Abstract
Alcohol dependence (AD) is an important contributory factor to the global burden of disease. The etiology of AD involves both environmental and genetic factors, and the disorder has a heritability of around 50%. The aim of the present study was to identify susceptibility genes for AD by performing a genome-wide association study (GWAS). The sample comprised 1,333 male in-patients with severe DSM-IV AD and 2,168 controls. These included 487 patients and 1,358 controls from a previous GWAS study by our group. All individuals were of German descent. Single marker tests and a polygenic score based analysis to assess the combined contribution of multiple markers with small effects were performed. The SNP rs1789891, which is located between the ADH1B and ADH1C genes, achieved genome-wide significance (p=1.27E–8; OR=1.46). Other markers from this region were also associated with AD, and conditional analyses indicated that these made a partially independent contribution. The SNP rs1789891 is in complete linkage disequilibrium with the functional Arg272Gln variant (p=1.24E–7, OR=1.31) of the ADH1C gene, which has been reported to modify the rate of ethanol oxidation to acetaldehyde in vitro. A polygenic score based approach produced a significant result (p=9.66E–9). This is the first GWAS of AD to provide genome-wide significant support for the role of the ADH gene cluster and to suggest a polygenic component to the etiology of AD. The latter result suggests that many more AD susceptibility genes still await identification.
Keywords: alcohol dehydrogenase, alcohol dependence, alcohol metabolism, genome-wide, GWAS, polygenic variation
Introduction
Alcohol dependence (AD) is a common and debilitating disorder, and ranks among the leading causes of the global burden of disease (World Health Organization 2009). Despite extensive research, it is unclear why only a proportion of all individuals exposed to alcohol become addicted. Elucidation of this point is important for the development of successful prevention and treatment strategies for AD. Previous research has shown that susceptibility to AD is partly genetic, and has a heritability of 40–60% (Enoch & Goldman 2002). The introduction of high-density SNP chips has enabled the systematic investigation of common variation in the human genome, and a large number of genome-wide association studies (GWAS) for a wide range of complex disorders have been published to date (Hindorff, Junkins, Hall et al. 2010). These investigations have implicated new variants, genes, and pathways. However, they have also demonstrated that sample size is crucial in detecting findings with genome-wide significance due to a combination of two effects. Firstly, effects of individual (common) variants are typically rather small and, secondly, as a consequence of the large number of simultaneously investigated markers on the arrays, the single marker analyses have to be corrected for the large number of tests (Yang, Benyamin, McEvoy et al. 2010), allowing only those markers with the strongest effects to surpass the level of genome-wide significance.
Recently, we reported a GWAS conducted in 487 male inpatients with DSM-IV AD and 1,358 population based control individuals (for a detailed description see Treutlein, Cichon, Ridinger et al. 2009). In brief, no marker surpassed the threshold of genome-wide significance. In a subsequent follow-up study of an independent sample of 1,024 male AD inpatients and 996 controls, we genotyped the 121 top SNPs from the GWAS as well as an additional 19 SNPs suggested by combining the GWAS results with gene expression data from rats subjected to chronic alcohol exposure. In a combined analysis of the GWAS and follow-up data, two tightly linked intergenic SNPs located close to the peroxisomal trans-2-enoyl-CoA reductase (PECR) gene achieved genome-wide significance.
The aims of the present study were: i) to enlarge our previous GWAS sample in order to increase the power to detect new susceptibility genes by raising them above the threshold of genome-wide significance, and ii) to test, for the first time, for the presence of a polygenic component to the development of AD. A polygenic component is defined as the presence of a multitude of small genetic effects (with individual effect sizes below the threshold for detection at the level of single markers within a GWAS), being measured as a summed effect. The latter method has recently been applied successfully to test for a polygenic contribution to schizophrenia and bipolar disorder (Purcell, Wray, Stone et al. 2009). For a detailed explanation of this approach see Purcell et al. (2009). In simple terms, the polygenic score approach uses information concerning association findings for several ten thousand SNPs from a discovery sample to calculate a summary score for each individual in a target sample. Subsequent analyses are performed to test whether the cases in the target sample have different scores to the controls in the target sample. Therefore, a GWAS is first carried out in a discovery sample. Following this step, the polygenic score for each individual in the target sample is calculated by counting the number of reference alleles (e.g. risk alleles or minor alleles) of this subject on any of the included loci. This number is weighted by multiplication with the log(OR) of the respective marker in the discovery sample. The sum of all of these products is then calculated and divided by the number of included loci. Finally, the association of this score with disease status is tested in the target sample by its inclusion as a predictor in a logistic regression model. The finding of the presence of a polygenic component to AD would indicate that many more individual genetic variants of small effect still await detection.
Materials and Methods
Subject recruitment
For the purposes of the present study, the sample from the previous GWAS (subsequently referred to as “GWAS1”) was enlarged by the inclusion of an additional 900 patients and the GWAS data of a further 862 German controls (subsequently referred to as “GWAS2”) Thus when in the text the “present study” is mentioned this refers to the pooled GWAS1 and GWAS2 data
All patients were male and of self reported German ancestry. All fulfilled DSM-IV criteria for AD and had been recruited from consecutive admissions to the psychiatry- and addiction medicine departments of German psychiatric hospitals participating in the German addiction research network (for a detailed description see Treutlein et al. 2009). The 900 additional patients included in GWAS2 were drawn from the 1,024 patients used to follow-up the 140 top SNPs from GWAS1 in our previous study (mean age at interview=43.3. SD=8.4 (GWAS2), mean=41.9. SD=8.8 (overall sample); mean age at onset =30.2, SD=7.8 (GWAS2) mean=27.0, SD=8.0 (overall sample).
