Abstract
A novel locus for autosomal recessive nonsyndromic hearing impairment (ARNSHI), DFNB96, was mapped to 1p36.31-p36.13. A whole genome linkage scan was performed using DNA samples from a consanguineous family from Pakistan with ARNSHI. A maximum two-point LOD score of 3.2 was obtained at marker rs8627 (chr1:8.34Mb) at θ=0 and a significant maximum multipoint LOD score of 3.8 was achieved at 15 contiguous markers from rs630075 (9.3 Mb) through rs10927583 (15.13 Mb). The 3-unit support interval and the region of homozygosity were both delimited by markers rs3817914 (6.42 Mb) and rs477558 (18.09 Mb) and contain 11.67 Mb. Of the 125 genes within the DFNB96 interval, the previously identified ARNSHI gene for DFNB36, ESPN and two genes that cause Bartter syndrome, CLCNKA and CLCNKB, were sequenced, but no potentially causal variants were identified.
Keywords: 1p36.31-p36.13, autosomal recessive nonsyndromic hearing impairment, CLCNKA, CLCNKB, DFNB96, ESPN
Description
Although >90 autosomal recessive nonsyndromic hearing impairment (ARNSHI) loci have been mapped and 41 ARNSHI genes have been identified, hundreds of ARNSHI genes remain to be discovered, and the knowledge on the functionality of these genes should aid in improving current diagnostic and treatment protocols for hearing impairment (HI). Here a new locus is reported, DFNB96 which maps to 1p36.31-p36.13 region with a maximum multipoint LOD score of 3.8. The DFNB96 locus was mapped to a region containing 11.67 Mb using DNA samples from a consanguineous Pakistani family which segregates ARNSHI. Upon study approval from the Institutional Review Boards of Quaid-I-Azam University and the Baylor College of Medicine and Affiliated Hospitals, informed consent was obtained from participating family members. Family 4514, consanguineous kindred from Sindh province, Pakistan, clearly segregates ARNSHI (Figure 1a). No possible cause of environmental HI such as perinatal events, infections, ototoxic drug use and trauma was elucidated. Careful physical examination was performed to rule out syndromic or vestibular disease. Audiograms from two HI individuals IV-1 and IV-6 revealed bilateral severe-to-profound HI that is pre-lingual by clinical history (Figure 2).
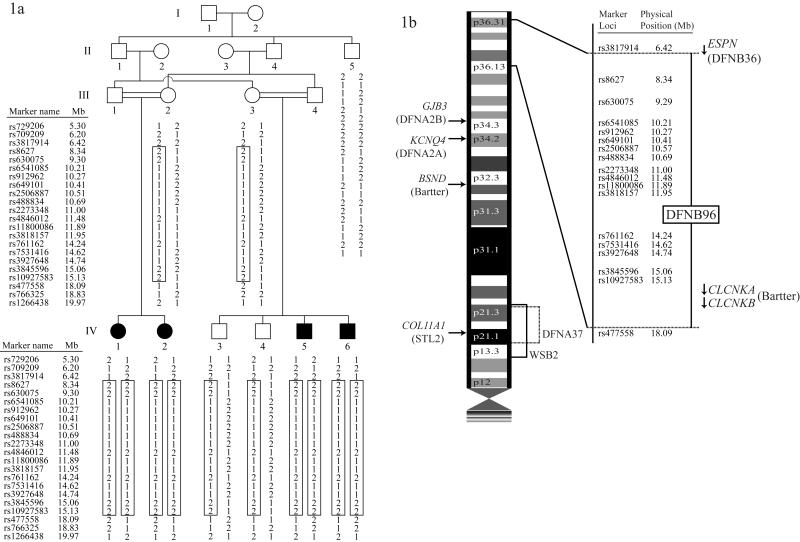
Figure1.
Panel 1a. Pedigree drawing and haplotype of family 4514. Filled symbols denote individuals with ARNSHI, while clear symbols represent hearing individuals. The haplotype segregating with ARNSHI is shown in a box, with paternal haplotypes shown on the left-side and materal haplotypes to the right. The region of homozygosity in individuals with ARNSHI is delimited by markers rs3817914 (chr1:6.42Mb) and rs477558 (chr1:18.09Mb).
