Abstract
Reversible protein S-glutathiolation has emerged as an important mechanism of post-translational modification. Under basal conditions several proteins remain adducted to glutathione, and physiological glutathiolation of proteins has been shown to regulate protein function. Enzymes that promote glutathiolation (e.g., glutathione-S-transferase-P) or those that remove glutathione from proteins (e.g., glutaredoxin) have been identified. Modification by glutathione has been shown to affect protein catalysis, ligand binding, oligomerization and protein-protein interactions. Conditions associated with oxidative or nitrosative stress, such as ischemia-reperfusion, hypertension and tachycardia increase protein glutathiolation via changes in the glutathione redox status (GSH/GSSG) or through the formation of sulfenic acid (SOH) or nitrosated (SNO) cysteine intermediates. These “activated” thiols promote reversible S-glutathiolation of key proteins involved in cell signaling, energy production, ion transport, and cell death. Hence, S-glutathiolation is ideally suited for integrating and mounting fine-tuned responses to changes in the redox state. S-glutathiolation also provides a temporary glutathione “cap” to protect protein thiols from irreversible oxidation and it could be an important mechanism of protein “encryption” to maintain proteins in a functionally silent state until they are needed during conditions of stress. Current evidence suggests that the glutathiolation-deglutathiolation cycle integrates and interacts with other post-translational mechanisms to regulate signal transduction, metabolism, inflammation, and apoptosis.
Keywords: glutathionylation, protein-mixed disulfides, nitric oxide, oxidative stress, heart, vasculature
Introduction
Glutathione (GSH) is the most abundant and versatile low molecular weight thiol in the cell. The cysteine residue of the peptide participates in several types of reactions including displacement, nucleophilic addition, and thiol-disulfide exchange. Because of its ability to participate in these reactions, GSH is viewed as the first line of defense against oxidants and is considered the ultimate radical sink [1, 2]. Several properties of GSH enable it to function as an efficient antioxidant. The high water solubility of GSH favors aqueous phase reactions and unlike cystine, oxidized glutathione (GSSG) remains in solution even at relatively high concentrations. The γ-glutamyl linkage of GSH is resistant to attack by intracellular peptidases, allowing the peptide to accumulate in cells at high concentrations (1–10 mM). GSH provides reducing equivalents to glutathione peroxidases for peroxide reduction and it reduces dehydroascorbate to ascorbic acid [3]. It functions as a coenzyme in the metabolism of methylglyoxal by glyoxalase [4]. Frequently, it participates in thiol transfer reactions catalyzed by thiol transferases and it can also form conjugates with endogenous and xenobiotic electrophiles. This reaction is catalyzed by enzymes of the glutathione-S-transferase (GST) family [5]. Conjugation with GSH decreases the overall chemical reactivity of electrophiles and increases their water solubility so that they could be actively transported across the cell membrane as glutathione conjugates [6] and extruded safely in the urine as mercapturic acid derivatives [7].
Glutathione is synthesized by a sequence of reactions that are catalyzed by glutamate cysteine ligase and glutathione synthetase [8]. It is also transported between different organs. When oxidized to GSSG, it is either extruded from the cell without metabolism (as GSSG) [9] or reduced back to GSH via glutathione reductase using reducing equivalents (NADPH) derived from the pentose shunt. The processes that extrude and reduce GSSG maintain the cytosolic GSH:GSSG ratio between 1000:1 and 100:1 [10]. This reducing environment ensures that most of the protein disulfides in the cytosol are in the reduced state. However, the redox state in the cell is not uniform. In contrast to the cytoplasm, the GSH:GSSG ratio in the endoplasmic reticulum (ER) lumen is maintained between 1:1 to 5:1 [11, 12]. This is believed to facilitate the induction of disulfide bonds in secretory proteins. Similarly, mitochondria can maintain a GSH:GSSG ratio different from that in the cytosol and therefore thiol-disulfide reactions can occur in the mitochondria that are independent of the cytosolic redox state [13].
Protein-mixed disulfides
That glutathione can form stable adducts with protein sulfhydryls was first proposed in the early 1920s by Sir Frederick Gowland Hopkins [14], who is credited with the discovery of glutathione in 1921 [15]. He suggested then that glutathione has “actual functions in the chemical dynamics of the cell” [15]. Proteins bound to glutathione, previously called protein-glutathione mixed-disulfides and now variably referred to as either glutathiolated or glutathionylated proteins, have been detected in several cells and tissues under multiple physiological and pathological conditions [16–18]. Under basal conditions 4–27% of the total red cell glutathione is bound to proteins [19]. In most other cells, a significant portion of cytosolic proteins remain bound to glutathione under non-stressed conditions [20–22], and approximately 50% of glutathione in the ER is protein-bound [23]. In addition, the levels of glutathiolated proteins in mitochondria vary as a function of respiratory state [13]. The levels of protein glutathiolation vary also between tissues. In the liver, for example, mixed disulfides account for 30% of the total glutathione pool [24], whereas in cultured myocytes 17–37% of the glutathione is bound to proteins [25]. It is likely that proteins with reactive, solvent-accessible cysteines dominate the pool of proteins that are glutathiolated under basal conditions. Hence, a relative few members of the proteome having reactive cysteine residues may provide a “sink” for large amounts of glutathione.
The glutathiolation/deglutathiolation cycle
The addition and removal of glutathione to proteins is regulated in a manner quite different from that of other post-translational modifications. For example, phosphorylation regulates protein function via tightly regulated, target-specific protein kinases and phosphatases that add phosphate groups to proteins or remove them from their serine, threonine, or tyrosine residues. This results in the transduction and propagation of signals that promote specific responses initiated by extracellular and intracellular stimuli. Inorganic phosphate does not bind spontaneously to proteins. In contrast, the addition of glutathione to protein can proceed without enzymatic catalysis and therefore proteins could be glutathiolated more diffusely. Nevertheless, thiolation reactions are not indiscriminate. Specificity for S-glutathiolation is imparted by thiol reactivity (i.e., cysteinyl pKR), sulfhydryl accessibility, and the reactivity of instigating intermediate species (see below). In addition, basic peptide environments conferred by lysines, arginines, or histidines vicinal to cysteine residues favor the addition of glutathione to proteins.
In principle, any solvent-exposed protein sulfhydryl in the thiolate anion (S−) form could initiate glutathiolation via nucleophilic attack of GSH resulting in the formation of a glutathiolated protein. This may be a primary mechanism that regulates basal or physiological levels of glutathione adducted to proteins. However, in most studies, glutathiolation has been found to occur in conditions of oxidative or nitrosative stress. The most straightforward mechanism by which glutathione could add to proteins is through the generation of GSSG and direct thiol-disulfide exchange. However, the intracellular concentration of GSSG in most cells is low and tightly regulated [26, 27]: GSSG resulting from the oxidation of GSH is reduced back to GSH by glutathione reductase [28]; and unreduced GSSG is actively extruded from the cell [29]. Glutathiolation by thiol-disulfide exchange may, however, be favored by transient and local increases in the intracellular GSSG concentration. For instance, the GSH/GSSG ratio in the ER is between 1 and 5 [11, 12], suggesting that protein glutathiolation in this organelle could be mediated by direct thiol-disulfide exchange. Mitochondria seem to lack mechanisms for the active extrusion of GSSG or glutathione conjugates across the membrane [30, 31], although they do maintain stringent control over thiol status [32]. For example, several mitochondrial proteins, e.g., glutathione reductase, glutaredoxins 2 and 5, participate in maintaining glutathionyl and protein-cysteinyl thiols in a reduced state [32]. Nevertheless, the mechanisms of protein glutathiolation in specific subcellular domains may be different, and it remains to be firmly established whether GSSG has organelle-specific roles in inducing protein glutathiolation.
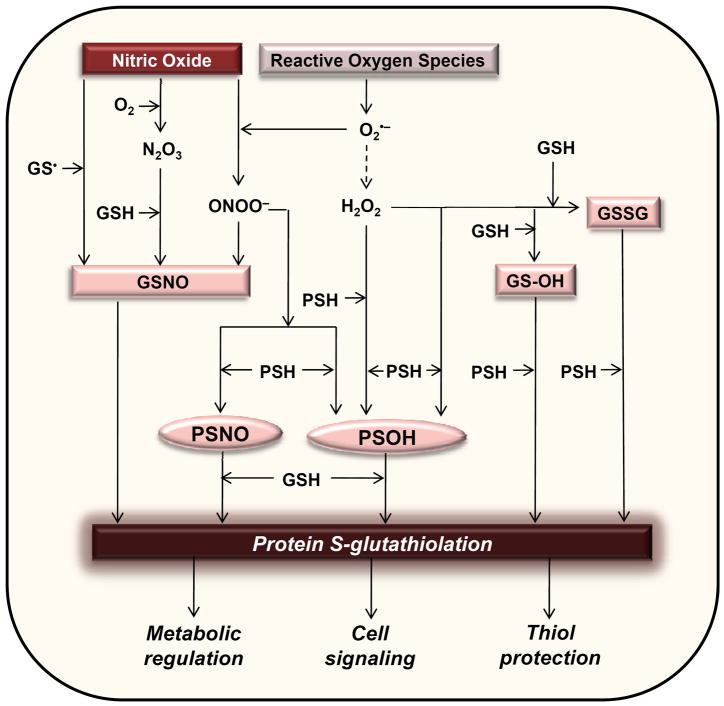
In most cases, it appears that a thiol-activation step precedes the addition of glutathione to proteins. Both glutathione and protein thiols can be activated by oxidants to form intermediates that can induce protein glutathiolation. Oxygen and nitrogen-derived species such as hydrogen peroxide (H2O2) and peroxynitrite (ONOO−/ONOOH) formed under pathological conditions can react with protein thiols via a two-electron oxidation to form sulfenic acids (PSOH) (Fig. 1). The sulfenylated cysteine can then react with GSH to form protein-mixed disulfides. A symmetrical set of reactions could also occur in which the thiolate of GSH is activated either by radical abstraction or sulfenic acid formation. Indeed, several proteins, such as hemoglobin [33], peroxiredoxins [34], the 20S proteasome [35, 36], branched-chain aminotransferase [37], β-actin [38], inhibitor of nuclear factor κB kinase subunit β (IKK-β) [39] and aldose reductase (AR) [40], have been shown to be glutathiolated via a sulfenic acid intermediate. Thus, proteins can be glutathiolated as a result of activation of protein cysteine residues by sulfenylation.
Figure 1. Mechanisms of protein S-glutathiolation.
Nitric oxide and reactive oxygen species activate protein or glutathionyl thiols resulting in S-nitrosocysteine (GSNO or PSNO) or sulfenylated (GSOH or PSOH) intermediates. In addition, the cellular abundance of GSSG is regulated by oxidative stress. These “activated” intermediate species react readily to form protein-glutathione adducts. Protein glutathiolation can regulate cellular function by modulating the activity of key metabolic and signaling enzymes. The glutathione adduct also forms a molecular “cap” that may protect protein thiols from advanced protein oxidation.