The control subjects for GWAS2 were psychiatrically healthy individuals who had been randomly selected from a Munich community sample, and who had been screened using the Composite International Diagnostic Interview. This control sample was distinct from the control sample used in the follow-up study. In view of the limited financial resources available for genotyping, this sample was chosen since GWAS data were already available and the controls were from the same region as the majority of our patients. The mean age at interview was 51.9, SD=13.2 for this control sample and mean=50.5, SD=12.1 for the overall control sample. All of the GWAS phenotypes and genotypes are stored in a comprehensive database at the Institute for Medical Biometry, Informatics, and Epidemiology in Bonn (IMBIE). The study was approved by the respective local ethics committees and was carried out in accordance with the Declaration of Helsinki. All participants provided written informed consent.
Genotyping and Data Analysis
The 900 GWAS2 patient samples were individually genotyped using Illumina Human610Quad or Illumina Human660w Quad BeadChips (Illumina, Inc., San Diego, CA, USA). The controls had been genotyped using Illumina HumanHap550 Bead Chips.
Software and databases
PLINK 1.07 (Purcell, Neale, Todd-Brown et al. 2007) and R 2.7.2 (R Development Core Team 2010) were used for quality control (QC) and the association statistics. SMARTPCA (Patterson, Price & Reich 2006) was used for principal component analysis (PCA). Haploview 4.2 (Barrett, Fry, Maller et al. 2004) was used for linkage disequilibrium (LD) assessment. For the purposes of replication, GWAS datasets were retrieved from the dbGAP database (Mailman, Feolo, Jin et al. 2007). These datasets had been generated within the following studies: (i) ‘Alcohol Research using Australian twins and their families’ (subsequently referred to as OZ-ALC); (ii) ‘CIDR: Collaborative Study on the Genetics of Alcoholism’ (COGA; subsample of European ancestry only); and (iii) ‘Study of Addiction: Genetics and Environment’ (SAGE; subsample of European ancestry only).
Quality control (QC)
Prior to QC, the genotype data were pooled with the preQC data from GWAS1. QC was then performed on the pooled data. The following criteria were applied: sample call rate (CR) >=0.98; and conformity between reported sex and genotypic sex. In the case of duplicates or cryptic relatedness (IBS across autosomal markers >=1.6), the sample with the lower call rate was removed. Outliers were identified using PCA.
Only SNPs present on all of the applied genotyping platforms were included in the analysis. To remain in the dataset, the SNPs had to fulfill the following criteria: CR >=0.98; minor allele frequency (MAF) >=0.01; conformity with Hardy-Weinberg equilibrium (HWE; p>=1E–6) in any subsample; HWE conformity across all samples; and no significant difference in allele frequency between subsamples for controls. Cluster plots of our top findings were then assessed visually, and SNPs with poor cluster quality were removed. In 900 patients (genotyped in the follow-up study of GWAS1 with sequenom and in GWAS2 with illumina chip), replication of 140 SNPs showed 99.8% identity.
Single marker test statistics
A logistic regression approach was used in the performance of single marker association tests for autosomal SNPs. Correction for population stratification (PS) was performed using the first two principal components (PC) resulting from a PCA across independent autosomal markers (pair-wise r2<0.05 within a sliding window of 200 SNPs width, conformity of SNPs with HWE (p>0.01), and a minor allele frequency (MAF) >=0.05). These two PCs where included as covariates in the logistic regression model. For the conditional analysis of markers other than rs1789891, this SNP was included as an additional covariate in the regression model, using the allelic dosage of A alleles as a predictor.
Polygenic score based analysis
For the score based analysis, only pairwise independent autosomal SNPs (r2<0.25) with MAF>0.02 and p<0.5 were used. The total GWAS sample was divided at random into two halves. The first half was used as the discovery sample (667 patients; 1,084 controls) and the second half was used as the target sample (666 patients; 1,084 controls). On the basis of the association results of single marker tests in the discovery sample, a polygenic score was calculated for the subjects in the target sample. For a detailed description of the method see Purcell et al. (2009). To build the polygenic score for an individual, the number of minor alleles of this subject on any of the included loci was multiplied by the log(OR) of the respective marker. The sum of all of these products was then calculated and finally divided by the number of included loci. Testing for association between disease status and polygenic score was performed in the target sample using a logistic regression approach and the following two models: a) using disease status as a response variable, and the polygenic score and the first two principal components from the PCA as predictors; and b) the same model but with exclusion of the polygenic score from the predictor list. These two models were then compared to determine whether inclusion of the polygenic score in the model significantly improved fit. In a second step, the score based analysis was recalculated by exchanging the target and discovery samples in order to cross-validate the results. Since the size of the discovery sample is essential for obtaining an accurate prediction (Purcell et al. 2009), the total German sample was used as a discovery sample for the subsequent score based analyses of the COGA, SAGE, and OZ-ALC samples.
Local imputation of functional locus Arg272Gln (rs1693482)
The SNP rs1693482 was imputed locally using BEAGLE 3.3 (Browning & Browning 2007) and default settings. Genotype data from the 60 Hapmap (The International HapMap Consortium 2005) CEU founders (release 23a) were used as a reference panel in order to allow comparison of our results with previously reported findings for SNP rs1693482, which was not present in the overlap of the applied genotyping platforms.
