Abstract
Sweet Screens: The promiscuity of sialyl- and fucosyltransferases has been exploited to develop a high-throughput screening (HTS) platform wherein transfer of a fluorescent analog of the natural donor substrate to a glycoprotein acceptor results in a robust fluorescence polarization readout. The use of this method to screen 16,000 compounds against 5 different glycosyltransferases has identified a panel of interesting inhibitors.
Keywords: Carbohydrates, Fucose, Glycoproteins, Glycosyltransferase, Sialic Acid
Sialyl- and fucosyltransferases decorate glycans of glycoproteins and glycolipids with terminal sialic acid and fucose residues, creating diverse structures recognized as ligands by pathogens and mammalian glycan binding proteins.[1] Their synthesis is carried out by the regulated expression of 20 sialyltransferase (ST) and 14 fucosyltransferase (FUT) genes that are highly conserved in mammalian species, each of which exhibits high specificity for their donor and acceptor substrates, giving rise to terminal sequences expressed in a cell type specific manner. Although phenotypes of knock-out mice have validated several of these enzymes as drug targets for the treatment of autoimmune disorders (ST6Gal I and ST3Gal I) and chronic inflammation (FUT7),[1b] no small molecule inhibitors have yet emerged.
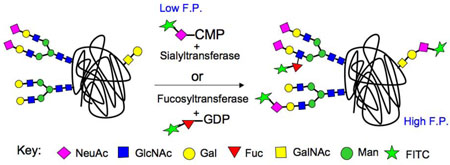
A limitation in identifying glycosyltransferase (GT) inhibitors has been the lack of simple and robust assays for high throughput screening (HTS) of compound libraries. Although a number of approaches have been used to screen for GT inhibitors, they comprise non-homogeneous multi-step formats, or are designed for screening a few specific enzymes.[2] Here we report a fluorescence polarization (FP) based assay system that is broadly applicable to members of the ST and FUT families. Since STs and FUTs are well documented to utilize nucleotide sugar donor substrates modified to contain bulky substituents on the sialic acid at C-9 or fucose C-6,[3] respectively, we reasoned that transfer of a sugar with a fluorescent tag to a suitable glycoprotein acceptor substrate would form the basis of a homogeneous FP assay due to the large difference in molecular size between the fluorescent substrate and glycoprotein products (Figure 1).[4]
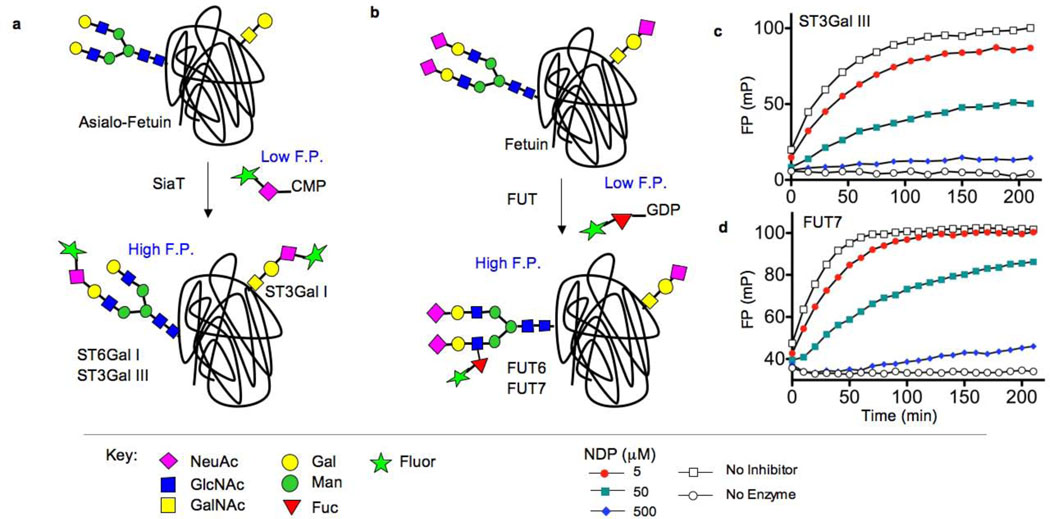
Figure 1. Fluorescence polarization-based assays for STs and FUTs (Option 1).