Panel 1b. Chromosome 1p displaying the genetic interval for DFNB96. The locations of NSHI gene ESPN and syndromic genes CLCNKA and CLCKNB with their direction of transcription denoted by an arrow are also displayed.
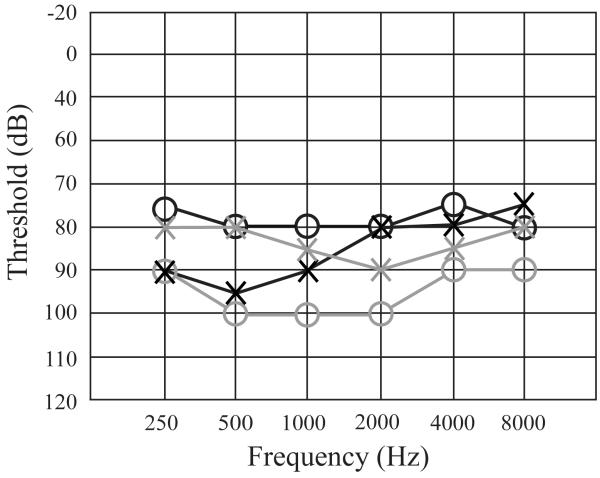
Figure2. Audiograms of individuals IV-1 and IV-6 of family 4514.
Air conduction testing is marked using circles for the right ear and crosses for the left ear. Black markings are for individual IV-1 while gray markings are for individual IV-6. Testing was performed for individual IV-1 at age 24 and for IV-6 at age 38. Hearing impairment for both individuals was bilateral and severe-to-profound involving all frequencies.
Standard DNA extraction from venous blood was performed for nine family members, four of whom have HI (Figure 1a). The GJB2 gene (MIM 121011) was sequenced in HI individuals and was negative for GJB2 variants. DNA samples from the nine family members were used to perform a whole genome linkage scan at the Center for Inherited Disease Research (CIDR) using the Infinium iSelect array which has ~6,000 SNP markers. No Mendelian inconsistencies in the genotype data were identified through PEDCHECK1. Likewise double recombination events over short genetic distances, which are most likely due to genotyping error, were not detected with MERLIN2 software.
Linkage analysis was performed using a completely penetrant autosomal recessive mode inheritance with a disease allele frequency of 0.001. Marker allele frequencies were estimated using observed and reconstructed genotypes of founders from 60 Pakistani families that underwent a genome scan at the same time. Using MLINK of the FASTLINK package3, the maximum two-point LOD score of 3.2 was achieved at marker rs8627 (chr1:8.34Mb) at θ=0 (Table 1). Genetic map distances according to the Rutgers combined linkage-physical map of the human genome Build 36 version4 were used to carry out the multipoint analysis. For markers which are not on the Rutgers map, the physical map position from the human reference sequence (Build 36) was used to interpolate the genetic map position. Multipoint linkage analysis was performed using ALLEGRO1.2c5 on chromosome 1p36 region. A significant maximum LOD score of 3.8 was obtained at 15 adjacent markers from rs630075 (9.29 Mb) to rs10927583 (15.13 Mb). The observed LOD score of 3.8 is greater than a LOD of 3.3 which is the criterion for genome-wide significance for parametric linkage studies6. The 3-unit support interval lies between SNP marker loci rs3817914 (6.42Mb) and rs477558 (18.09Mb) (Table 1). When haplotypes were reconstructed using SimWalk27, the region of homozygosity was found to be bounded by the same markers that flank the 3-unit support interval (Figure 1a). The upper and lower boundary of homozygosity was delimited by historic recombination events between the markers rs3817914 and rs8627 and markers rs10927583 and rs477558, respectively.
Table1.