Results
Effect of quality control (QC)
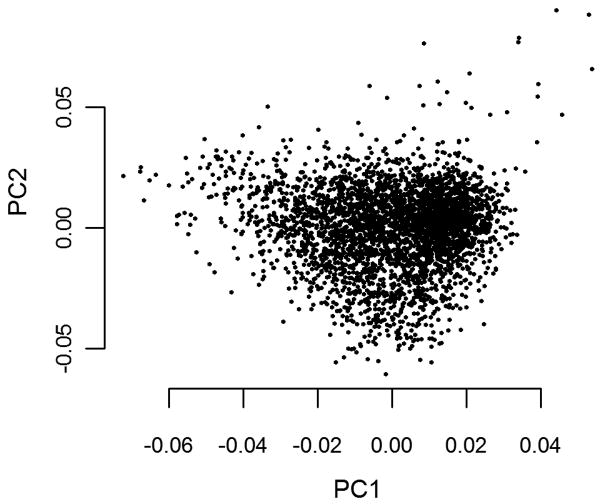
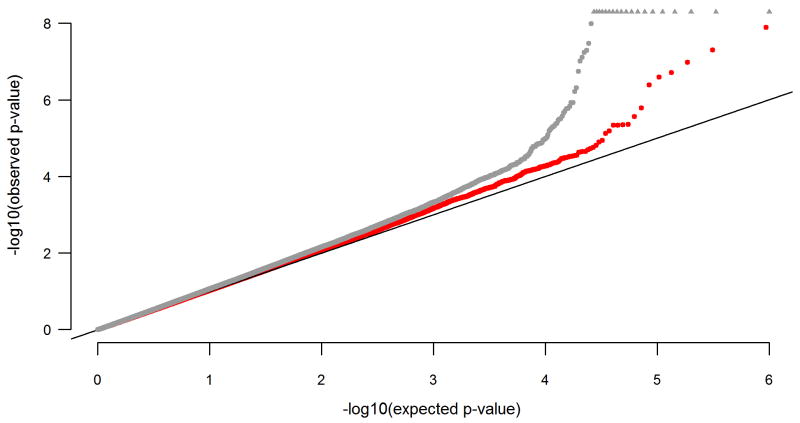
Thirty of the 900 GWAS2 cases were excluded from further analyses due to: failed genotyping/low call rate (n=13); a genome-wide average identity by state (IBS) score of above 1.60, indicating cryptic relatedness (n=1); discrepancies between reported- and genotype-derived sex (n=2); or outlier status (n=14). Figure 1 shows the results of the PCA following the exclusion of outliers. Of the 502,276 SNPs present in the SNP set content of all samples after pooling with the GWAS1 data, 500,467 were autosomal. Of these, 92.5% passed QC. The final data set comprised 463,044 autosomal SNPs from a total of 1,333 cases and 2,168 controls. The mean CR of the final set of SNPs (cases and controls combined) was 99.94% (SD=0.12%). To visualize the outcome of the QC steps, the pre- and postQC association p-values are depicted as a quantile-quantile plot (Figure 2).
Figure 1.
Scatter plot showing the first two principal components resulting from principal component analysis (PCA) after the removal of outliers.
Figure 2.
Quantile-quantile-plot of expected and observed association test p-values prior to- (gray) and following (red) quality control (QC). P-values<1E–8 are denoted with triangles at the top of the plot.
Statistical analysis of pooled data
Single marker tests
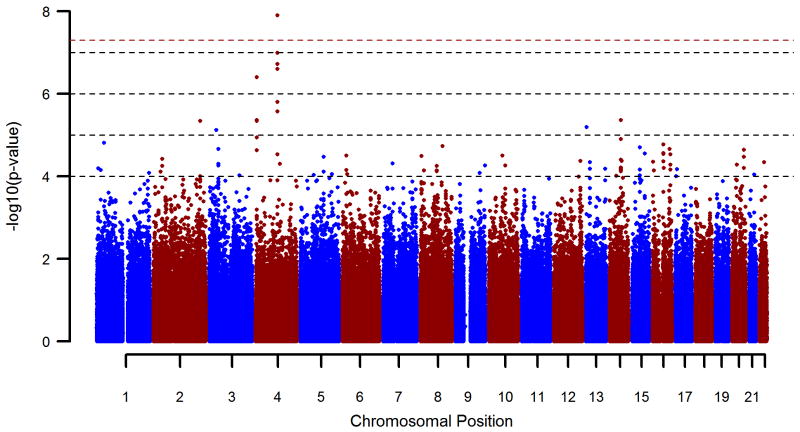
Figure 3 provides a genome-wide overview of the single marker test results. Inflation factor λ after QC was 1.037. Table 1 shows all single marker association findings with p<1E–5. The SNP rs1789891, which is located between the ADH1B and ADH1C genes, achieved genome-wide significance (p=1.27E–8; OR=1.46). Five other SNPs in the region of rs1789891 showed p<1E–5. A conditional analysis was performed on these SNPs by including rs1789891 as a covariate in the regression model. This revealed rs1789891-independent effects for the SNPs rs1789924 (p=8.13E–3), rs2851300 (p=5.44E–3), and rs4699748 (p=1.44E–2).
Figure 3.
Manhattan plot of pooled GWAS sample logistic regression p-values of all tested autosomal markers. The red dashed line shows the level of genome wide significance (p=5E-8).
Table 1.