(a) Sialyltransferase (SiaT) assay based on transfer of FITCNeuAc from CMP-FITCNeuAc to N-linked (ST6Gal I, ST3Gal III) and O-linked (ST3Gal I) glycans of asialo-fetuin. (b) Fucosyltransferase (FUT) assay based on transfer of a fluorescent fucose analog from GDP-FLUORFuc to an N-glycan on fetuin. (c) Time course of ST3Gal III-mediated transfer of FITCNeuAc to asialo-fetuin produces increased fluorescence polarization over time, and is inhibited by the known competitive inhibitor, CDP. (d) Time-course of the FUT7 fluorescence polarization assay using GDP-FLUORFuc, fetuin, and FUT7, and inhibition by the known competitive inhibitor GDP.
To test this assay strategy we synthesized fluorescein-containing analogs of the corresponding nucleotide sugar substrates, CMP-NeuAc and GDP-Fucose, respectively (Schemes 1 and 2, and Supplementary Methods). Fetuin (~50 kDa) was chosen as the glycoprotein acceptor substrate since it is commercially available and contains well characterized N-linked and O-linked glycans that are recognized as acceptor substrates by nearly all STs and FUTs.[5] As shown in Figure 1c and Figure S1, transfer of FITCNeuAc to asialo-fetuin by the ST3Gal III, ST6Gal I, or ST3Gal I STs results in a time-dependent increase in FP, which can be inhibited in a dose-dependent manner by the competitive inhibitor, CDP. Similarly, transfer of FLUOR-Fuc by either FUT6 or FUT7 to native fetuin also yields a robust increase in FP, which is inhibitable by GDP (Figure 1d and Figure S1). A powerful aspect of this assay is that it is catalytic in nature, as evidenced by the lack of FP signal when the acceptor substrate is withheld (Figure S2), and therefore allows for the identification of both donor and acceptor substrate site inhibitors (Figure S3) and requires minimal amounts of difficult to express enzymes.
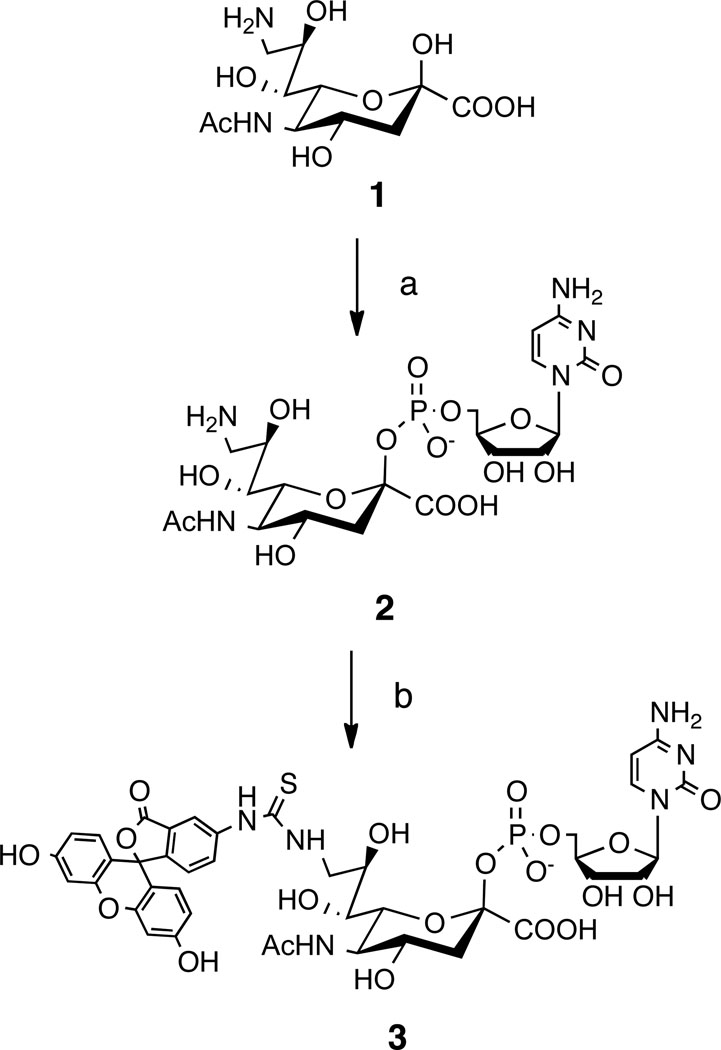
Scheme 1.