Protein glutathiolation reactions are also stimulated by nitric oxide (NO). In particular, S-nitros(yl)ation of cysteines appears to be a key step in NO-mediated glutathiolation. For instance, actin [38, 41, 42], AR [43], adenine nucleotide translocator (ANT) [38], ATP synthase [38, 42], sarco/endoplasmic reticulum Ca2+ ATPase (SERCA) [44, 45], p21ras [46], glyceraldehyde-3-phosphate dehydrogenase (GAPDH) [47], branched chain amino acid aminotransferase (BCAT) [48], and ryanodine receptor 2 (RyR2) [49] can be glutathiolated in an NO-dependent manner, and many of these proteins undergo S-nitrosation as the intermediate step leading to glutathiolation. The involvement of NO in glutathiolation of SERCA [44], AR [40], ANT [38] and ATP synthase [42] has been shown to occur in vivo as well. Although the specific mechanisms by which NO induces protein glutathiolation are not entirely clear, one possibility is that S-nitrosoglutathione (GSNO) rather than NO itself mediates protein glutathiolation. Alternatively, because cellular nitrosocysteines are nearly all associated with proteins rather than glutathione and peptidyl thiols [50], it is likely that the formation of PSNO could also promote protein glutathiolation (Fig. 1). Additionally, NO-mediated protein glutathiolation could involve the formation of an N-hydroxysulfenamide-like intermediate generated from the protein nucleophilic attack on the nitrogen rather than the sulfur of GSNO. This type of reaction has been shown to occur in vitro for aldose reductase [51]; however, the general applicability of this mechanism remains to be assessed. GSNO can also glutathiolate proteins by forming more reactive intermediates such as glutathionesulfenic acid (GSOH) that yields GSH thiosulfinate [GS(O)SG] upon dimerization [52]. The GS(O)SG formed is an active glutathiolating agent in GSNO solutions, and, with some proteins, it is a more potent glutathiolating agent than GSNO. Thus, protein glutathiolation can be induced through the formation of potent sulfenic acid, nitrosated, hydroxysulfenamide, and thiosulfinate intermediates.
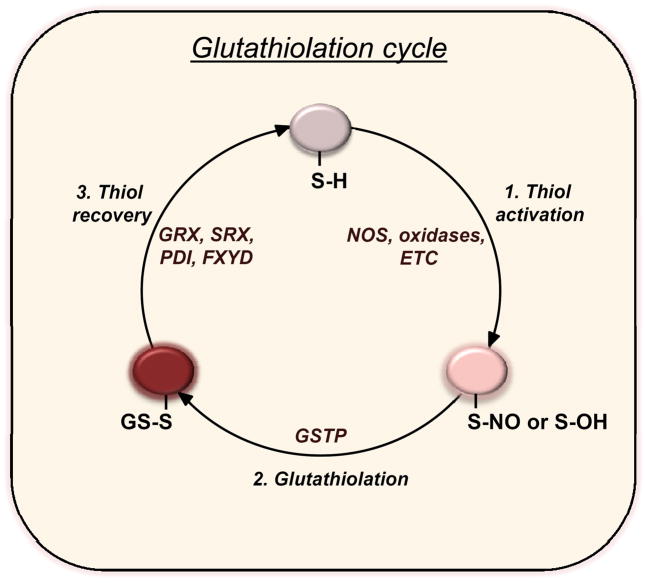
The formation of glutathiolated proteins can also be enhanced by enzyme catalysis. Several studies now suggest that the transfer of glutathione to low pK cysteines is catalyzed by enzymes such as glutaredoxin (GRX) and glutathione-S-transferase pi (GSTP) [53]. Both GRX1 and GRX2 have been shown to promote GS− transfer to proteins [54] and it has been shown that GSH-loaded GSTP glutathiolates sulfenylated peroxiredoxin (Prx) after hydroperoxide detoxification [55]. Similar redox switching by GSTP was shown for AR both with the purified enzyme and during myocardial ischemia-reperfusion [56]. During ischemia AR becomes activated by sulfenylation of C298 [57], but upon reperfusion, the sulfenylated residue becomes glutathiolated—a reaction that is accelerated by GSTP. These studies indicate that, at least for some proteins, glutathiolation reactions are catalyzed enzymatically (Fig. 2). However, it remains unclear whether enzymatic glutathiolation could be generalized to other proteins and whether enzymes other than GRX and GSTP participate in such reactions.
Figure 2. Regulation of the S-glutathiolation cycle.
Steps in the glutathiolation cycle include: 1) Thiol activation: nitric oxide synthases (NOS), cellular oxidases, or the electron transport chain (ETC) generate reactive nitrogen or oxygen species that promote the formation of nitrosated or sulfenylated proteins; 2) Glutathiolation: GSH can react non-enzymatically with the activated thiols, or the glutathiolation reaction can be accelerated in the presence of GSH-loaded GSTP; 3) Thiol recovery: GSH can reduce glutathiolated proteins non-enzymatically, or the reaction can be catalyzed via glutaredoxin (GRX), sulfiredoxin (SRX), protein disulfide isomerase (PDI), or other proteins with reactive cysteines such as FXYD protein.
Although the addition of glutathione to most proteins does not require enzyme catalysis, protein de-glutathiolation appears to be more extensively regulated. Several enzymes including GRX, sulfiredoxin (SRX) and protein-disulfide isomerase (PDI) catalyze the de-glutathiolation of proteins [58, 59] (Fig. 2). Glutaredoxin is more efficient at reducing protein-mixed sulfides than PDI [60], and it has been shown to reactivate thiolated proteins [61] and suppress protein thiolation in general [62], suggesting that it most commonly regulates the “off” state of glutathiolation-based cell signaling. In addition, other proteins with reactive cysteines could catalyze de-glutathiolation via trans-thiolation reactions. For example, the FXYD protein reverses inhibition of the Na+-K+ pump by de-glutathiolating the β-subunit of the pump, and, in the process, becomes glutathiolated itself [63]. Such trans-thiolation reactions are likely to be a common and important mode of regulation of the glutathiolation/deglutathiolation cycle of other proteins as well.
Specificity of protein glutathiolation
Protein sulfhydryls display wide heterogeneity in their susceptibility for oxidation or glutathiolation. Most thiols (SH) do not react at physiologically significant rates with hydroperoxides or other reactive species [64]. Yet proteins, such as Prx and thioredoxin, contain a cysteine that is in the thiolate form that readily reacts with H2O2 to form sulfenic acid [65, 66]; the sulfenylated residue then could facilitate glutathiolation reactions [55]. In addition, divalent metal ions function as allosteric effectors to control cysteinyl modifications. An example of this type of regulation is PKC that binds Zn2+ in a way that renders sulfhydryls in the regulatory site susceptible to oxidation [67]. In metalloproteins, the bound Zn2+ commonly functions as a Lewis acid that reduces the pKa of Cys [68], leading to enhanced susceptibility for reaction with reactive species. Hence, the propensity of a particular cysteine to be in the thiolate anion form or cysteinyl interactions with metals could enhance the specificity of glutathiolation reactions for some proteins.
The fact that S-glutathiolation reactions are favored by basic amino acids in the vicinity of the protein cysteine residue and, conversely, that acidic amino acids prevent S-thiolation [69, 70] has led to a search for a consensus motif that favors protein glutathiolation reactions. However, such a motif could be influenced by the tertiary or quaternary structures of proteins, making it difficult to identify a simple consensus sequence. It is likely that, in many proteins, the specificity of glutathiolation is conveyed through motifs exclusive to “intermediate” modifications. For example, cysteinyl modifications that are intermediate in the glutathiolation cycles of many proteins, e.g., S-nitrosation, are suggested to have motifs that determine specificity. The most salient example of this is the putative “SNO motif” [71], comprised of flanking acidic (Asp, Glu) and basic (Arg, His, Lys) residues that coordinate thiolate anion formation and NO transfer [72, 73]. Therefore such motifs could indirectly regulate the specificity of “downstream” glutathiolation reactions.
Protein modifications that favor glutathiolation could also be stabilized to promote S-thiolation reactions and prevent further thiol oxidation. For instance, the sulfenic acid form of protein tyrosine phosphatase 1B (PTP1B) is rapidly converted to a sulfenyl-amide (Cys-S-N-R), which prevents further oxidation to sulfinic and sulfonic acids and promotes glutathiolation and thiol recovery [74]. Sulfenic acids could also be stabilized non-covalently by interactions with surrounding amino acids. Glutathione reductase and NADH peroxidase form stabilized sulfenic acids [66] that could favor thiolation reactions. AR forms an apparently stable protein-sulfenic acid at Cys298 during ischemia, and, upon reperfusion, this sulfenylated site is glutathiolated enzymatically by GSTP [56]. Hence, stabilized sulfenic acids could impart specificity to protein glutathiolation. Specificity of glutathiolation could also be enhanced by enzymatic catalysis, in which binding to the active site of GSTP or GRX determines which proteins are glutathiolated. These enzymes may also induce glutathiolation of cysteine residues that are normally unreactive. For example, the sterically hindered cysteine residue at the active site of peroxiredoxin VI is inaccessible to free glutathione, but GS− transfer to this site is catalyzed by GSTP [75]. Nevertheless, further investigations are required to identify mechanisms that promote selective glutathiolation of proteins and to identify structural features of glutathiolating enzymes that determine substrate specificity.
Physiological response regulation by protein glutathiolation
Emerging evidence suggests that several physiological processes depend upon protein glutathiolation under conditions not associated with generalized oxidative stress. For example, arterial relaxation in response to acetylcholine increases NO-mediated glutathiolation of several proteins including actin, indicating that proteins are glutathiolated as a course of normal organ function [38]. This further suggests that glutathiolation of proteins under non-stressed conditions may be of functional relevance. For instance, glutathiolation of actin decreases spontaneous and tropomyosin-assisted polymerization of G-actin [76]; therefore changes in actin glutathiolation could mediate changes in cell shape and contractility. Glutathiolation appears also to be an important regulator of the calcium homeostasis. Stimulation of calcium uptake by NO occurs via peroxynitrite-mediated glutathiolation of SERCA [44]. In addition, it has been reported that stimulation with angiotensin (Ang) II causes inhibition of the sarcolemmal Na+-K+ pump [77]. This inhibition was found to be associated with protein kinase C (PKC)-dependent activation of NADPH oxidase and concomitant glutathiolation of the β1-subunit of the pump, which was reversed by GRX-1. A similar glutathiolation-linked inhibition of the β1-subunit of the Na+-K+-pump was observed upon stimulation of adenylyl cyclase by forskolin, which involved also the activation of PKC and NADPH oxidase [78]. Collectively, these findings suggest that physiological changes in NO and ROS production could alter protein glutathiolation events and that glutathiolation may be an important intermediary step in redox-sensitive signaling events.
