In addition to modulating catatlytic activity itself, it is intriguing to speculate that phosphorylation of Y330 may also recruit the binding of a protein/s (e.g. SH2 and/or PTB domain-containing proteins) to the C tail which would then add an entirely new element to the regulation of PKA activity. While additional experimentation will be required to determine the contributions of this phosphorylation to the spectrum of cellular events mediated by growth factors, the current results provide the mechanistic basis for crosstalk between growth factor RTKs and protein kinase A, two major signal transduction pathways.
Supplementary Material
Acknowledgments
This work was funded by a Vermont Cancer Center/Lake Champlain Cancer Research Organization Pilot Award (P.B.D), through the Vermont Genetics Network NIH grant P20 RR16462 from the INBRE Program of the National Center for Research Resources (B.A.B), NIH grant HL68891 (W.R.D.) and Totman Trust for Biomedical Research (W.R.D.).
References
- Ballif BA, Carey GR, Sunyaev SR, Gygi SP. Large-scale identification and evolution indexing of tyrosine phosphorylation sites from murine brain. J Proteome Res. 2008;7:311–8. doi: 10.1021/pr0701254. [DOI] [PubMed] [Google Scholar]
- Barbier AJ, Poppleton HM, Yigzaw Y, Mullenix JB, Wiepz GJ, Bertics PJ, Patel TB. Transmodulation of epidermal growth factor receptor function by cyclic AMP-dependent protein kinase. J Biol Chem. 1999;274:14067–73. doi: 10.1074/jbc.274.20.14067. [DOI] [PubMed] [Google Scholar]
- Batkin M, Schvartz I, Shaltiel S. Snapping of the carboxyl terminal tail of the catalytic subunit of PKA onto its core: characterization of the sites by mutagenesis. Biochemistry. 2000;39:5366–73. doi: 10.1021/bi000153z. [DOI] [PubMed] [Google Scholar]
- Bornfeldt KE, Krebs EG. Crosstalk between protein kinase A and growth factor receptor signaling pathways in arterial smooth muscle. Cell Signal. 1999;11:465–77. doi: 10.1016/s0898-6568(99)00020-0. [DOI] [PubMed] [Google Scholar]
- Bornfeldt KE, Raines EW, Graves LM, Skinner MP, Krebs EG, Ross R. Platelet-derived growth factor. Distinct signal transduction pathways associated with migration versus proliferation. Ann N Y Acad Sci. 1995;766:416–30. doi: 10.1111/j.1749-6632.1995.tb26691.x. [DOI] [PubMed] [Google Scholar]
- Cheng X, Ma Y, Moore M, Hemmings BA, Taylor SS. Phosphorylation and activation of cAMP-dependent protein kinase by phosphoinositide-dependent protein kinase. Proc Natl Acad Sci U S A. 1998;95:9849–54. doi: 10.1073/pnas.95.17.9849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chestukhin A, Litovchick L, Schourov D, Cox S, Taylor SS, Shaltiel S. Functional malleability of the carboxyl-terminal tail in protein kinase A. J Biol Chem. 1996;271:10175–82. doi: 10.1074/jbc.271.17.10175. [DOI] [PubMed] [Google Scholar]
- Ciardiello F, Bianco R, Damiano V, De Lorenzo S, Pepe S, De Placido S, Fan Z, Mendelsohn J, Bianco AR, Tortora G. Antitumor activity of sequential treatment with topotecan and anti-epidermal growth factor receptor monoclonal antibody C225. Clin Cancer Res. 1999;5:909–16. [PubMed] [Google Scholar]
- Ciardiello F, Tortora G. Interactions between the epidermal growth factor receptor and type I protein kinase A: biological significance and therapeutic implications. Clin Cancer Res. 1998;4:821–8. [PubMed] [Google Scholar]
- deBlaquiere J, Walker F, Michelangeli VP, Fabri L, Burgess AW. Platelet-derived growth factor stimulates the release of protein kinase A from the cell membrane. J Biol Chem. 1994;269:4812–8. [PubMed] [Google Scholar]