Synthesis of CMP-FITCNeuAc 3. Reagents and conditions: a) CTP, CMP-NeuAc Synthetase, 85%; b) FITC, MeOH, H2O, NaHCO3, 58%.
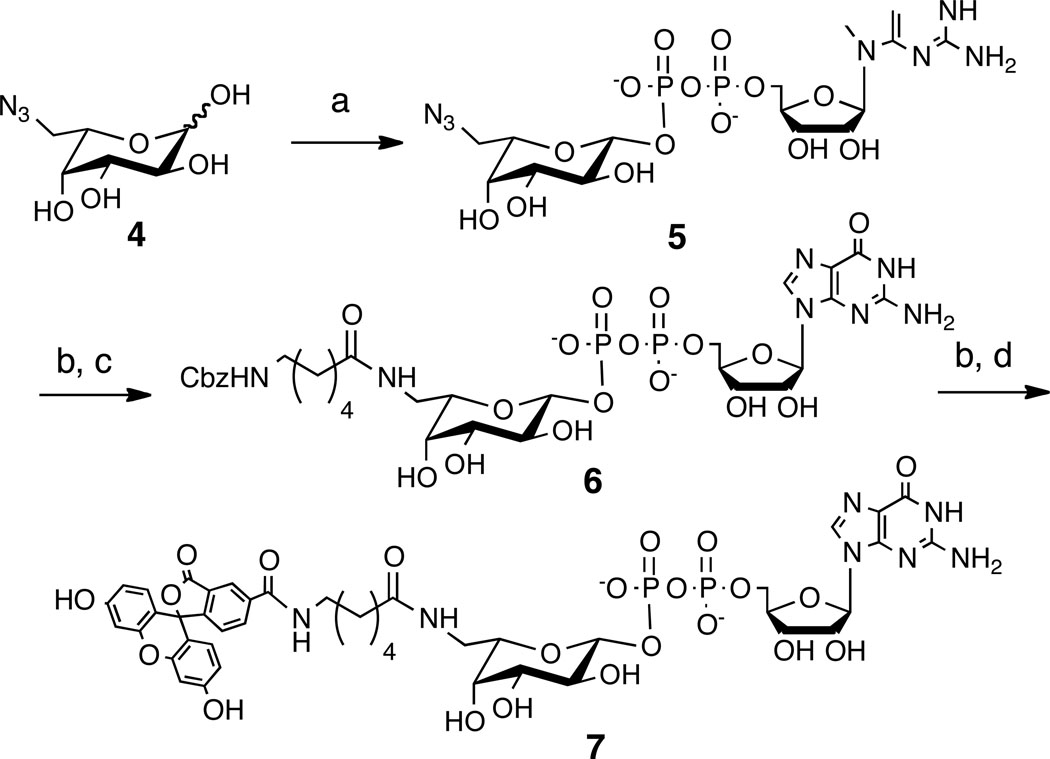
Scheme 2.
Synthesis of GDP-FLOURFuc 7. Reagents and Conditions: a) FKP, ATP, GTP, Pyrophosphatase, Ref. [6]; b) H2, Pd/C 20 mM NH4OH, 95–99%; c) N-Cbz-Aminocaproic Acid NHS Ester, DMF, H2O, NaHCO3, 90%; d) 5-Carboxyfluorescein NHS Ester, DMF, H2O, NaHCO3, 81%.