Stress-induced changes in glutathiolation
Although the glutathiolation status of some proteins is regulated under physiological conditions, changes in protein glutathiolation often occur in disease conditions associated with increased oxidative stress (see Table 1). Modification of proteins involved in intermediary metabolism, contraction and ion transport can regulate cardiovascular response to injury. For example, during tachycardia stimulation of NADPH oxidase results in an increase in the glutathiolation of RyR2, which might be an important mechanism for increasing the rate of calcium release during conditions of heightened cardiac activity [79]. In the heart, oxidative stress imposed by ischemia-reperfusion (I-R) leads to a 15-fold increase in glutathiolated proteins, resulting in the modification of GAPDH [80], AR [57] and actin [76] as well as components of complex I [81]. Changes in the activity of these proteins due to glutathiolation may be important in regulating cardiac metabolism and contractility during I-R. Moreover, changes in protein glutathiolation could significantly affect other myocardial responses to stress. For example, the hearts of GRX-null mice demonstrate attenuated hypertrophy in response to Ang II [82].
Table 1.
Glutathiolated proteins in cardiovascular tissues.
| Protein | Function | Site | Condition | Cell or tissue | Response | Refs |
|---|---|---|---|---|---|---|
| Actin | Structure, polymerization | C10, C374 | acetycholine-induced relaxation, ischemia | Aorta, heart, VSMCs | Impaired polymerization | [38, 76, 123] |
| Aldose reductase | Glucose metabolism | C298 | Reperfusion, L-arginine feeding | Heart, aorta | Inhibition of activity | [56, 124, 125] |
| ATP synthase α-subunit | ATP production | C163 | iNOS overexpression | Heart | Inhibition of activity | [13, 38] |
| ANT | Nucleotide transport; MPT | C57 | iNOS overexpression | Heart | Facilitates ADP/ATP translocation; prevents MPT | [38, 111] |
| SERCA | Ca2+ transport | C674 | acetycholine-induced relaxation | Aorta, cardiac myocytes | Activation, Increased SR Ca2+ uptake | [92, 126] |
| p21ras | Cell growth | C118 | oxLDL, AngII | ECs, VSMCs, myocytes | ERK activation, insulin resistance, hypertrophy | [127–130] |
| Na+/K+ pump (β1 subunit) | Membrane potential, excitation-contraction coupling, energy metabolism | C46 | Present basally; increased by Ang II, forskolin, paraquat, uncoupled NOS | Heart, myocytes | Inhibition of pump activity | [131–134] |
| GAPDH | Glycolysis, mRNA stability | C149(152) | Ischemia-reperfusion | Heart, ECs | Inhibition of enzyme activity; enhanced ET-1 mRNA stability | [135, 136] |
| Complex I – NQR | Electron transport, redox signaling | C206 | Ischemia-reperfusion | Heart | Regulation of mitochondrial superoxide production | [137, 138] |
| Complex II | Electron transport | C90 | Present basally (removed upon reperfusion) | Heart | Enhanced electron transport activity; Protection from complex II nitration | [139, 140] |
| eNOS | NO production | C689, C908 | Hypertension | Aorta, ECs | Superoxide production | [89] |
| Caspase-3 | Cell death | C184, C220 | Present basally (removed with TNF-α treatment) | ECs | Protection from cleavage and cell death | [141] |
| Kir6.1 | Vascular tone | C176 | Oxidative stress | Vascular mesenteric rings | Channel inhibition, impairment of KATP channel-mediated vasodilation | [142, 143] |
| RyR2 | Ca2+ transport | C36, C822, C917, C1582, C2330, C3158, C3602* | Tachycardia, exercise | Heart | Increased Ca2+ release | [144–149] |
| Caspase-1 | Initiation of inflammation | C362, C397 | High superoxide production | Macrophages | Decreased cytokine production | [150] |
From human RyR2 sequence: Residues correspond to equivalent cysteines identified to be glutathiolated in RyR1.
Protein glutathiolation reactions may be important also in regulating atherogenesis, which is associated with extensive oxidative stress and vascular inflammation. Although little is known about the role of protein glutathiolation in atherosclerosis, it has been reported that high density lipoprotein (HDL)-associated paraoxonase 1 (PON1) can undergo S-glutathiolation under oxidative stress and that this modification causes reversible inactivation of the enzyme [83]. Because PON1 is involved in the removal of oxidized lipids [84], its inactivation by glutathiolation could exacerbate the atherogenic effects of oxidative stress. In this regard it has been shown that oxidized low density lipoprotein (oxLDL) promotes protein S-glutathiolation in macrophages [85] and that insulin resistance induced by oxLDL is mediated by S-glutathiolation of p21ras [86]. In addition, recent work showing that serum levels of S-glutathiolated proteins are increased in smokers [87] and patients with arteriosclerosis obliterans [88] suggest that measurement of glutathiolated proteins in the plasma may be of diagnostic value in assessing systemic oxidative stress.
Protein glutathiolation may be also an important component of the mechanisms regulating hypertension. It has been recently demonstrated that endothelial nitric oxide synthase (eNOS) is S-glutathiolated in the vessels of spontaneously hypertensive rats [89]. Glutathiolation of eNOS was found to reversibly decrease NOS activity and increase superoxide production, and these changes are associated with impaired endothelial vasodilation. Because eNOS plays a central role in regulating several cardiovascular functions, it was suggested that S-glutathiolation of eNOS could cause profound changes in cellular and vascular function and could mediate redox signaling under oxidative stress [89]. S-glutathiolation has also been observed in HL60 cells in response to treatment with PABA/NO, which is a direct nitrogen monoxide (NO) donor [90]. The glutathione metabolite of PABA/NO was found to inactivate SERCA. This led to an increase in intracellular calcium release resulting in the auto-regulation of eNOS through S-glutathiolation. Based on these findings it was suggested that glutathiolation of eNOS serves as a physiological regulator that acutely controls NO production, whereas SERCA, by regulating [Ca2+]i could provide a tonic or steady-state regulation of NO levels. Although this evidence provides robust support to the concept that dysregulation of protein glutathiolation reactions may be an important mechanism underlying cardiovascular disease, additional work is required to understand how coordinate changes in glutathiolation of multiple proteins regulate (patho)physiological processes.
Regulation of protein function
Given that under basal conditions a substantial amount of GSH is bound to proteins and that oxidative stress increases protein glutathiolation, it appears that a primary function of glutathiolation is to prevent irreversible oxidation of cysteine side chains or their aberrant modification by reactive nitrogen species and other electrophilic molecules such as products of lipid peroxidation. A temporary glutathione “cap” could be effective in protecting proteins under stress. A similar protective mechanism has been proposed for protein S-nitros(yl)ation [91]. However, for this type of protection to work the cap must be removed when favorable conditions return. As discussed above, the glutathiolation-deglutathiolation cycle is tightly regulated by enzymatic catalysis as well as tonic changes in the redox state of the cell (Fig. 2). Nevertheless, addition of glutathione to a protein is not an innocuous event. It could radically alter the structure of the protein, leading to changes in its function or activity, or its binding to ligands or other proteins. Whereas the binding of glutathione at the active site of proteins such as AR inhibits their catalytic activity [51], modification of regulatory sites by glutathiolation activates proteins such as SERCA [92] and p21ras [93]. In some cases, protein modification by glutathione does not affect the activity of the protein although it prevents stimulation by activating ligands; e.g., glutathiolation does not affect the basal deactylase activity of sirtuin 1 (SIRT1), but it inhibits the stimulation of SIRT1 by resveratrol [94]. In vascular smooth muscle cells, glutathiolation of myosin heavy chain (MHC) IIB promotes its interaction with Axl [95]. The interaction with MHC-IIB increases Axl phosphorylation; an event that might be important for direct cell migration during vascular injury.
Glutathiolation could also alter the quaternary structure of proteins. By modifying surface cysteines, addition of glutathione could prevent protein oligomerization, or, by promoting inter-protein thiol disulfide exchange, stimulate the formation of cystine-linked protein oligomers. Such effects of glutathiolation have been widely reported in the literature. For instance, it has been shown that glutathiolation of superoxide dismutase 1 (SOD1) at the dimer interface destabilizes the protein leading to its dissociation into monomers [96]. This may affect the initiation of SOD1 aggregation in amyotrophic lateral sclerosis (ALS). Similarly, glutathiolation of the catalytic cysteine of 1-cys-D-peroxiredoxin causes the dissociation of the non-covalent homodimer, whereas inter-protein thiol-disulfide exchange promoted by glutathiolation causes oligomerization of thiamet oligopeptidase [97]. Thus, post-translational modification by glutathione could variably affect protein structure leading to profound changes in function, activity and binding.
In addition to preventing oxidative modification of proteins, constitutive glutathiolation could contribute to protein encryption. A particularly intriguing example of this phenomenon has been reported for tissue factor (TF) [98], which was found to be constitutively glutathiolated and thereby maintained in a state of low functionality. However, upon vascular injury protein-disulfide isomerase (PDI) released from adherent platelets and the disrupted vessel wall was found to activate TF by catalyzing the isomerization of mixed disulfide to an intramolecular disulfide [98]. Thus, protein glutathiolation could be a mechanism for maintaining proteins in a functionally silent (encrypted) state until they are needed during conditions of stress or injury.
A similar mechanism has been reported for other proteins as well. For instance, the 20S proteasome purified from stationary phase cells is natively S-glutathiolated; however, de-glutathiolation by glutaredoxin or thioredoxins increases its chymotrypsin-like and post-acidic cleavage activities [35]. Similarly, in non-infected cells, interferon regulatory factor 3 (IRF3) is natively glutathiolated; however, upon viral infection the protein undergoes de-glutathiolation, which is catalyzed by GRX [99]. This change in the glutathiolation status of IRF3 was found to be necessary for its efficient interaction with CREB binding protein (CBP) and for the transcriptional activation of interferon genes.
Activation by deglutathiolation has also been reported for tumor necrosis factor (TNF) receptor associated factor 6 (TRAF6), which is an intermediate signaling molecule in signal transduction by members of the interleukin-6 (IL-6)/toll-like receptor (TLR) family. In HEK293 and HeLa cells, TRAF6 is glutathiolated under basal conditions; however, upon IL-1 stimulation it undergoes GRX-catalyzed de-glutathiolation, which was found to be essential for its auto-polyubiquitination and subsequent activation [100]. Likewise, in endothelial cells, caspase-3 is extensively glutathiolated under basal conditions. Glutathiolated caspase-3 is inactive and resistant to TNF-α-induced cleavage. Stimulation with TNF-α, however, leads to the activation of GRX, which in turn causes de-glutathiolation and activation of caspase-3 [101]. Thus, glutathiolation may be a pervasive, redox-sensitive mechanism of protein encryption that keeps select proteins functionally silent until they are needed to trigger coagulation, inflammation or cell death.