Best SNPs (p<1.0E–5) of the pooled GWAS 1 + 2.
| SNP | Chromosome | Position | Reference Allele | Minor Allele Frequency | Odds ratio | 95%-CI of Odds Ratio | Na | Pb | Pcondc |
|---|---|---|---|---|---|---|---|---|---|
| rs1344694 | 2 | 216601882 | T | .35 | 1.27 | 1.15–1.41 | 3,483 | 4.59E–6 | |
| rs9825310 | 3 | 32215123 | A | .47 | 1.25 | 1.14–1.38 | 3,480 | 7.55E–6 | |
| rs7694974 | 4 | 4738538 | A | .48 | 1.26 | 1.14–1.39 | 3,497 | 4.40E–6 | |
| rs6818421 | 4 | 4740463 | A | .49 | 1.26 | 1.14–1.39 | 3,497 | 4.61E–6 | |
| rs1000579 | 4 | 4770395 | G | .43 | 1.30 | 1.17–1.43 | 3,500 | 4.01E–7 | |
| rs1789891d | 4 | 100469442 | A | .16 | 1.46 | 1.28–1.67 | 3,500 | 1.27E–8 | NA |
| rs169482e | 4 | 100482988 | T | .42 | 1.31 | 1.19–1.45 | 3,501 | 1.24E–7 | 6.22E–3 |
| rs1789924 | 4 | 100493309 | T | .42 | 1.30 | 1.18–1.44 | 3,501 | 1.92E–7 | 8.13E-3 |
| rs2851300 | 4 | 100498847 | T | .42 | 1.31 | 1.19–1.45 | 3,500 | 1.04E–7 | 5.44E-3 |
| rs1372679 | 4 | 100523553 | T | .10 | 1.48 | 1.26–1.74 | 3,501 | 1.61E–6 | 7.31E-2 |
| rs1372680 | 4 | 100523568 | A | .10 | 1.47 | 1.25–1.72 | 3,501 | 2.71E–6 | 9.24E-2 |
| rs4699748 | 4 | 100540466 | A | .11 | 1.49 | 1.28–1.73 | 3,501 | 2.51E–7 | 1.44E-2 |
| rs4770403 | 13 | 22653127 | A | .19 | 1.33 | 1.17–1.50 | 3,501 | 6.43E–6 | |
| rs2810114 | 14 | 70465357 | G | 28 | 1.29 | 1.16–1.44 | 3,486 | 4.39E–6 |
Markers in the ADH gene cluster region are indicated in bold font.
Number of non-missing individuals for each SNP.
Logistic regression p-value including correction for population stratification using 1st two principal components
logistic regression p-value of top SNPs in the ADH gene cluster after conditioning on rs1789891
top finding of present study
based on imputed data
Comparison with AD GWAS data from the COGA, SAGE, and OZ-ALC samples obtained through dbGAP (Mailman et al. 2007) showed that the SNP rs1789891 had also shown nominal significance in the COGA GWAS (p=1.47E–2), and that the same allele had been associated with AD. No significant association with this variant was found in the SAGE- or the OZ-ALC sample (Table 2).
Table 2. Test results of the top markers (p<1E–5) from the present study in the three data sets retrieved from dbGAP for replication purposes.
| SNP | Chromosome | Position | Reference allele | Odds ratio COGA | p-value COGA | p-value SAGE | p-value OZ-ALC |
|---|---|---|---|---|---|---|---|
| rs1344694 | 2 | 216601882 | T | 0.91 | 0.293 | 0.415 | 0.736 |
| rs9825310 | 3 | 32215123 | A | 1.28 | 0.003 | 0.178 | 0.745 |
| rs7694974 | 4 | 4738538 | A | 0.88 | 0.136 | 0.435 | NAa |
| rs6818421 | 4 | 4740463 | A | 0.88 | 0.126 | 0.435 | NAa |
| rs1000579 | 4 | 4770395 | G | 0.92 | 0.344 | 0.352 | 0.495 |
| rs1789891 | 4 | 100469442 | A | 1.31 | 0.015 | 0.309 | 0.906 |
| rs1789924 | 4 | 100493309 | T | 1.10 | 0.271 | 0.851 | 0.632 |
| rs2851300 | 4 | 100498847 | T | 1.09 | 0.307 | 0.878 | NAa |
| rs1372679 | 4 | 100523553 | T | 1.33 | 0.035 | 0.230 | NAa |
| rs1372680 | 4 | 100523568 | A | 1.28 | 0.068 | 0.268 | 0.883 |
| rs4699748 | 4 | 100540466 | A | 1.28 | 0.047 | 0.246 | NAa |
| rs4770403 | 13 | 22653127 | A | 0.90 | 0.323 | 0.095 | 0.065 |
| rs2810114 | 14 | 70465357 | G | 0.92 | 0.337 | 0.278 | NAa |
Nominally significant p-values are denoted in bold font. For the COGA sample, the OR estimators are shown.
data for this SNP were not available in the retrieved dbGAP data set
Two further SNPs located around ADH1C showed p<1E–5 in the present study and yielded nominal significance with the same allele in the COGA sample. These were rs1372679 (p=1.61E–6, OR=1.48; pCOGA=3.53E–2, ORCOGA=1.33), and rs4699748 (p=2.51E–7, OR=1.49; pCOGA=4.66E–2, ORCOGA=1.28; Table 2). The SNP rs9825310 (p=7.55E–6, OR=1.25), which is located near GPD1L on chromosome 3p22.3, also showed nominally significant association with the same allele in the COGA sample (pCOGA=2.95E–3, ORCOGA= 1.28). An analysis was performed to test for association in our GWAS for the top SNPs (p<1E–5) reported by the COGA (Edenberg, Koller, Xuei et al. 2010) and SAGE (Bierut, Agrawal, Bucholz et al. 2010) GWAS. This yielded no significant results.
Polygenic score based analysis
Genotype scores derived from each half of the German GWAS sample differed significantly between patients and controls of the respective remaining half (p=1.28E–6 and p=9.66E–9), and genotype scores derived from the total German sample differed between patients and controls in the COGA (p=3.92E–2) as well as in the SAGE (p=1.13E–4) sample (scores based on ∼ 84,000 SNPs).