To assess the performance of the assay in a real HTS setting, the Maybridge Hitfinder collection of 16,000 compounds, was screened at 10 µM against each of the five GT targets. The results of these screens were successful in two respects. First, on a plate-to-plate basis, Z’ was consistently above 0.8 for each enzyme (Table S1). Second, inhibitors were identified for all targets demonstrating that small molecule inhibitors for this family of enzymes can indeed be discovered (Table S1). This is especially encouraging given the size of the library screened, and the low substrate concentrations used for screening. It was striking, however, that the hit rates varied considerably amongst enzymes (Table S1), which is likely due to library bias rather than assay design, since each enzyme assay was set up similarly, with substrate concentrations well below KM, and reactions allowed to proceed to ~75% transfer of the donor substrate.
To validate hits from the screening campaign, a simple secondary assay was employed. Reactions were carried out identically as in the primary screen, but instead of reading FP, the reactions were quenched, loaded onto an SDS-PAGE gel, and in-gel fluorescence was measured (Figure S4). This method, along with analysis of the total fluorescence of any putative hit, allows for rapid removal of false positives occurring from fluorescent compound interference or other HTS artifacts, which, we should note, were remarkably low (~ 6% of the analyzed hits) (Figure S4).
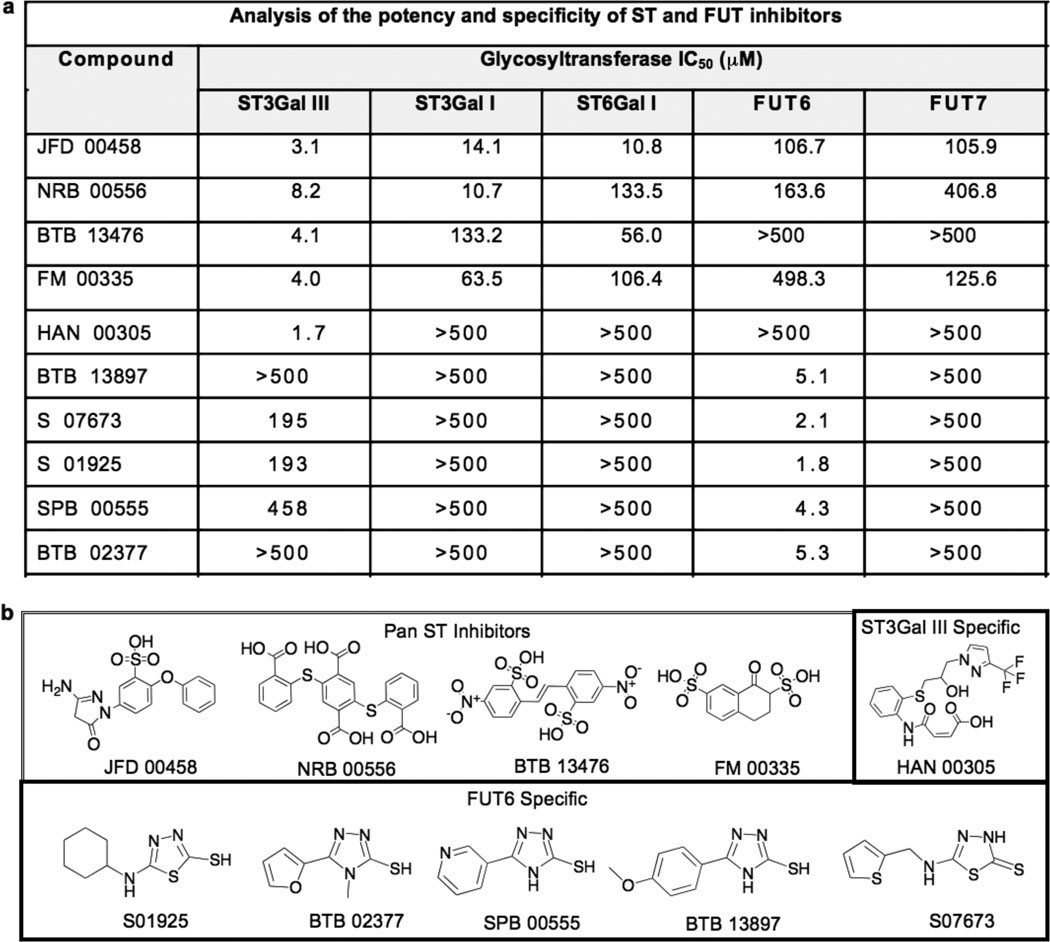
Of the confirmed hits, structural analysis and specificity profiles revealed several interesting inhibitor classes. As shown in Figure 2, a group of negatively charged pan ST inhibitors showed either weak (e.g. JFD00458) or negligible activity against the FTs. One inhibitor (HAN 00305) exhibited >250 fold higher potency for ST3Gal III than all the other STs and FTs, and in contrast, a group of structurally related inhibitors of FUT6 showed negligible inhibition of the other GTs.