S-glutathiolation of proteins involved in intermediary metabolism
Many structural and catalytic protein components of the metabolic machinery undergo reversible S-glutathiolation, suggesting that protein glutathiolation regulates metabolic activity under physiological conditions. Several metabolic scenarios could be envisioned under which glutathiolation could regulate metabolism to enhance adaptation and survival under conditions of oxidative stress. For instance, inhibition of AR by glutathiolation [51] could increase the flux of glucose through glycolysis and the pentose phosphate pathway leading to an increase in energy production and NADPH synthesis, or partial inhibition of glycolysis by glutathiolation of GAPDH during ischemia [80] could stimulate the shunt or alter the stability of endothelin-1 (ET-1) mRNA [102]. Alternatively, increased protein glutathiolation could protect metabolic enzymes such as GAPDH and AR from irreversible oxidation temporarily only to activate them when favorable conditions return.
The protective role of glutathiolation against protein oxidation may be particularly significant in the mitochondrion. Mitochondria do not synthesize GSH, which they import from the cytosol [103]. Moreover, no GSSG transporting protein has been identified in the mitochondrial membrane. Thus, unlike the host cell, mitochondria cannot extrude GSSG. As a result, GSSG that is generated in the mitochondrion stays within the organelle until it is reduced back to GSH by mitochondrial glutathione reductase. This conservation of GSSG within the mitochondrial matrix favors uncatalyzed [13] or GRX-2 catalyzed protein S-glutathiolation [104]. It is therefore not surprising that many mitochondrial proteins such as the subunits of complex I [81] and complex IV [105], aconitase [106], pyruvate dehydrogenase [107], isocitrate dehydrogenase [108], ANT [38, 109], and ATP synthase [13, 38] undergo glutathiolation upon experimentally induced oxidative or nitrosative stress. Notably, glutathiolation of mitochondrial proteins such as complex I has also been observed in the post-ischemic heart [81], suggesting that proteins in mitochondria undergo glutathiolation in vivo. In contrast to complex I, however, glutathiolation of complex II is decreased in the post-ischemic heart and this has been associated with elevated superoxide levels [110]. A regulatory role of protein glutathiolation in mitochondria is consistent with recent observations that inhibition of mitochondrial permeability transition (MPT) by carbon monoxide (CO) was accompanied by S-glutathiolation of ANT [111]. Because both HO-1-derived CO [112] and iNOS-derived NO [113] prevent MPT in the heart, future investigations might provide unique insights into the mechanisms by which protein glutathiolation regulates metabolic activity and cell death pathways in the heart.
Interactions between protein glutathiolation and other signal transduction pathways
Glutathiolation of structural and metabolic proteins could significantly affect their function or, indirectly, the physiological processes they are involved in; but the most widespread impact of protein glutathiolation is likely to be due to the modification of proteins involved in cell signaling. By modifying specific components of the signaling network, glutathiolation could simultaneously affect multiple downstream events and gene regulation programs. Indeed key regulators of cell signaling such as IKK-α [114] and β [115], transcription factors such as NF-κB [116], activator protein-1 (AP-1) [69], p53 [117] and kinases and phosphatases such as protein-tyrosine phosphatase 1B (PTP1B) [118] and phosphatase and tensin homolog (PTEN) [119] have been found to be susceptible to glutathiolation. Most of the work has been with isolated proteins. However, both in vitro and intracellular studies have been reported for the NF-κB pathway. In vitro, the p50 subunit of NF-κB is directly modified by GSSG at GSH/GSSH ratios between 1 and 0.1 [116], whereas in embryonic fibroblasts, increasing glutathione by N-acetylcysteine during hypoxia results in the glutathiolation of p65 and enhanced hypoxic apoptosis due to NF-κB inactivation [120]. In addition, intracellular generation of H2O2 by overexpression of NADPH oxidase 1 (Nox1) or exogenous addition of peroxide has been reported to induce S-glutathiolation of IKK-β via the formation of a sulfenic acid intermediate [39]. It was found that modification of IKK-β by H2O2 was abolished by the overexpression of GRX-1, which allowed the activation of IKK-β by TNF-α even in the presence of the peroxide. Conversely, knockdown of GRX1 sensitized the cells to inhibition of IKK-β by H2O2, leading to enhanced repression of NF-κB transcriptional activity. These data suggest that glutathiolation might be a physiological mechanism that regulates the activation of the NF-κB pathway; although, as yet, there is no direct evidence for NF-κB regulation by glutathiolation in cardiovascular tissues.
Signaling pathways other than NF-κB that regulate cell survival have also been shown to be sensitive to S-glutathiolation. Significant levels of glutathiolated p53 have been detected in the brain of Alzheimer’s patients [121] and in solid tumors [117]. In tumors, the level of glutathiolation was increased upon DNA damage or oxidant stress, suggesting that the shielding of the cysteine residues of p53 by glutathiolation could compromise apoptosis during tumorigenesis [117]. The involvement of glutathiolation in the regulation of apoptosis by Fas/FasL [122] is even more intriguing. It has been reported that stimulation with FasL induces glutathiolation of Fas; however, this is not mediated by an increase in ROS generation by NADPH oxidase. Instead, glutathiolation is induced after caspase-mediated degradation of GRX-1. In support of this mechanism, it was shown that overexpression of GRX1 attenuated S-glutathiolation of Fas and partially protected against FasL-induced apoptosis [122].
Summary and Perspective
Accumulating evidence supports the view that covalent attachment of glutathione is a unique and pervasive mechanism of protein post-translational modification. Although the proclivity of GSH to form protein adducts (mixed disulfides) has been appreciated since the discovery of glutathione itself, recent work has led to the identification of several specific glutathiolation reactions that regulate protein function during physiological signal transduction and during tissue stress, injury or disease. This work suggests that protein glutathiolation is an effective mechanism for preventing irreversible oxidation of protein thiols during conditions of oxidative stress and that transient glutathiolation is an intermediate step in the reduction and recovery of sulfenylated or nitrosated proteins.
A large number of proteins have been reported to be modified by glutathione. This modification could change the catalytic or structural properties of proteins, affect protein oligomerization, or alter ligand binding. In some cases, glutathiolation has been found to be a mechanism for maintaining proteins in an inactive state, causing them to remain dormant until activated by well-regulated deglutathiolation reactions. Specific proteins, such as GRX, PDI and SRX, have been shown to catalyze de-glutathiolation reactions; in some instances, the process of glutathiolation itself has been found to be catalyzed by GRX and GSTP. Taken together, this evidence could be used to build a compelling case to support the notion that protein glutathiolation is a bona-fide post translational mechanism for regulating protein function and cell signaling. Nevertheless, the relative non-specific reactivity of glutathione with multiple proteins is difficult to overlook. Hence, a major challenge for future investigations in this area is to reconcile indiscriminate protein-mixed disulfide formation with specific glutathiolation reactions. In addition, it will be important to develop a more coherent understanding of the coordinate regulation of multiple proteins by S-glutathiolation. The properties of this post-translational modification are ideally suited to mount a fine-tuned response to changes in the redox state, thereby exerting integrated control over several processes. Additional work is also required to identify modes of convergence with other post-translational mechanisms, i.e., to understand how glutathiolation affects other modifications such as S-nitrosation or phosphorylation. Because protein glutathiolation controls mechanisms that transect critical areas of redox regulation and signal transduction, future discoveries in this field are likely to be even more fascinating than the knowledge gathered so far.
Highlights.
S-glutathiolation regulates cardiovascular function.
S-glutathiolation protects protein thiols from irreversible oxidation.
S-glutathiolation regulates intermediary metabolism and cell signaling.
S-glutathiolation can maintain proteins in an encrypted state.
Enzymes regulate the glutathiolation-deglutathiolation cycle.
Acknowledgments
Work in the authors’ laboratories is supported in part by grants from the NIH (RR024489, HL55477, and HL59378).