Discussion
The SNP showing genome-wide significance in the present study is located in the region harboring the ADH gene cluster. This is the most consistently reported region in linkage analyses (Prescott, Sullivan, Kuo et al. 2006), and has shown evidence of association in a number of candidate gene studies of AD (Birley, James, Dickson et al. 2009). Existing knowledge of the biochemical function of the ADHs and the results of animal studies provide further strong support for the hypothesis that the ADHs are involved in the etiology of AD. The ADHs are involved in the enzymatic degradation of ingested ethanol, a process which results in the toxic intermediate acetaldehyde (Edenberg 2007). This is normally metabolized rapidly to acetate, in a reaction that is catalyzed by the aldehyde dehydrogenases (ALDHs). Accumulation of acetaldehyde in the blood causes the so called “flushing reaction”, which leads to the avoidance of alcohol consumption. This is the rationale for the use of the ALDH inhibitor disulfiram in the treatment of AD (Edenberg 2007; Petersen 1992; Suh, Pettinati, Kampman et al. 2006). A naturally occurring reduction in ALDH2 activity due to a functional mutation (Glu504Lys, rs671) in this gene, which is highly prevalent in Asians, protects a substantial proportion of this population from AD (Ball 2008; Eng, Luczak & Wall 2007; Enomoto, Takase, Yasuhara et al. 1991). Similar protective mechanisms have been suggested for variations in the ADH genes (Edenberg 2007; Li, Zhao & Gelernter 2011; Macgregor, Lind, Bucholz et al. 2008; Osier, Pakstis, Soodyall et al. 2002).
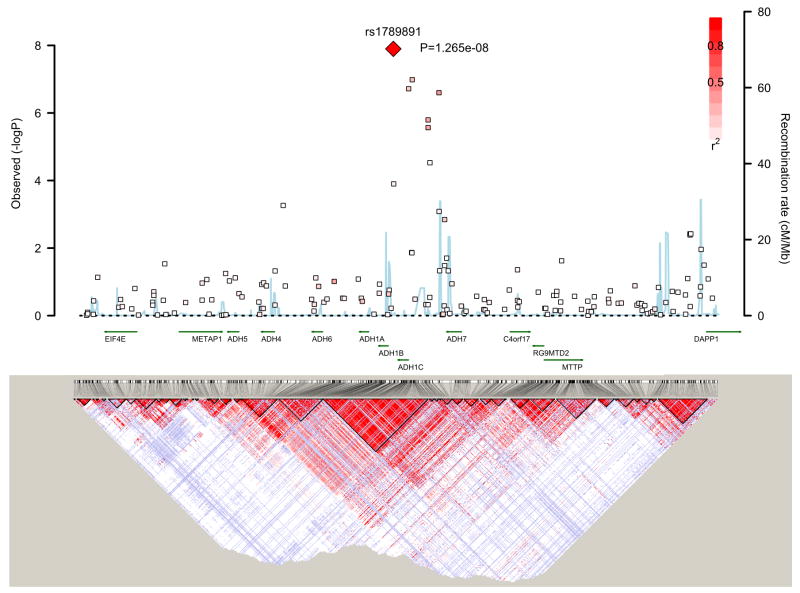
Seven ADH genes are known to exist in humans. Of these, class I gene products (ADH1A, ADH1B, and ADH1C) account for most of the capacity of the liver for ethanol oxidation. High LD has been reported for several variants across these genes (Edenberg 2007). The top SNP of the present study is located between the ADH1B and ADH1C genes. This is in complete LD (D′=1.0, r2=0.27, HapMap rel 24 CEU) with rs1693482 (Arg272Gln), a common functional amino acid substitution in the ADH1C gene. An Australian study found that Arg272Gln was associated with “amount of alcohol consumption” (Macgregor et al. 2008). This finding is compatible with the hypothesis that increased ADH activity may be protective against AD. Since Arg272Gln was not present in our data set, it was necessary to impute this marker. Based on the imputed genotypes, the higher-function 272Arg variant (Edenberg 2007) corresponding to the protective allele C of rs1789891 in the present GWAS was indeed found to be the protective allele in our sample. However, the association with Arg272Gln, (p=1.24E-7, OR=1.31) was not stronger than the association with rs1789891 (p=1.27E-8; OR=1.46). It therefore remains uncertain whether the observed association was due to the effect of the Arg272Gln. The SNP rs1789891 is located intergenically, in a region in which many SNPs are in high LD (Figure 4). Previous studies have shown that several variants in the ADH cluster, which are in LD with one another (Edenberg 2007), contribute independently to the risk of AD (Macgregor et al. 2008). The present findings are consistent with this observation. Apart from Arg272Gln, six of the 13 top SNPs with p<1E–5 were located within or around the ADH1C gene and were in LD with one another (D′ between 0.61 and 1.0) and the conditional analysis showed small rs1789891-independent effects (Table 1).Even in the absence of definite functional evidence for the association with rs1789891, this finding is relevant as it receives independent support from the COGA study, which found association with AD for the same allele (p=1.47E–2, OR=1.31).
Figure 4.
Illustration of p-values in a ±500kb interval around the most significant finding for rs1789891 on chromosome 4q including gene annotation using the SNAP proxy tool (http://www.broadinstitute.org/mpg/snap/). Linkage disequilibrium maps are from the International HapMap Project (http://hapmap.ncbi.nlm.nih.gov/).