Figure 2. Novel inhibitors of STs and FTs identified from high-throughput screening.
a) Potency and selectivity of validated inhibitors was assessed by titration analysis using the fluorescence polarization assay. Results are the average of at least two independent experiments carried out in triplicate with standard deviations less than 10%. b) Structures of validated sialyltransferase and fucosyltransferase hits include a related set of pan sialyltransferase inhibitors, an ST3Gal III specific inhibitor, and a set of FUT6 specific inhibitors with a related pharmacophore.
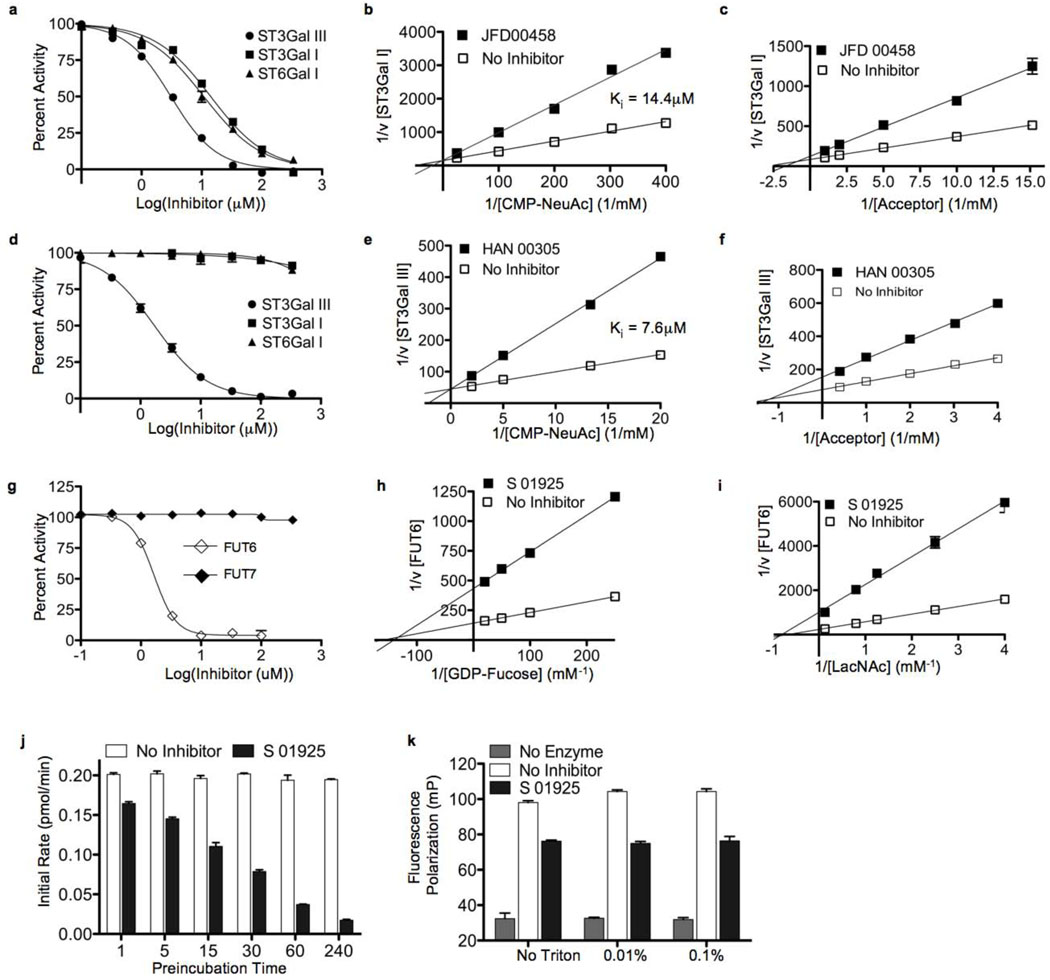
Representative inhibitors of each of these groups were selected for kinetics analysis using the natural nucleotide sugar to better understand the mechanism of inhibition and confirm that they were not interfering solely with the binding of the fluorescent substituents on the donor substrates used in the screen. As expected, based upon the presence of a negatively charged sulfonic acid moiety, the pan-ST inhibitor JFD00458 was competitive with the common donor substrate CMP-NeuAc (Figure 3a–c). However, the extremely selective ST3Gal III inhibitor, HAN00305, was also found to be competitive with the donor substrate site (Figure 3d–f), highlighting the fact that although similarities may exist at the CMP-NeuAc binding site of the STs, there are also significant differences that allow for selective inhibition. A group of inhibitors with a common pharmacophore were highly selective for FUT6, as exemplified by S01925 (Figure 3g), which showed non-competitive inhibition with respect to both donor and acceptor substrates (Figure 3h,i). Further studies showed that the degree of inhibition was time-dependent, suggesting that S01925 was an irreversible inhibitor (Figure 3j). Although promiscuous aggregators would also exhibit these characteristics, these molecules are selective, and moreover, they are completely detergent insensitive (Figure 3k).[6]
Figure 3. Mechanism of inhibition of ST and FUT inhibitors.