Footnotes
Disclosures: None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kaplowitz N, Aw TY, Ookhtens M. The regulation of hepatic glutathione. Annu Rev Pharmacol Toxicol. 1985;25:715–44. doi: 10.1146/annurev.pa.25.040185.003435. [DOI] [PubMed] [Google Scholar]
- 2.Forman HJ, Zhang H, Rinna A. Glutathione: overview of its protective roles, measurement, and biosynthesis. Mol Aspects Med. 2009;30:1–12. doi: 10.1016/j.mam.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Winkler BS, Orselli SM, Rex TS. The redox couple between glutathione and ascorbic acid: a chemical and physiological perspective. Free Radic Biol Med. 1994;17:333–49. doi: 10.1016/0891-5849(94)90019-1. [DOI] [PubMed] [Google Scholar]
- 4.Vander Jagt DL, Daub E, Krohn JA, Han LP. Effects of pH and thiols on the kinetics of yeast glyoxalase I. An evaluation of the random pathway mechanism. Biochemistry. 1975;14:3669–75. doi: 10.1021/bi00687a024. [DOI] [PubMed] [Google Scholar]
- 5.Hayes JD, Flanagan JU, Jowsey IR. Glutathione transferases. Annu Rev Pharmacol Toxicol. 2005;45:51–88. doi: 10.1146/annurev.pharmtox.45.120403.095857. [DOI] [PubMed] [Google Scholar]
- 6.Vatsyayan R, Lelsani PC, Awasthi S, Singhal SS. RLIP76: a versatile transporter and an emerging target for cancer therapy. Biochem Pharmacol. 2010;79:1699–705. doi: 10.1016/j.bcp.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boyland E, Chasseaud LF. The role of glutathione and glutathione S-transferases in mercapturic acid biosynthesis. Adv Enzymol Relat Areas Mol Biol. 1969;32:173–219. doi: 10.1002/9780470122778.ch5. [DOI] [PubMed] [Google Scholar]
- 8.Botta D, White CC, Vliet-Gregg P, Mohar I, Shi S, McGrath MB, et al. Modulating GSH synthesis using glutamate cysteine ligase transgenic and gene-targeted mice. Drug Metab Rev. 2008;40:465–77. doi: 10.1080/03602530802186587. [DOI] [PubMed] [Google Scholar]
- 9.Ballatori N, Krance SM, Marchan R, Hammond CL. Plasma membrane glutathione transporters and their roles in cell physiology and pathophysiology. Mol Aspects Med. 2009;30:13–28. doi: 10.1016/j.mam.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gilbert HF. Redox control of enzyme activities by thiol/disulfide exchange. Methods Enzymol. 1984;107:330–51. doi: 10.1016/0076-6879(84)07022-1. [DOI] [PubMed] [Google Scholar]
- 11.Townsend DM. S-glutathionylation: indicator of cell stress and regulator of the unfolded protein response. Mol Interv. 2007;7:313–24. doi: 10.1124/mi.7.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dixon BM, Heath SH, Kim R, Suh JH, Hagen TM. Assessment of endoplasmic reticulum glutathione redox status is confounded by extensive ex vivo oxidation. Antioxid Redox Signal. 2008;10(5):963–72. doi: 10.1089/ars.2007.1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia J, Han D, Sancheti H, Yap LP, Kaplowitz N, Cadenas E. Regulation of mitochondrial glutathione redox status and protein glutathionylation by respiratory substrates. J Biol Chem. 2010;285:39646–54. doi: 10.1074/jbc.M110.164160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hopkins FG. Glutathione: Its Influence in the Oxidation of Fats and Proteins. Biochem J. 1925;19:787–819. doi: 10.1042/bj0190787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hopkins FG. On an Autoxidisable Constituent of the Cell. Biochem J. 1921;15:286–305. doi: 10.1042/bj0150286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hill BG, Bhatnagar A. Role of glutathiolation in preservation, restoration and regulation of protein function. IUBMB Life. 2007;59:21–6. doi: 10.1080/15216540701196944. [DOI] [PubMed] [Google Scholar]
- 17.Biswas S, Chida AS, Rahman I. Redox modifications of protein-thiols: emerging roles in cell signaling. Biochem Pharmacol. 2006;71:551–64. doi: 10.1016/j.bcp.2005.10.044. [DOI] [PubMed] [Google Scholar]
- 18.Shackelford RE, Heinloth AN, Heard SC, Paules RS. Cellular and molecular targets of protein S-glutathiolation. Antioxid Redox Signal. 2005;7:940–50. doi: 10.1089/ars.2005.7.940. [DOI] [PubMed] [Google Scholar]
- 19.Kleinman WA, Komninou D, Leutzinger Y, Colosimo S, Cox J, Lang CA, et al. Protein glutathiolation in human blood. Biochem Pharmacol. 2003;65:741–6. doi: 10.1016/s0006-2952(02)01560-5. [DOI] [PubMed] [Google Scholar]
- 20.Klatt P, Lamas S. Regulation of protein function by S-glutathiolation in response to oxidative and nitrosative stress. Eur J Biochem. 2000;267:4928–44. doi: 10.1046/j.1432-1327.2000.01601.x. [DOI] [PubMed] [Google Scholar]
- 21.Thomas JA, Poland B, Honzatko R. Protein sulfhydryls and their role in the antioxidant function of protein S-thiolation. Arch Biochem Biophys. 1995;319:1–9. doi: 10.1006/abbi.1995.1261. [DOI] [PubMed] [Google Scholar]
- 22.Ziegler DM. Role of reversible oxidation-reduction of enzyme thiols-disulfides in metabolic regulation. Annu Rev Biochem. 1985;54:305–29. doi: 10.1146/annurev.bi.54.070185.001513. [DOI] [PubMed] [Google Scholar]
- 23.Bass R, Ruddock LW, Klappa P, Freedman RB. A major fraction of endoplasmic reticulum-located glutathione is present as mixed disulfides with protein. J Biol Chem. 2004;279:5257–62. doi: 10.1074/jbc.M304951200. [DOI] [PubMed] [Google Scholar]
- 24.Isaacs J, Binkley F. Glutathione dependent control of protein disulfide-sulfhydryl content by subcellular fractions of hepatic tissue. Biochim Biophys Acta. 1977;497:192–204. doi: 10.1016/0304-4165(77)90152-0. [DOI] [PubMed] [Google Scholar]
- 25.Lewko WM. Glutathione levels in cultured heart cells. Influence of buthionine sulfoximine, an inhibitor of glutathione synthesis. Biochem Pharmacol. 1987;36:219–23. doi: 10.1016/0006-2952(87)90692-7. [DOI] [PubMed] [Google Scholar]
- 26.Akerboom TP, Bilzer M, Sies H. The relationship of biliary glutathione disulfide efflux and intracellular glutathione disulfide content in perfused rat liver. J Biol Chem. 1982;257:4248–52. [PubMed] [Google Scholar]
- 27.Alpert AJ, Gilbert HF. Detection of oxidized and reduced glutathione with a recycling postcolumn reaction. Anal Biochem. 1985;144:553–62. doi: 10.1016/0003-2697(85)90153-8. [DOI] [PubMed] [Google Scholar]
- 28.Meister A, Anderson ME. Glutathione. Annu Rev Biochem. 1983;52:711–60. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 29.Ballatori N, Krance SM, Marchan R, Hammond CL. Plasma membrane glutathione transporters and their roles in cell physiology and pathophysiology. Mol Aspects Med. 2009;30:13–28. doi: 10.1016/j.mam.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ravindranath V, Reed DJ. Glutathione depletion and formation of glutathione-protein mixed disulfide following exposure of brain mitochondria to oxidative stress. Biochem Biophys Res Commun. 1990;169:1075–9. doi: 10.1016/0006-291x(90)92004-j. [DOI] [PubMed] [Google Scholar]
- 31.Hill BG, Awe SO, Vladykovskaya E, Ahmed Y, Liu SQ, Bhatnagar A, et al. Myocardial ischaemia inhibits mitochondrial metabolism of 4-hydroxy-trans-2-nonenal. Biochem J. 2009;417:513–24. doi: 10.1042/BJ20081615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koehler CM, Beverly KN, Leverich EP. Redox pathways of the mitochondrion. Antioxid Redox Signal. 2006;8:813–22. doi: 10.1089/ars.2006.8.813. [DOI] [PubMed] [Google Scholar]
- 33.Regazzoni L, Panusa A, Yeum KJ, Carini M, Aldini G. Hemoglobin glutathionylation can occur through cysteine sulfenic acid intermediate: electrospray ionization LTQ-Orbitrap hybrid mass spectrometry studies. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:3456–61. doi: 10.1016/j.jchromb.2009.05.020. [DOI] [PubMed] [Google Scholar]
- 34.Greetham D, Grant CM. Antioxidant activity of the yeast mitochondrial one-Cys peroxiredoxin is dependent on thioredoxin reductase and glutathione in vivo. Mol Cell Biol. 2009;29:3229–40. doi: 10.1128/MCB.01918-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silva GM, Netto LE, Discola KF, Piassa-Filho GM, Pimenta DC, Barcena JA, et al. Role of glutaredoxin 2 and cytosolic thioredoxins in cysteinyl-based redox modification of the 20S proteasome. Febs J. 2008;275:2942–55. doi: 10.1111/j.1742-4658.2008.06441.x. [DOI] [PubMed] [Google Scholar]
- 36.Demasi M, Silva GM, Netto LE. 20 S proteasome from Saccharomyces cerevisiae is responsive to redox modifications and is S-glutathionylated. J Biol Chem. 2003;278:679–85. doi: 10.1074/jbc.M209282200. [DOI] [PubMed] [Google Scholar]
- 37.Conway ME, Coles SJ, Islam MM, Hutson SM. Regulatory control of human cytosolic branched-chain aminotransferase by oxidation and S-glutathionylation and its interactions with redox sensitive neuronal proteins. Biochemistry. 2008;47:5465–79. doi: 10.1021/bi800303h. [DOI] [PubMed] [Google Scholar]
- 38.West MB, Hill BG, Xuan YT, Bhatnagar A. Protein glutathiolation by nitric oxide: an intracellular mechanism regulating redox protein modification. FASEB J. 2006;20:1715–7. doi: 10.1096/fj.06-5843fje. [DOI] [PubMed] [Google Scholar]
- 39.Reynaert NL, van der Vliet A, Guala AS, McGovern T, Hristova M, Pantano C, et al. Dynamic redox control of NF-kappaB through glutaredoxin-regulated S-glutathionylation of inhibitory kappaB kinase beta. Proc Natl Acad Sci USA. 2006;103:13086–91. doi: 10.1073/pnas.0603290103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaiserova K, Tang XL, Srivastava S, Bhatnagar A. Role of nitric oxide in regulating aldose reductase activation in the ischemic heart. J Biol Chem. 2008;283:9101–12. doi: 10.1074/jbc.M709671200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dailianis S, Patetsini E, Kaloyianni M. The role of signalling molecules on actin glutathionylation and protein carbonylation induced by cadmium in haemocytes of mussel Mytilus galloprovincialis (Lmk) J Exp Biol. 2009;212:3612–20. doi: 10.1242/jeb.030817. [DOI] [PubMed] [Google Scholar]