This marker was also among the top SNPs in a GWAS of conduct disorder symptoms, which involved 909 patients with attention deficit disorder aged between five and 17 years and their parents (p=3.44E–5 (Sonuga-Barke, Lasky-Su, Neale et al. 2008); risk allele A [http://www.genome.gov/gwastudies]). Conduct disorder and AD are frequently comorbid, and twin studies support the hypothesis that conduct problems in children and antisocial behavior in adults are correlated genetically with AD (Pickens, Svikis, McGue et al. 1995; Slutske, Heath, Dinwiddie et al. 1998).
Thus one possible hypothesis is that the ADH variant contributes to the development of both conduct disorder and AD in the sense of a shared etiology. Another explanation is that the association finding originates from one of the two disorders, and that the association with the other disorder is simply the result of comorbidity. If the association arises from conduct disorder, the association would only be expected in patients with this disorder. Unfortunately, we were unable to test these alternative hypotheses by stratifying our sample for co-morbid conduct disorder. Further studies are required to disentangle this issue.
A total of 13 markers in the present study reached a value of p<1E–5, including rs1789891. Of the seven variants that are not located at the ADH1C locus, four SNPs are located in genes (see Table 1; for functional annotation of these genes see supporting Table S1).
Comparison of the 13 top SNPs from the present study and the COGA, SAGE, and OZ-ALC, showed that four of these 13 SNPs had shown nominal significance in the COGA study (Table 2). Three of these SNPs were in the ADH gene cluster, including the SNP with genome wide significance (Table 1). The fourth SNP, rs9825310, is located intergenically on chromosome 3. The closest RefSeq gene to rs9825310 is the glycerol-3-phosphate dehydrogenase 1-like (GPD1L) gene (OMIM MIM ID *611778), which is located at a distance of ∼30kb. In humans, mutations in the GPD1L gene have been reported to cause Brugada syndrome 2, which is a progressive disorder of myocardial conduction (OMIM MIM ID #611777). Interestingly, this gene has received support for the phenotype “alcohol stimulated activity” in the mouse (Palmer, Lessov-Schlaggar, Ponder et al. 2006), as it is located in the QTL Etact2 on mouse chromosome 9 (http://bioinfo.mc.vanderbilt.edu/ERGR/qtlds.php?qtl=Etact2).
A possible explanation for why consistent findings were only observed in the COGA sample and not in the two other large data sets is that of these three previous studies, the COGA study had the most similar recruitment scheme to ours, as patients were recruited through alcohol treatment programs. This enriches the sample with the most severe AD cases, diagnosed in accordance with DSM. In an attempt to increase the power of the present study, we increased the homogeneity of the patient sample by including male patients with severe AD only. Thus our findings are confined to this specific patient group. Nevertheless, genes identified through systematic screening in clinically homogeneous samples comprised of severely ill patients can provide promising candidates for studies of more heterogeneous samples.
As with GWAS of other neuropsychiatric phenotypes published to date, there is limited consistency among the top findings for AD. This is likely to reflect the small genetic effects conferred by these variants and the very limited power of each individual study to detect them consistently at a genome-wide significant level.
Although the hypothesis that many variants with small effect contribute in concert to AD is well established, no study has demonstrated that the sum of a multitude of non-significant effects contributes to the development of AD. Our group is the first to have found supportive evidence for such a contribution. This finding shows that many more genetic variants still await detection. We were able to show that these variants which may confer vulnerability through many different biological mechanisms, act across different samples. Since the effect sizes of individual risk alleles involved in polygenic susceptibility will be low, it will be difficult to distinguish true risk effects from false positives at the level of individual genes. Pathway-based analysis involving the incorporation of risk alleles from the multiple genes that contribute to a particular pathway may overcome this limitation, and allow the translation into a meaningful biological context.
In summary, in view of the a priori evidence for the validity of the ADH association finding and the independent replication, the present study has demonstrated the potential of the GWAS approach, which could lead to many more valuable findings if pursued in sufficiently large and homogeneous samples. The finding of a polygenic component is consistent with findings for other psychiatric disorders, and suggests the existence of a continuum of genetic effects, and that a host of susceptibility genes still await identification.
Supplementary Material
Acknowledgments
Funding Support: This work was supported by grants U01 HL089856, R01 MH087590 and R01 MH081862 from the National Heart, Lung, and Blood Institute (NIH/NHLBI) (to MM), by grant FKZ 01GS08152 from the National Genome Research Network (NGFNplus – see under www.ngfn-alkohol.de and Spanagel et al., 2010 - to MR, MMN) of the German Federal Ministry of Education and Research (BMBF), and by grants FKZ 01GS0117/NGFN and FKZ EB 01011300. KM was supported by grant 01EB0410 from the Bundesministerium für Bildung und Forschung. MMN and SC received support from the Alfried Krupp von Bohlen und Halbach-Stiftung.
Additional Contributions: We thank Marina Füg and Christine Hohmeyer for their expert technical assistance. We also thank Thomas G. Schulze, MD, Georg-August-University Göttingen, for helpful discussions.
dbGAP datasets: Funding support for the [CIDR-COGA Study] was provided through [the Center for Inherited Disease Research (CIDR) and the Collaborative Study on the Genetics of Alcoholism (COGA)]. The [CIDR-COGA Study] is a genome-wide association study funded as part of the [Collaborative Study on the Genetics of Alcoholism (COGA)]. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the [Collaborative Study on the Genetics of Alcoholism (COGA)]. Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for the collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/sites/entrez?Db=gap through dbGaP accession number phs000125.v1.p1.
Funding support for the Study of Addiction: Genetics and Environment (SAGE) was provided through the NIH Genes, Environment, and Health Initiative [GEI] (U01 HG004422). SAGE is one of the genome-wide association studies funded as part of the Gene Environment Association Studies (GENEVA) under GEI. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the GENEVA Coordinating Center (U01 HG004446). Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for the collection of datasets and samples was provided by the Collaborative Study on the Genetics of Alcoholism (COGA; U10 AA008401), the Collaborative Genetic Study of Nicotine Dependence (COGEND; P01 CA089392), and the Family Study of Cocaine Dependence (FSCD; R01 DA013423). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, the National Institute on Drug Abuse, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000092.v1.p1 through dbGaP accession number phs000092.v1.p.