a) Inhibition by the pan sialyltransferase inhibitor JFD 00458 was assessed for all three sialyltransferases using the fluorescence polarization assay. Percent activity is relative to DMSO only (100%) and no enzyme (0%). Kinetic analysis of JFD 00458 inhibition of ST3Gal I was assessed with: b) CMP-NeuAc as the varied donor substrate, and c) Galβ1–3GalNAc as the varied acceptors substrate. d) The ST3Gal III specific inhibitor HAN 00305, was assessed for potency against all three sialyltransferases. To assess the mechanism of inhibition, kinetics were performed with: e) CMP-NeuAc as the varied donor substrate, and f) with Galβ1–4GlcNAc as the varied acceptor substrate. g) A representative inhibitor of FUT6, S 01925, exhibits selective inhibition as assessed using the fluorescence polarization assay. Kinetic analysis of the mode of inhibition of FUT6 by S 01925 shows that it is non-competitive with respect to: h) the donor substrate, GDP-Fucose, and i) the acceptor substrate, Galβ1–4GlcNAc. j) Inhibition of FUT6 increases with time of pre-incubation of S01925, and k) is not reversible by detergents that disrupt non-specific aggregators. All experiments are representative of three independent experiments.
The successful identification of inhibitors of STs and FUTs from a pilot screening campaign highlights the ability of this simple assay to identify inhibitors of high potency and selectivity. Although the results presented here were for a set of five exemplary STs and FUTs, based on literature precedence,[3] the assay should be applicable to most members of the mammalian ST and FUT families, as well as certain pathogenically important bacterial enzymes such as the FUTs of H. pylori[7] and the Cst-II ST of C. jejuni.[8] We suggest that this approach may also work with other families of GTs that can transfer donor substrates with unnatural fluorescent substituents. Metabolic engineering studies utilizing GlcNAc and GalNAc analogs with unnatural N-acyl groups suggest the potential for designing suitable donor substrates for these GT families.[9]
Due to the catalytic nature of the assay and the sensitivity of fluorescence polarization measurements the above assay has minimal reagent requirements, making the transition to large-scale screens feasible. Ultimately it will be important to obtain inhibitors that are cell permeable since GTs are Golgi-resident enzymes. Thus, cell-based secondary assays will be an important addition to large-scale campaigns to identify inhibitors with potential for in vivo applications.