- 42.Townsend DM, Findlay VJ, Fazilev F, Ogle M, Fraser J, Saavedra JE, et al. A glutathione S-transferase pi-activated prodrug causes kinase activation concurrent with S-glutathionylation of proteins. Mol Pharmacol. 2006;69:501–8. doi: 10.1124/mol.105.018523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baba SP, Wetzelberger K, Hoetker JD, Bhatnagar A. Posttranslational glutathiolation of aldose reductase (AKR1B1): a possible mechanism of protein recovery from S-nitrosylation. Chem Biol Interact. 2009;178:250–8. doi: 10.1016/j.cbi.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Adachi T, Weisbrod RM, Pimentel DR, Ying J, Sharov VS, Schoneich C, et al. S-Glutathiolation by peroxynitrite activates SERCA during arterial relaxation by nitric oxide. Nat Med. 2004;10:1200–7. doi: 10.1038/nm1119. [DOI] [PubMed] [Google Scholar]
- 45.Cohen RA, Adachi T. Nitric-oxide-induced vasodilatation: regulation by physiologic s-glutathiolation and pathologic oxidation of the sarcoplasmic endoplasmic reticulum calcium ATPase. Trends Cardiovasc Med. 2006;16:109–14. doi: 10.1016/j.tcm.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 46.Clavreul N, Adachi T, Pimental DR, Ido Y, Schoneich C, Cohen RA. S-glutathiolation by peroxynitrite of p21ras at cysteine-118 mediates its direct activation and downstream signaling in endothelial cells. FASEB J. 2006;20:518–20. doi: 10.1096/fj.05-4875fje. [DOI] [PubMed] [Google Scholar]
- 47.Mohr S, Hallak H, de Boitte A, Lapetina EG, Brune B. Nitric oxide-induced S-glutathionylation and inactivation of glyceraldehyde-3-phosphate dehydrogenase. J Biol Chem. 1999;274:9427–30. doi: 10.1074/jbc.274.14.9427. [DOI] [PubMed] [Google Scholar]
- 48.Coles SJ, Easton P, Sharrod H, Hutson SM, Hancock J, Patel VB, et al. S-Nitrosoglutathione inactivation of the mitochondrial and cytosolic BCAT proteins: S-nitrosation and S-thiolation. Biochemistry. 2009;48:645–56. doi: 10.1021/bi801805h. [DOI] [PubMed] [Google Scholar]
- 49.Donoso P, Sanchez G, Bull R, Hidalgo C. Modulation of cardiac ryanodine receptor activity by ROS and RNS. Front Biosci. 2011;16:553–67. doi: 10.2741/3705. [DOI] [PubMed] [Google Scholar]
- 50.Lancaster JR., Jr Protein cysteine thiol nitrosation: maker or marker of reactive nitrogen species-induced nonerythroid cellular signaling? Nitric Oxide. 2008;19:68–72. doi: 10.1016/j.niox.2008.04.028. [DOI] [PubMed] [Google Scholar]
- 51.Chandra A, Srivastava S, Petrash JM, Bhatnagar A, Srivastava SK. Modification of aldose reductase by S-nitrosoglutathione. Biochemistry. 1997;36:15801–9. doi: 10.1021/bi9714722. [DOI] [PubMed] [Google Scholar]
- 52.Huang KP, Huang FL. Glutathionylation of proteins by glutathione disulfide S-oxide. Biochem Pharmacol. 2002;64:1049–56. doi: 10.1016/s0006-2952(02)01175-9. [DOI] [PubMed] [Google Scholar]
- 53.Townsend DM, Manevich Y, He L, Hutchens S, Pazoles CJ, Tew KD. Novel role for glutathione S-transferase pi. Regulator of protein S-Glutathionylation following oxidative and nitrosative stress. J Biol Chem. 2009;284:436–45. doi: 10.1074/jbc.M805586200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gallogly MM, Starke DW, Leonberg AK, Ospina SM, Mieyal JJ. Kinetic and mechanistic characterization and versatile catalytic properties of mammalian glutaredoxin 2: implications for intracellular roles. Biochemistry. 2008;47:11144–57. doi: 10.1021/bi800966v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Manevich Y, Feinstein SI, Fisher AB. Activation of the antioxidant enzyme 1-CYS peroxiredoxin requires glutathionylation mediated by heterodimerization with pi GST. Proc Natl Acad Sci USA. 2004;101:3780–5. doi: 10.1073/pnas.0400181101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wetzelberger K, Baba SP, Thirunavukkarasu M, Ho YS, Maulik N, Barski OA, et al. Postischemic deactivation of cardiac aldose reductase: role of glutathione S-transferase P and glutaredoxin in regeneration of reduced thiols from sulfenic acids. J Biol Chem. 2010;285:26135–48. doi: 10.1074/jbc.M110.146423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaiserova K, Srivastava S, Hoetker JD, Awe SO, Tang XL, Cai J, et al. Redox activation of aldose reductase in the ischemic heart. J Biol Chem. 2006;281:15110–20. doi: 10.1074/jbc.M600837200. [DOI] [PubMed] [Google Scholar]
- 58.Mieyal JJ, Gallogly MM, Qanungo S, Sabens EA, Shelton MD. Molecular mechanisms and clinical implications of reversible protein S-glutathionylation. Antioxid Redox Signal. 2008;10:1941–88. doi: 10.1089/ars.2008.2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park JW, Mieyal JJ, Rhee SG, Chock PB. Deglutathionylation of 2-Cys peroxiredoxin is specifically catalyzed by sulfiredoxin. J Biol Chem. 2009;284:23364–74. doi: 10.1074/jbc.M109.021394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xiao R, Lundstrom-Ljung J, Holmgren A, Gilbert HF. Catalysis of thiol/disulfide exchange. Glutaredoxin 1 and protein-disulfide isomerase use different mechanisms to enhance oxidase and reductase activities. J Biol Chem. 2005;280:21099–106. doi: 10.1074/jbc.M411476200. [DOI] [PubMed] [Google Scholar]
- 61.Barrett WC, DeGnore JP, Konig S, Fales HM, Keng YF, Zhang ZY, et al. Regulation of PTP1B via glutathionylation of the active site cysteine 215. Biochemistry. 1999;38:6699–705. doi: 10.1021/bi990240v. [DOI] [PubMed] [Google Scholar]
- 62.Xing K, Lou MF. The possible physiological function of thioltransferase in cells. FASEB J. 2003;17:2088–90. doi: 10.1096/fj.02-1164fje. [DOI] [PubMed] [Google Scholar]
- 63.Bibert S, Liu CC, Figtree GA, Garcia A, Hamilton EJ, Marassi FM, et al. FXYD Proteins Reverse Inhibition of the Na+-K+ Pump Mediated by Glutathionylation of Its β1 Subunit. J Biol Chem. 2011;286:18562–72. doi: 10.1074/jbc.M110.184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Forman HJ, Fukuto JM, Torres M. Redox signaling: thiol chemistry defines which reactive oxygen and nitrogen species can act as second messengers. Am J Physiol Cell Physiol. 2004;287:C246–56. doi: 10.1152/ajpcell.00516.2003. [DOI] [PubMed] [Google Scholar]
- 65.Ellis HR, Poole LB. Roles for the two cysteine residues of AhpC in catalysis of peroxide reduction by alkyl hydroperoxide reductase from Salmonella typhimurium. Biochemistry. 1997;36:13349–56. doi: 10.1021/bi9713658. [DOI] [PubMed] [Google Scholar]
- 66.Poole LB, Karplus PA, Claiborne A. Protein sulfenic acids in redox signaling. Annu Rev Pharmacol Toxicol. 2004;44:325–47. doi: 10.1146/annurev.pharmtox.44.101802.121735. [DOI] [PubMed] [Google Scholar]
- 67.Gopalakrishna R, Jaken S. Protein kinase C signaling and oxidative stress. Free Radic Biol Med. 2000;28:1349–61. doi: 10.1016/s0891-5849(00)00221-5. [DOI] [PubMed] [Google Scholar]
- 68.Dudev T, Lim C. Factors governing the protonation state of cysteines in proteins: an Ab initio/CDM study. J Am Chem Soc. 2002;124:6759–66. doi: 10.1021/ja012620l. [DOI] [PubMed] [Google Scholar]
- 69.Klatt P, Molina EP, De Lacoba MG, Padilla CA, Martinez-Galesteo E, Barcena JA, et al. Redox regulation of c-Jun DNA binding by reversible S-glutathiolation. FASEB J. 1999;13:1481–90. doi: 10.1096/fasebj.13.12.1481. [DOI] [PubMed] [Google Scholar]
- 70.Mallis RJ, Poland BW, Chatterjee TK, Fisher RA, Darmawan S, Honzatko RB, et al. Crystal structure of S-glutathiolated carbonic anhydrase III. FEBS Lett. 2000;482:237–41. doi: 10.1016/s0014-5793(00)02022-6. [DOI] [PubMed] [Google Scholar]
- 71.Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol. 2005;6:150–66. doi: 10.1038/nrm1569. [DOI] [PubMed] [Google Scholar]
- 72.Perez-Mato I, Castro C, Ruiz FA, Corrales FJ, Mato JM. Methionine adenosyltransferase S-nitrosylation is regulated by the basic and acidic amino acids surrounding the target thiol. J Biol Chem. 1999;274:17075–9. doi: 10.1074/jbc.274.24.17075. [DOI] [PubMed] [Google Scholar]
- 73.Stamler JS, Toone EJ, Lipton SA, Sucher NJ. (S)NO signals: translocation, regulation, and a consensus motif. Neuron. 1997;18:691–6. doi: 10.1016/s0896-6273(00)80310-4. [DOI] [PubMed] [Google Scholar]
- 74.Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, et al. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–73. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 75.Ralat LA, Manevich Y, Fisher AB, Colman RF. Direct evidence for the formation of a complex between 1-cysteine peroxiredoxin and glutathione S-transferase pi with activity changes in both enzymes. Biochemistry. 2006;45:360–72. doi: 10.1021/bi0520737. [DOI] [PubMed] [Google Scholar]
- 76.Chen FC, Ogut O. Decline of contractility during ischemia-reperfusion injury: actin glutathionylation and its effect on allosteric interaction with tropomyosin. Am J Physiol Cell Physiol. 2006;290:C719–27. doi: 10.1152/ajpcell.00419.2005. [DOI] [PubMed] [Google Scholar]
- 77.Figtree GA, Liu CC, Bibert S, Hamilton EJ, Garcia A, White CN, et al. Reversible oxidative modification: a key mechanism of Na+-K+ pump regulation. Circ Res. 2009;105:185–93. doi: 10.1161/CIRCRESAHA.109.199547. [DOI] [PubMed] [Google Scholar]
- 78.White CN, Liu CC, Garcia A, Hamilton EJ, Chia KK, Figtree GA, et al. Activation of cAMP-dependent signaling induces oxidative modification of the cardiac Na+-K+ pump and inhibits its activity. J Biol Chem. 2010;285:13712–20. doi: 10.1074/jbc.M109.090225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sanchez G, Pedrozo Z, Domenech RJ, Hidalgo C, Donoso P. Tachycardia increases NADPH oxidase activity and RyR2 S-glutathionylation in ventricular muscle. J Mol Cell Cardiol. 2005;39:982–91. doi: 10.1016/j.yjmcc.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 80.Eaton P, Wright N, Hearse DJ, Shattock MJ. Glyceraldehyde phosphate dehydrogenase oxidation during cardiac ischemia and reperfusion. J Mol Cell Cardiol. 2002;34:1549–60. doi: 10.1006/jmcc.2002.2108. [DOI] [PubMed] [Google Scholar]