Funding support for the [CIDR-OZ-ALC GWAS] was provided through [the Center for Inherited Disease Research (CIDR) and the National Institute on Alcohol Abuse and Alcoholism (NIAAA). The [CIDR-OZ-ALC GWAS] is a genome-wide association study funded as part of the NIAAA grant 5 R01 AA013320-04. Assistance with phenotype harmonization and genotype cleaning, as well as with general study coordination, was provided by the [CIDR-OZ-ALC GWAS]. Assistance with data cleaning was provided by the National Center for Biotechnology Information. Support for the collection of datasets and samples was provided by the MARC: Risk Mechanisms in Alcoholism and Comorbidity (MARC; P60 AA011998-11). Funding support for genotyping, which was performed at the Johns Hopkins University Center for Inherited Disease Research, was provided by the NIH GEI (U01HG004438), the National Institute on Alcohol Abuse and Alcoholism, and the NIH contract “High throughput genotyping for studying the genetic contributions to human disease” (HHSN268200782096C). The datasets used for the analyses described in this manuscript were obtained from dbGaP at http://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000181.v1.p1 through dbGaP accession number phs000181.v1.p1.
Wolfgang Gaebel has received symposia support from Janssen-Cilag GmbH, Neuss, Lilly Deutschland GmbH, Bad Homburg and Servier, Munich. He is a member of the Scientific Advisory Board of Lundbeck International Neuroscience Foundation (LINF), Denmark.
Footnotes
Financial Disclosure and possible conflicts of interest: The other coauthors have no biomedical financial interests or potential conflicts of interest to declare.
Author contributions: MaR, MMN, and SC were responsible for the study concept and design. MoR, NW, MSo, PZ, WM, RM, WG, ND, NS, SL, MI, KM, and FK contributed to the acquisition of samples. JF, MM, and MSt performed the quality control procedures and the statistical analyses. BMM, SH, and PH assisted with the data analysis and the interpretation of findings. JF, MaR, JT, MMN, SC, and CS drafted the manuscript. MMN and SC provided a critical revision of the manuscript for important intellectual content. All of the authors have critically reviewed the content of the manuscript, and have approved submission of the final version for publication.
References
- Ball D. Addiction science and its genetics. Addiction. 2008;103:360–367. doi: 10.1111/j.1360-0443.2007.02061.x. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2004;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bierut LJ, Agrawal A, Bucholz KK, Doheny KF, Laurie C, Pugh E, Fisher S, Fox L, Howells W, Bertelsen S, Hinrichs AL, Almasy L, Breslau N, Culverhouse RC, Dick DM, Edenberg HJ, Foroud T, Grucza RA, Hatsukami D, Hesselbrock V, Johnson EO, Kramer J, Krueger RF, Kuperman S, Lynskey M, Mann K, Neuman RJ, Nöthen MM, Nurnberger JI, Porjesz B, Ridinger M, Saccone NL, Saccone SF, Schuckit MA, Tischfield JA, Wang JC, Rietschel M, Goate AM, Rice JP. A genome-wide association study of alcohol dependence. Proc Natl Acad Sci U S A. 2010;107:5082–5087. doi: 10.1073/pnas.0911109107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birley AJ, James MR, Dickson PA, Montgomery GW, Heath AC, Martin NG, Whitfield JB. ADH single nucleotide polymorphism associations with alcohol metabolism in vivo. Human Molecular Genetics. 2009;18:1533–1542. doi: 10.1093/hmg/ddp060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Rapid and Accurate Haplotype Phasing and Missing-Data Inference for Whole-Genome Association Studies By Use of Localized Haplotype Clustering. The American Journal of Human Genetics. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ. The genetics of alcohol metabolism: role of alcohol dehydrogenase and aldehyde dehydrogenase variants. Alcohol Res Health. 2007;30:5–13. [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Koller DL, Xuei X, Wetherill L, McClintick JN, Almasy L, Bierut LJ, Bucholz KK, Goate A, Aliev F, Dick D, Hesselbrock V, Hinrichs A, Kramer J, Kuperman S, Nurnberger JI, Rice JP, Schuckit MA, Taylor R, Todd Webb B, Tischfield JA, Porjesz B, Foroud T. Genome-wide association study of alcohol dependence implicates a region on chromosome 11. Alcohol Clin Exp Res. 2010;34:840–852. doi: 10.1111/j.1530-0277.2010.01156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng MY, Luczak SE, Wall TL. ALDH2, ADH1B, and ADH1C genotypes in Asians: a literature review. Alcohol Res Health. 2007;30:22–27. [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, Goldman D. Problem drinking and alcoholism: diagnosis and treatment. Am Fam Physician. 2002;65:441–448. [PubMed] [Google Scholar]
- Enomoto N, Takase S, Yasuhara M, Takada A. Acetaldehyde metabolism in different aldehyde dehydrogenase-2 genotypes. Alcohol Clin Exp Res. 1991;15:141–144. doi: 10.1111/j.1530-0277.1991.tb00532.x. [DOI] [PubMed] [Google Scholar]
- Hindorff LA, Junkins HA, Hall PN, Mehta JP, Manolio TA. A Catalog of Genome-Wide Association Studies. [Accessed 6 December 2010];2010 Available at: http://www.genome.gov/gwastudies.