- Deming PB, Campbell SL, Baldor LC, Howe AK. Protein kinase A regulates 3-phosphatidylinositide dynamics during platelet-derived growth factor-induced membrane ruffling and chemotaxis. J Biol Chem. 2008;283:35199–211. doi: 10.1074/jbc.M804448200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dostmann WR, Nickl C, Thiel S, Tsigelny I, Frank R, Tegge WJ. Delineation of selective cyclic GMP-dependent protein kinase Ialpha substrate and inhibitor peptides based on combinatorial peptide libraries on paper. Pharmacol Ther. 1999;82:373–87. doi: 10.1016/s0163-7258(98)00063-1. [DOI] [PubMed] [Google Scholar]
- Fishman D, Galitzki L, Priel E, Segal S. Epidermal growth factor regulates protein kinase A activity in murine fibrosarcoma cells: differences between metastatic and nonmetastatic tumor cell variants. Cancer Res. 1997;57:5410–5. [PubMed] [Google Scholar]
- Gould CM, Kannan N, Taylor SS, Newton AC. The chaperones Hsp90 and Cdc37 mediate the maturation and stabilization of protein kinase C through a conserved PXXP motif in the C-terminal tail. J Biol Chem. 2009;284:4921–35. doi: 10.1074/jbc.M808436200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves LM, Bornfeldt KE, Raines EW, Potts BC, Macdonald SG, Ross R, Krebs EG. Protein kinase A antagonizes platelet-derived growth factor-induced signaling by mitogen-activated protein kinase in human arterial smooth muscle cells. Proc Natl Acad Sci U S A. 1993;90:10300–4. doi: 10.1073/pnas.90.21.10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves LM, Bornfeldt KE, Sidhu JS, Argast GM, Raines EW, Ross R, Leslie CC, Krebs EG. Platelet-derived growth factor stimulates protein kinase A through a mitogen-activated protein kinase-dependent pathway in human arterial smooth muscle cells. J Biol Chem. 1996;271:505–11. doi: 10.1074/jbc.271.1.505. [DOI] [PubMed] [Google Scholar]
- Guo A, Villen J, Kornhauser J, Lee KA, Stokes MP, Rikova K, Possemato A, Nardone J, Innocenti G, Wetzel R, Wang Y, MacNeill J, Mitchell J, Gygi SP, Rush J, Polakiewicz RD, Comb MJ. Signaling networks assembled by oncogenic EGFR and c-Met. Proc Natl Acad Sci U S A. 2008;105:692–7. doi: 10.1073/pnas.0707270105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe AK, Baldor LC, Hogan BP. Spatial regulation of the cAMP-dependent protein kinase during chemotactic cell migration. Proc Natl Acad Sci U S A. 2005;102:14320–5. doi: 10.1073/pnas.0507072102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR, Miller WT. Receptor tyrosine kinases: mechanisms of activation and signaling. Curr Opin Cell Biol. 2007;19:117–23. doi: 10.1016/j.ceb.2007.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalvy S, Renault MA, Leen LL, Belloc I, Bonnet J, Gadeau AP, Desgranges C. Autocrine expression of osteopontin contributes to PDGF-mediated arterial smooth muscle cell migration. Cardiovasc Res. 2007;75:738–47. doi: 10.1016/j.cardiores.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Kannan N, Haste N, Taylor SS, Neuwald AF. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc Natl Acad Sci U S A. 2007;104:1272–7. doi: 10.1073/pnas.0610251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp BE, Graves DJ, Benjamini E, Krebs EG. Role of multiple basic residues in determining the substrate specificity of cyclic AMP-dependent protein kinase. J Biol Chem. 1977;252:4888–94. [PubMed] [Google Scholar]
- Kennedy EJ, Yang J, Pillus L, Taylor SS, Ghosh G. Identifying critical non-catalytic residues that modulate protein kinase A activity. PLoS One. 2009;4:e4746. doi: 10.1371/journal.pone.0004746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor KL, Mercurio AM. Protein kinase A regulates Rac and is required for the growth factor-stimulated migration of carcinoma cells. J Biol Chem. 2001;276:47895–900. doi: 10.1074/jbc.M107235200. [DOI] [PubMed] [Google Scholar]