Supplementary Material
Footnotes
We thank Dr. R. Brossmer for an initial sample of CMP-FITCNeuAc, Dr. L. Comstock for the FKP expression plasmid, Dr. C. Rademacher for bioinformatics and data analysis, and Dr. M. O’Reilly for assistance in our screening efforts. This work was supported by the NIH (T32 AI007606, R01AI050143, MH-084512).
Literature Cited
- 1.a) Varki A, Cummings RD, Esko JD, Freeze HH, Stanley P, Bertozzi CR, Hart GW, Etzler ME, editors. Essentials of Glycobiology. 2nd Edition. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2009. [PubMed] [Google Scholar]; b) Lowe JB, Marth JD. Annu Rev Biochem. 2003;72:643–691. doi: 10.1146/annurev.biochem.72.121801.161809. [DOI] [PubMed] [Google Scholar]
- 2.a) Ahsen O, Voigtmann U, Klotz M, Nifantiev N, Schottelius A, Ernst A, Müller-Tiemann B, Parczyk K. Anal Biochem. 2008;372:96–105. doi: 10.1016/j.ab.2007.08.029. [DOI] [PubMed] [Google Scholar]; b) Gross B, Kraybill B, Walker S. J Am Chem Soc. 2005;127:14588–14589. doi: 10.1021/ja0555217. [DOI] [PubMed] [Google Scholar]; c) Gross BJ, Swoboda JG, Walker S. J Am Chem Soc. 2008;130:440–441. doi: 10.1021/ja078125s. [DOI] [PubMed] [Google Scholar]; d) Hang HC, Yu C, Ten Hagen KG, Tian E, Winans KA, Tabak LA, Bertozzi CR. Chem Biol. 2004;11:337–345. doi: 10.1016/j.chembiol.2004.02.023. [DOI] [PubMed] [Google Scholar]; e) Helm J, Hu Y, Chen L, Gross B, Walker S. J Am Chem Soc. 2003;125:11168–11169. doi: 10.1021/ja036494s. [DOI] [PubMed] [Google Scholar]; f) Lee L, Mitchell M, Huang S, Fokin V, Sharpless K, Wong C. J Am Chem Soc. 2003;125:9588–9589. doi: 10.1021/ja0302836. [DOI] [PubMed] [Google Scholar]
- 3.a) Gross H, Sticher U, Brossmer R. Anal Biochem. 1990;186:127–134. doi: 10.1016/0003-2697(90)90585-w. [DOI] [PubMed] [Google Scholar]; b) Srivastava G, Kaur K, Hindsgaul O, Palcic M. J Biol Chem. 1992;267:22356–22361. [PubMed] [Google Scholar]; c) Maeda T, Nishimura S. Chemistry. 2008;14:478–487. doi: 10.1002/chem.200700760. [DOI] [PubMed] [Google Scholar]
- 4.Li Z, Mehdi S, Patel I, Kawooya J, Judkins M, Zhang W, Diener K, Lozada A, Dunnington D. J Biomol Screen. 2000;5:31–38. doi: 10.1177/108705710000500107. [DOI] [PubMed] [Google Scholar]
- 5.a) Green ED, Adelt G, Baenziger JU, Wilson S, Van Halbeek H. J Biol Chem. 1988;263:18253–18268. [PubMed] [Google Scholar]; b) Spiro RG, Bhoyroo VD. J Biol Chem. 1974;249:5704–5717. [PubMed] [Google Scholar]
- 6.Feng B, Shoichet B. Nat Protoc. 2006;1:550–553. doi: 10.1038/nprot.2006.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W, Hu T, Frantom P, Zheng T, Gerwe B, Del Amo D, Garret S, Seidel Rr, Wu P. Proc Natl Acad Sci U S A. 2009;106:16096–16101. doi: 10.1073/pnas.0908248106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu H, Cheng J, Ding L, Khedri Z, Chen Y, Chin S, Lau K, Tiwari VK, Chen X. J Am Chem Soc. 2009;131:18467–18477. doi: 10.1021/ja907750r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laughlin S, Agard N, Baskin J, Carrico I, Chang P, Ganguli A, Hangauer M, Lo A, Prescher J, Bertozzi C. Methods Enzymol. 2006;415:230–250. doi: 10.1016/S0076-6879(06)15015-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.