- Li D, Zhao H, Gelernter J. Strong Association of the Alcohol Dehydrogenase 1B Gene (ADH1B) with Alcohol Dependence and Alcohol-Induced Medical Diseases. Biol Psychiatry. 2011 doi: 10.1016/j.biopsych.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macgregor S, Lind PA, Bucholz KK, Hansell NK, Madden PAF, Richter MM, Montgomery GW, Martin NG, Heath AC, Whitfield JB. Associations of ADH and ALDH2 gene variation with self report alcohol reactions, consumption and dependence: an integrated analysis. Human Molecular Genetics. 2008;18:580–593. doi: 10.1093/hmg/ddn372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L, Popova N, Pretel S, Ziyabari L, Lee M, Shao Y, Wang ZY, Sirotkin K, Ward M, Kholodov M, Zbicz K, Beck J, Kimelman M, Shevelev S, Preuss D, Yaschenko E, Graeff A, Ostell J, Sherry ST. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39 doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osier MV, Pakstis AJ, Soodyall H, Comas D, Goldman D, Odunsi A, Okonofua F, Parnas J, Schulz LO, Bertranpetit J, Bonne-Tamir B, Lu RB, Kidd JR, Kidd KK. A global perspective on genetic variation at the ADH genes reveals unusual patterns of linkage disequilibrium and diversity. Am J Hum Genet. 2002;71:84–99. doi: 10.1086/341290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer AA, Lessov-Schlaggar CN, Ponder CA, McKinnon CS, Phillips TJ. Sensitivity to the locomotor-stimulant effects of ethanol and allopregnanolone: a quantitative trait locus study of common genetic influence. Genes Brain Behav. 2006;5:506–517. doi: 10.1111/j.1601-183X.2005.00198.x. [DOI] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population Structure and Eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen EN. The pharmacology and toxicology of disulfiram and its metabolites. Acta Psychiatr Scand Suppl. 1992;369:7–13. doi: 10.1111/j.1600-0447.1992.tb03309.x. [DOI] [PubMed] [Google Scholar]
- Pickens RW, Svikis DS, McGue M, LaBuda MC. Common genetic mechanisms in alcohol, drug, and mental disorder comorbidity. Drug Alcohol Depend. 1995;39:129–138. doi: 10.1016/0376-8716(95)01151-n. [DOI] [PubMed] [Google Scholar]
- Prescott CA, Sullivan PF, Kuo PH, Webb BT, Vittum J, Patterson DG, Thiselton DL, Myers JM, Devitt M, Halberstadt LJ, Robinson VP, Neale MC, van den Oord EJ, Walsh D, Riley BP, Kendler KS. Genomewide linkage study in the Irish affected sib pair study of alcohol dependence: evidence for a susceptibility region for symptoms of alcohol dependence on chromosome 4. Mol Psychiatry. 2006;11:603–611. doi: 10.1038/sj.mp.4001811. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, Sullivan PF, Sklar P. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. 2009;460:748–752. doi: 10.1038/nature08185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: 2010. [Google Scholar]
- Slutske WS, Heath AC, Dinwiddie SH, Madden PA, Bucholz KK, Dunne MP, Statham DJ, Martin NG. Common genetic risk factors for conduct disorder and alcohol dependence. J Abnorm Psychol. 1998;107:363–374. doi: 10.1037//0021-843x.107.3.363. [DOI] [PubMed] [Google Scholar]
- Sonuga-Barke EJS, Lasky-Su J, Neale BM, Oades R, Chen W, Franke B, Buitelaar J, Banaschewski T, Ebstein R, Gill M, Anney R, Miranda A, Mulas F, Roeyers H, Rothenberger A, Sergeant J, Steinhausen HC, Thompson M, Asherson P, Faraone SV. Does parental expressed emotion moderate genetic effects in ADHD? An exploration using a genome wide association scan. Am J Med Genet B Neuropsychiatr Genet. 2008;147B:1359–1368. doi: 10.1002/ajmg.b.30860. [DOI] [PubMed] [Google Scholar]
- Spanagel R, Bartsch D, Brors B, Dahmen N, Deussing J, Eils R, Ende G, Gallinat J, Gebicke-Haerter P, Heinz A, Kiefer F, Jäger W, Mann K, Matthäus F, Nöthen M, Rietschel M, Sartorius A, Schütz G, Sommer WH, Sprengel R, Walter H, Wichmann E, Wienker T, Wurst W, Zimmer A. An integrated genome research network for studying the genetics of alcohol addiction. Addict Biol. 2010;15:369–379. doi: 10.1111/j.1369-1600.2010.00276.x. [DOI] [PubMed] [Google Scholar]
- Suh JJ, Pettinati HM, Kampman KM, O'Brien CP. The status of disulfiram: a half of a century later. J Clin Psychopharmacol. 2006;26:290–302. doi: 10.1097/01.jcp.0000222512.25649.08. [DOI] [PubMed] [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, Fehr C, Scherbaum N, Steffens M, Ludwig KU, Frank J, Wichmann HE, Schreiber S, Dragano N, Sommer WH, Leonardi-Essmann F, Lourdusamy A, Gebicke-Haerter P, Wienker TF, Sullivan PF, Nöthen MM, Kiefer F, Spanagel R, Mann K, Rietschel M. Genome-wide association study of alcohol dependence. Arch Gen Psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Global health risks: Mortality and burden of disease attributable to selected major risks. Geneva: WHO; 2009. [Google Scholar]
- Yang J, Benyamin B, McEvoy BP, Gordon S, Henders AK, Nyholt DR, Madden PA, Heath AC, Martin NG, Montgomery GW, Goddard ME, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nat Genet. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.