- Piiper A, Lutz MP, Cramer H, Elez R, Kronenberger B, Dikic I, Muller-Esterl W, Zeuzem S. Protein kinase A mediates cAMP-induced tyrosine phosphorylation of the epidermal growth factor receptor. Biochem Biophys Res Commun. 2003;301:848–54. doi: 10.1016/s0006-291x(03)00055-x. [DOI] [PubMed] [Google Scholar]
- Pursiheimo JP, Jalkanen M, Tasken K, Jaakkola P. Involvement of protein kinase A in fibroblast growth factor-2-activated transcription. Proc Natl Acad Sci U S A. 2000;97:168–73. doi: 10.1073/pnas.97.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikova K, Guo A, Zeng Q, Possemato A, Yu J, Haack H, Nardone J, Lee K, Reeves C, Li Y, Hu Y, Tan Z, Stokes M, Sullivan L, Mitchell J, Wetzel R, Macneill J, Ren JM, Yuan J, Bakalarski CE, Villen J, Kornhauser JM, Smith B, Li D, Zhou X, Gygi SP, Gu TL, Polakiewicz RD, Rush J, Comb MJ. Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell. 2007;131:1190–203. doi: 10.1016/j.cell.2007.11.025. [DOI] [PubMed] [Google Scholar]
- Romano RA, Kannan N, Kornev AP, Allison CJ, Taylor SS. A chimeric mechanism for polyvalent trans-phosphorylation of PKA by PDK1. Protein Sci. 2009;18:1486–97. doi: 10.1002/pro.146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JD. A-kinase-anchoring proteins and cytoskeletal signalling events. Biochem Soc Trans. 2003;31:87–9. doi: 10.1042/bst0310087. [DOI] [PubMed] [Google Scholar]
- Stork PJ, Schmitt JM. Crosstalk between cAMP and MAP kinase signaling in the regulation of cell proliferation. Trends Cell Biol. 2002;12:258–66. doi: 10.1016/s0962-8924(02)02294-8. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Buechler JA, Yonemoto W. cAMP-dependent protein kinase: framework for a diverse family of regulatory enzymes. Annu Rev Biochem. 1990;59:971–1005. doi: 10.1146/annurev.bi.59.070190.004543. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Kim C, Cheng CY, Brown SH, Wu J, Kannan N. Signaling through cAMP and cAMP-dependent protein kinase: diverse strategies for drug design. Biochim Biophys Acta. 2008;1784:16–26. doi: 10.1016/j.bbapap.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SS, Kim C, Vigil D, Haste NM, Yang J, Wu J, Anand GS. Dynamics of signaling by PKA. Biochim Biophys Acta. 2005;1754:25–37. doi: 10.1016/j.bbapap.2005.08.024. [DOI] [PubMed] [Google Scholar]
- Walsh DA, Glass DB, Mitchell RD. Substrate diversity of the cAMP-dependent protein kinase: regulation based upon multiple binding interactions. Curr Opin Cell Biol. 1992;4:241–51. doi: 10.1016/0955-0674(92)90039-f. [DOI] [PubMed] [Google Scholar]
- Yang F, Liu Y, Bixby SD, Friedman JD, Shokat KM. Highly efficient green fluorescent protein-based kinase substrates. Anal Biochem. 1999;266:167–73. doi: 10.1006/abio.1998.2885. [DOI] [PubMed] [Google Scholar]
- Yang J, Kennedy EJ, Wu J, Deal MS, Pennypacker J, Ghosh G, Taylor SS. Contribution of non-catalytic core residues to activity and regulation in protein kinase A. J Biol Chem. 2009;284:6241–8. doi: 10.1074/jbc.M805862200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemoto W, Garrod SM, Bell SM, Taylor SS. Identification of phosphorylation sites in the recombinant catalytic subunit of cAMP-dependent protein kinase. J Biol Chem. 1993;268:18626–32. [PubMed] [Google Scholar]
- Zaccolo M, Pozzan T. Discrete microdomains with high concentration of cAMP in stimulated rat neonatal cardiac myocytes. Science. 2002;295:1711–5. doi: 10.1126/science.1069982. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.