- 81.Chen J, Chen CL, Rawale S, Chen CA, Zweier JL, Kaumaya PT, et al. Peptide-based antibodies against glutathione-binding domains suppress superoxide production mediated by mitochondrial complex I. J Biol Chem. 2010;285:3168–80. doi: 10.1074/jbc.M109.056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bachschmid MM, Xu S, Maitland-Toolan KA, Ho YS, Cohen RA, Matsui R. Attenuated cardiovascular hypertrophy and oxidant generation in response to angiotensin II infusion in glutaredoxin-1 knockout mice. Free Radic Biol Med. 2010;49:1221–9. doi: 10.1016/j.freeradbiomed.2010.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Rozenberg O, Aviram M. S-Glutathionylation regulates HDL-associated paraoxonase 1 (PON1) activity. Biochem Biophys Res Commun. 2006;351:492–8. doi: 10.1016/j.bbrc.2006.10.059. [DOI] [PubMed] [Google Scholar]
- 84.Ahmed Z, Ravandi A, Maguire GF, Emili A, Draganov D, La Du BN, et al. Apolipoprotein A-I promotes the formation of phosphatidylcholine core aldehydes that are hydrolyzed by paraoxonase (PON-1) during high density lipoprotein oxidation with a peroxynitrite donor. J Biol Chem. 2001;276:24473–81. doi: 10.1074/jbc.M010459200. [DOI] [PubMed] [Google Scholar]
- 85.Wang Y, Qiao M, Mieyal JJ, Asmis LM, Asmis R. Molecular mechanism of glutathione-mediated protection from oxidized low-density lipoprotein-induced cell injury in human macrophages: role of glutathione reductase and glutaredoxin. Free Radic Biol Med. 2006;41:775–85. doi: 10.1016/j.freeradbiomed.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 86.Clavreul N, Bachschmid MM, Hou X, Shi C, Idrizovic A, Ido Y, et al. S-glutathiolation of p21ras by peroxynitrite mediates endothelial insulin resistance caused by oxidized low-density lipoprotein. Arterioscler Thromb Vasc Biol. 2006;26:2454–61. doi: 10.1161/01.ATV.0000242791.28953.4c. [DOI] [PubMed] [Google Scholar]
- 87.Muscat JE, Kleinman W, Colosimo S, Muir A, Lazarus P, Park J, et al. Enhanced protein glutathiolation and oxidative stress in cigarette smokers. Free Radic Biol Med. 2004;36:464–70. doi: 10.1016/j.freeradbiomed.2003.11.018. [DOI] [PubMed] [Google Scholar]
- 88.Nonaka K, Kume N, Urata Y, Seto S, Kohno T, Honda S, et al. Serum levels of S-glutathionylated proteins as a risk-marker for arteriosclerosis obliterans. Circ J. 2007;71:100–5. doi: 10.1253/circj.71.100. [DOI] [PubMed] [Google Scholar]
- 89.Chen CA, Wang TY, Varadharaj S, Reyes LA, Hemann C, Talukder MA, et al. S-glutathionylation uncouples eNOS and regulates its cellular and vascular function. Nature. 2010;468:1115–8. doi: 10.1038/nature09599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Manevich Y, Townsend DM, Hutchens S, Tew KD. Diazeniumdiolate mediated nitrosative stress alters nitric oxide homeostasis through intracellular calcium and S-glutathionylation of nitric oxide synthetase. PLoS One. 2010;5:e14151. doi: 10.1371/journal.pone.0014151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kohr MJ, Sun J, Aponte A, Wang G, Gucek M, Murphy E, et al. Simultaneous measurement of protein oxidation and S-nitrosylation during preconditioning and ischemia/reperfusion injury with resin-assisted capture. Circ Res. 2011;108:418–26. doi: 10.1161/CIRCRESAHA.110.232173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Lancel S, Qin F, Lennon SL, Zhang J, Tong X, Mazzini MJ, et al. Oxidative posttranslational modifications mediate decreased SERCA activity and myocyte dysfunction in Galphaq-overexpressing mice. Circ Res. 2010;107:228–32. doi: 10.1161/CIRCRESAHA.110.217570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sethuraman M, Clavreul N, Huang H, McComb ME, Costello CE, Cohen RA. Quantification of oxidative posttranslational modifications of cysteine thiols of p21ras associated with redox modulation of activity using isotope-coded affinity tags and mass spectrometry. Free Radic Biol Med. 2007;42:823–9. doi: 10.1016/j.freeradbiomed.2006.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zee RS, Yoo CB, Pimentel DR, Perlman DH, Burgoyne JR, Hou X, et al. Redox regulation of sirtuin-1 by S-glutathiolation. Antioxid Redox Signal. 2010;13(7):1023–32. doi: 10.1089/ars.2010.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Cavet ME, Smolock EM, Menon P, Konishi A, Korshunov VA, Berk BC. Gas6-Axl pathway: the role of redox-dependent association of Axl with nonmuscle myosin IIB. Hypertension. 2010;56:105–11. doi: 10.1161/HYPERTENSIONAHA.109.144642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Wilcox KC, Zhou L, Jordon JK, Huang Y, Yu Y, Redler RL, et al. Modifications of superoxide dismutase (SOD1) in human erythrocytes: a possible role in amyotrophic lateral sclerosis. J Biol Chem. 2009;284:13940–7. doi: 10.1074/jbc.M809687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Demasi M, Piassa Filho GM, Castro LM, Ferreira JC, Rioli V, Ferro ES. Oligomerization of the cysteinyl-rich oligopeptidase EP24.15 is triggered by S-glutathionylation. Free Radic Biol Med. 2008;44:1180–90. doi: 10.1016/j.freeradbiomed.2007.12.012. [DOI] [PubMed] [Google Scholar]
- 98.Reinhardt C, von Bruhl ML, Manukyan D, Grahl L, Lorenz M, Altmann B, et al. Protein disulfide isomerase acts as an injury response signal that enhances fibrin generation via tissue factor activation. J Clin Invest. 2008;118:1110–22. doi: 10.1172/JCI32376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Prinarakis E, Chantzoura E, Thanos D, Spyrou G. S-glutathionylation of IRF3 regulates IRF3-CBP interaction and activation of the IFN beta pathway. EMBO J. 2008;27:865–75. doi: 10.1038/emboj.2008.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chantzoura E, Prinarakis E, Panagopoulos D, Mosialos G, Spyrou G. Glutaredoxin-1 regulates TRAF6 activation and the IL-1 receptor/TLR4 signalling. Biochem Biophys Res Commun. 2010;403:335–9. doi: 10.1016/j.bbrc.2010.11.029. [DOI] [PubMed] [Google Scholar]
- 101.Pan S, Berk BC. Glutathiolation regulates tumor necrosis factor-alpha-induced caspase-3 cleavage and apoptosis: key role for glutaredoxin in the death pathway. Circ Res. 2007;100:213–9. doi: 10.1161/01.RES.0000256089.30318.20. [DOI] [PubMed] [Google Scholar]
- 102.Rodriguez-Pascual F, Redondo-Horcajo M, Magan-Marchal N, Lagares D, Martinez-Ruiz A, Kleinert H, et al. Glyceraldehyde-3-phosphate dehydrogenase regulates endothelin-1 expression by a novel, redox-sensitive mechanism involving mRNA stability. Mol Cell Biol. 2008;28:7139–55. doi: 10.1128/MCB.01145-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Griffith OW, Meister A. Origin and turnover of mitochondrial glutathione. Proc Natl Acad Sci USA. 1985;82:4668–72. doi: 10.1073/pnas.82.14.4668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Beer SM, Taylor ER, Brown SE, Dahm CC, Costa NJ, Runswick MJ, et al. Glutaredoxin 2 catalyzes the reversible oxidation and glutathionylation of mitochondrial membrane thiol proteins: implications for mitochondrial redox regulation and antioxidant DEFENSE. J Biol Chem. 2004;279:47939–51. doi: 10.1074/jbc.M408011200. [DOI] [PubMed] [Google Scholar]
- 105.Fratelli M, Demol H, Puype M, Casagrande S, Eberini I, Salmona M, et al. Identification by redox proteomics of glutathionylated proteins in oxidatively stressed human T lymphocytes. Proc Natl Acad Sci USA. 2002 Mar 19;99:3505–10. doi: 10.1073/pnas.052592699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eaton P, Byers HL, Leeds N, Ward MA, Shattock MJ. Detection, quantitation, purification, and identification of cardiac proteins S-thiolated during ischemia and reperfusion. J Biol Chem. 2002;277:9806–11. doi: 10.1074/jbc.M111454200. [DOI] [PubMed] [Google Scholar]
- 107.Odin JA, Huebert RC, Casciola-Rosen L, LaRusso NF, Rosen A. Bcl-2-dependent oxidation of pyruvate dehydrogenase-E2, a primary biliary cirrhosis autoantigen, during apoptosis. J Clin Invest. 2001;108:223–32. doi: 10.1172/JCI10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kil IS, Park JW. Regulation of mitochondrial NADP+-dependent isocitrate dehydrogenase activity by glutathionylation. J Biol Chem. 2005;280:10846–54. doi: 10.1074/jbc.M411306200. [DOI] [PubMed] [Google Scholar]
- 109.Queiroga CS, Almeida AS, Martel C, Brenner C, Alves PM, Vieira HL. Glutathionylation of adenine nucleotide translocase induced by carbon monoxide prevents mitochondrial membrane permeabilization and apoptosis. J Biol Chem. 2010;285:17077–88. doi: 10.1074/jbc.M109.065052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Chen YR, Chen CL, Pfeiffer DR, Zweier JL. Mitochondrial complex II in the post-ischemic heart: oxidative injury and the role of protein S-glutathionylation. J Biol Chem. 2007;282:32640–54. doi: 10.1074/jbc.M702294200. [DOI] [PubMed] [Google Scholar]
- 111.Queiroga CS, Almeida AS, Martel C, Brenner C, Alves PM, Vieira HL. Glutathionylation of adenine nucleotide translocase induced by carbon monoxide prevents mitochondrial membrane permeabilization and apoptosis. J Biol Chem. 2010;285:17077–88. doi: 10.1074/jbc.M109.065052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang G, Hamid T, Keith RJ, Zhou G, Partridge CR, Xiang X, et al. Cardioprotective and antiapoptotic effects of heme oxygenase-1 in the failing heart. Circulation. 2010;121:1912–25. doi: 10.1161/CIRCULATIONAHA.109.905471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.West MB, Rokosh G, Obal D, Velayutham M, Xuan YT, Hill BG, et al. Cardiac myocyte-specific expression of inducible nitric oxide synthase protects against ischemia/reperfusion injury by preventing mitochondrial permeability transition. Circulation. 2008;118:1970–8. doi: 10.1161/CIRCULATIONAHA.108.791533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mukherjee TK, Mishra AK, Mukhopadhyay S, Hoidal JR. High concentration of antioxidants N-acetylcysteine and mitoquinone-Q induces intercellular adhesion molecule 1 and oxidative stress by increasing intracellular glutathione. J Immunol. 2007;178:1835–44. doi: 10.4049/jimmunol.178.3.1835. [DOI] [PubMed] [Google Scholar]
- 115.Kil IS, Kim SY, Park JW. Glutathionylation regulates IkappaB. Biochem Biophys Res Commun. 2008;373:169–73. doi: 10.1016/j.bbrc.2008.06.007. [DOI] [PubMed] [Google Scholar]
- 116.Pineda-Molina E, Klatt P, Vazquez J, Marina A, Garcia de Lacoba M, Perez-Sala D, et al. Glutathionylation of the p50 subunit of NF-kappaB: a mechanism for redox-induced inhibition of DNA binding. Biochemistry. 2001;40:14134–42. doi: 10.1021/bi011459o. [DOI] [PubMed] [Google Scholar]
- 117.Velu CS, Niture SK, Doneanu CE, Pattabiraman N, Srivenugopal KS. Human p53 is inhibited by glutathionylation of cysteines present in the proximal DNA-binding domain during oxidative stress. Biochemistry. 2007;46:7765–80. doi: 10.1021/bi700425y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Rinna A, Torres M, Forman HJ. Stimulation of the alveolar macrophage respiratory burst by ADP causes selective glutathionylation of protein tyrosine phosphatase 1B. Free Radic Biol Med. 2006;41:86–91. doi: 10.1016/j.freeradbiomed.2006.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Yu CX, Li S, Whorton AR. Redox regulation of PTEN by S-nitrosothiols. Mol Pharmacol. 2005;68:847–54. doi: 10.1124/mol.104.010504. [DOI] [PubMed] [Google Scholar]
- 120.Qanungo S, Starke DW, Pai HV, Mieyal JJ, Nieminen AL. Glutathione supplementation potentiates hypoxic apoptosis by S-glutathionylation of p65-NFkappaB. J Biol Chem. 2007;282:18427–36. doi: 10.1074/jbc.M610934200. [DOI] [PubMed] [Google Scholar]
- 121.Di Domenico F, Cenini G, Sultana R, Perluigi M, Uberti D, Memo M, et al. Glutathionylation of the pro-apoptotic protein p53 in Alzheimer’s disease brain: implications for AD pathogenesis. Neurochem Res. 2009;34:727–33. doi: 10.1007/s11064-009-9924-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Anathy V, Aesif SW, Guala AS, Havermans M, Reynaert NL, Ho YS, et al. Redox amplification of apoptosis by caspase-dependent cleavage of glutaredoxin 1 and S-glutathionylation of Fas. J Cell Biol. 2009;184:241–52. doi: 10.1083/jcb.200807019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hill BG, Ramana KV, Cai J, Bhatnagar A, Srivastava SK. Measurement and identification of S-glutathiolated proteins. Methods Enzymol. 2010;473:179–97. doi: 10.1016/S0076-6879(10)73009-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Baba SP, Wetzelberger K, Hoetker JD, Bhatnagar A. Posttranslational glutathiolation of aldose reductase (AKR1B1): a possible mechanism of protein recovery from S-nitrosylation. Chem Biol Interact. 2009;178:250–8. doi: 10.1016/j.cbi.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.West MB, Ramana KV, Kaiserova K, Srivastava SK, Bhatnagar A. L-Arginine prevents metabolic effects of high glucose in diabetic mice. FEBS Lett. 2008;582:2609–14. doi: 10.1016/j.febslet.2008.06.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Adachi T, Weisbrod RM, Pimentel DR, Ying J, Sharov VS, Schoneich C, et al. S-Glutathiolation by peroxynitrite activates SERCA during arterial relaxation by nitric oxide. Nat Med. 2004;10:1200–7. doi: 10.1038/nm1119. [DOI] [PubMed] [Google Scholar]
- 127.Clavreul N, Adachi T, Pimental DR, Ido Y, Schoneich C, Cohen RA. S-glutathiolation by peroxynitrite of p21ras at cysteine-118 mediates its direct activation and downstream signaling in endothelial cells. FASEB J. 2006;20:518–20. doi: 10.1096/fj.05-4875fje. [DOI] [PubMed] [Google Scholar]
- 128.Clavreul N, Bachschmid MM, Hou X, Shi C, Idrizovic A, Ido Y, et al. S-glutathiolation of p21ras by peroxynitrite mediates endothelial insulin resistance caused by oxidized low-density lipoprotein. Arterioscler Thromb Vasc Biol. 2006;26:2454–61. doi: 10.1161/01.ATV.0000242791.28953.4c. [DOI] [PubMed] [Google Scholar]
- 129.Adachi T, Pimentel DR, Heibeck T, Hou X, Lee YJ, Jiang B, et al. S-glutathiolation of Ras mediates redox-sensitive signaling by angiotensin II in vascular smooth muscle cells. J Biol Chem. 2004;279:29857–62. doi: 10.1074/jbc.M313320200. [DOI] [PubMed] [Google Scholar]
- 130.Pimentel DR, Adachi T, Ido Y, Heibeck T, Jiang B, Lee Y, et al. Strain-stimulated hypertrophy in cardiac myocytes is mediated by reactive oxygen species-dependent Ras S-glutathiolation. J Mol Cell Cardiol. 2006;41:613–22. doi: 10.1016/j.yjmcc.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 131.Bundgaard H, Liu CC, Garcia A, Hamilton EJ, Huang Y, Chia KK, et al. beta(3) adrenergic stimulation of the cardiac Na+-K+ pump by reversal of an inhibitory oxidative modification. Circulation. 2010;122:2699–708. doi: 10.1161/CIRCULATIONAHA.110.964619. [DOI] [PubMed] [Google Scholar]
- 132.White CN, Liu CC, Garcia A, Hamilton EJ, Chia KK, Figtree GA, et al. Activation of cAMP-dependent signaling induces oxidative modification of the cardiac Na+-K+ pump and inhibits its activity. J Biol Chem. 2010;285:13712–20. doi: 10.1074/jbc.M109.090225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Figtree GA, Liu CC, Bibert S, Hamilton EJ, Garcia A, White CN, et al. Reversible oxidative modification: a key mechanism of Na+-K+ pump regulation. Circ Res. 2009;105:185–93. doi: 10.1161/CIRCRESAHA.109.199547. [DOI] [PubMed] [Google Scholar]
- 134.Bibert S, Liu CC, Figtree GA, Garcia A, Hamilton EJ, Marassi FM, et al. FXYD Proteins Reverse Inhibition of the Na+-K+ Pump Mediated by Glutathionylation of Its β1 Subunit. J Biol Chem. 2011;286:18562–72. doi: 10.1074/jbc.M110.184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Eaton P, Wright N, Hearse DJ, Shattock MJ. Glyceraldehyde phosphate dehydrogenase oxidation during cardiac ischemia and reperfusion. J Mol Cell Cardiol. 2002;34:1549–60. doi: 10.1006/jmcc.2002.2108. [DOI] [PubMed] [Google Scholar]
- 136.Rodriguez-Pascual F, Redondo-Horcajo M, Magan-Marchal N, Lagares D, Martinez-Ruiz A, Kleinert H, et al. Glyceraldehyde-3-phosphate dehydrogenase regulates endothelin-1 expression by a novel, redox-sensitive mechanism involving mRNA stability. Mol Cell Biol. 2008;28:7139–55. doi: 10.1128/MCB.01145-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen J, Chen CL, Rawale S, Chen CA, Zweier JL, Kaumaya PT, et al. Peptide-based antibodies against glutathione-binding domains suppress superoxide production mediated by mitochondrial complex I. J Biol Chem. 2010;285:3168–80. doi: 10.1074/jbc.M109.056846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Chen CL, Zhang L, Yeh A, Chen CA, Green-Church KB, Zweier JL, et al. Site-specific S-glutathiolation of mitochondrial NADH ubiquinone reductase. Biochemistry. 2007;46:5754–65. doi: 10.1021/bi602580c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen YR, Chen CL, Pfeiffer DR, Zweier JL. Mitochondrial complex II in the post-ischemic heart: oxidative injury and the role of protein S-glutathionylation. J Biol Chem. 2007;282:32640–54. doi: 10.1074/jbc.M702294200. [DOI] [PubMed] [Google Scholar]
- 140.Chen CL, Chen J, Rawale S, Varadharaj S, Kaumaya PP, Zweier JL, et al. Protein tyrosine nitration of the flavin subunit is associated with oxidative modification of mitochondrial complex II in the post-ischemic myocardium. J Biol Chem. 2008;283:27991–8003. doi: 10.1074/jbc.M802691200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pan S, Berk BC. Glutathiolation regulates tumor necrosis factor-alpha-induced caspase-3 cleavage and apoptosis: key role for glutaredoxin in the death pathway. Circ Res. 2007;100:213–9. doi: 10.1161/01.RES.0000256089.30318.20. [DOI] [PubMed] [Google Scholar]
- 142.Yang Y, Shi W, Cui N, Wu Z, Jiang C. Oxidative stress inhibits vascular K(ATP) channels by S-glutathionylation. J Biol Chem. 2010;285:38641–8. doi: 10.1074/jbc.M110.162578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yang Y, Shi W, Chen X, Cui N, Konduru AS, Shi Y, et al. Molecular basis and structural insight of vascular K(ATP) channel gating by S-glutathionylation. J Biol Chem. 2011;286:9298–307. doi: 10.1074/jbc.M110.195123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sanchez G, Pedrozo Z, Domenech RJ, Hidalgo C, Donoso P. Tachycardia increases NADPH oxidase activity and RyR2 S-glutathionylation in ventricular muscle. J Mol Cell Cardiol. 2005;39:982–91. doi: 10.1016/j.yjmcc.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 145.Aracena P, Sanchez G, Donoso P, Hamilton SL, Hidalgo C. S-glutathionylation decreases Mg2+ inhibition and S-nitrosylation enhances Ca2+ activation of RyR1 channels. J Biol Chem. 2003;278:42927–35. doi: 10.1074/jbc.M306969200. [DOI] [PubMed] [Google Scholar]
- 146.Hidalgo C, Sanchez G, Barrientos G, Aracena-Parks P. A transverse tubule NADPH oxidase activity stimulates calcium release from isolated triads via ryanodine receptor type 1 S -glutathionylation. J Biol Chem. 2006;281:26473–82. doi: 10.1074/jbc.M600451200. [DOI] [PubMed] [Google Scholar]
- 147.Aracena P, Tang W, Hamilton SL, Hidalgo C. Effects of S-glutathionylation and S-nitrosylation on calmodulin binding to triads and FKBP12 binding to type 1 calcium release channels. Antioxid Redox Signal. 2005;7:870–81. doi: 10.1089/ars.2005.7.870. [DOI] [PubMed] [Google Scholar]
- 148.Aracena-Parks P, Goonasekera SA, Gilman CP, Dirksen RT, Hidalgo C, Hamilton SL. Identification of cysteines involved in S-nitrosylation, S-glutathionylation, and oxidation to disulfides in ryanodine receptor type 1. J Biol Chem. 2006;281:40354–68. doi: 10.1074/jbc.M600876200. [DOI] [PubMed] [Google Scholar]
- 149.Sanchez G, Escobar M, Pedrozo Z, Macho P, Domenech R, Hartel S, et al. Exercise and tachycardia increase NADPH oxidase and ryanodine receptor-2 activity: possible role in cardioprotection. Cardiovasc Res. 2008;77:380–6. doi: 10.1093/cvr/cvm011. [DOI] [PubMed] [Google Scholar]
- 150.Meissner F, Molawi K, Zychlinsky A. Superoxide dismutase 1 regulates caspase-1 and endotoxic shock. Nat Immunol. 2008;9:866–72. doi: 10.1038/ni.1633. [DOI] [PubMed] [Google Scholar]