Two-point and multipoint LOD scores for family 4514 at chromosome 1p36.31-p36.13
| Marker1 | Physical map position 2 |
Genetic map position 3 |
Multipoint LOD score |
Two-point LOD score at θ = |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0.00 | 0.01 | 0.05 | 0.10 | 0.20 | 0.30 | 0.40 | ||||
| rs729206 | 5,295,728 | 13.90 | − ∞ | − ∞ | −2.4 | −1.1 | −0.6 | −0.3 | −0.1 | −0.1 |
| rs709209 | 6,201,001 | 15.76 | − ∞ | − ∞ | −2.4 | −1.1 | −0.6 | −0.2 | −0.1 | −0.1 |
| rs3817914 | 6,423,619 | 16.21 | − ∞ | − ∞ | −2.5 | −1.2 | −0.7 | −0.2 | −0.1 | −0.1 |
| rs8627 | 8,335,522 | 19.58 | 3.7 | 3.2 | 3.1 | 2.8 | 2.5 | 1.8 | 1.1 | 0.5 |
| rs630075 | 9,293,498 | 21.16 | 3.8 | 2.5 | 2.4 | 2.2 | 1.9 | 1.3 | 0.7 | 0.3 |
| rs6541085 | 10,208,296 | 22.24 | 3.8 | 1.6 | 1.5 | 1.3 | 1.0 | 0.4 | −0.0 | −0.2 |
| rs912962 | 10,271,299 | 22.33 | 3.8 | 1.6 | 1.5 | 1.3 | 1.0 | 0.4 | −0.0 | −0.2 |
| rs649101 | 10,412,698 | 22.54 | 3.8 | 1.9 | 1.8 | 1.6 | 1.3 | 0.7 | 0.2 | −0.1 |
| rs2506887 | 10,507,259 | 22.68 | 3.8 | 1.1 | 1.0 | 0.8 | 0.6 | 0.0 | −0.3 | −0.3 |
| rs488834 | 10,690,489 | 23.03 | 3.8 | 2.1 | 2.1 | 1.9 | 1.6 | 1.1 | 0.5 | 0.2 |
| rs2273348 | 11,001,664 | 23.68 | 3.8 | 1.7 | 1.7 | 1.5 | 1.3 | 0.9 | 0.5 | 0.2 |
| rs4846012 | 11,480,513 | 24.86 | 3.8 | 2.7 | 2.6 | 2.6 | 2.0 | 1.4 | 0.8 | 0.3 |
| rs11800086 | 11,894,336 | 26.07 | 3.8 | 1.7 | 1.6 | 1.5 | 1.3 | 0.9 | 0.5 | 0.2 |
| rs3818157 | 11,949,895 | 26.22 | 3.8 | 1.4 | 1.4 | 1.2 | 1.6 | 0.7 | 0.4 | 0.1 |
| rs761162 | 14,243,724 | 31.08 | 3.8 | 1.4 | 1.3 | 1.1 | 0.8 | 0.3 | −0.1 | −0.2 |
| rs7531416 | 14,621,347 | 32.11 | 3.8 | 2.6 | 2.5 | 2.3 | 1.9 | 1.3 | 0.6 | 0.1 |
| rs3927648 | 14,742,313 | 32.49 | 3.8 | 2.4 | 2.3 | 2.1 | 1.8 | 1.2 | 0.7 | 0.2 |
| rs3845596 | 15,061,886 | 33.52 | 3.8 | 2.2 | 2.1 | 1.9 | 1.7 | 1.2 | 0.7 | 0.3 |
| rs10927583 | 15,131,012 | 33.74 | 3.8 | 2.0 | 1.9 | 1.7 | 1.5 | 0.9 | 0.5 | 0.1 |
| rs477558 | 18,092,414 | 39.60 | −3.3 | −2.1 | −0.6 | 0.1 | 0.2 | 0.3 | 0.2 | 0.1 |
| rs766325 | 18,829,045 | 42.28 | −3.0 | −2.2 | −1.0 | −0.4 | −0.2 | −0.1 | −0.1 | −0.1 |
| rs1266438 | 19,968,763 | 44.67 | −2.7 | −2.4 | −1.1 | −0.5 | −0.3 | −0.2 | −0.1 | −0.1 |
Markers in bold denote marker limits based on the 3-unit support interval and the homozygous region
Physical map positions in base pairs from Build 36 of the human reference sequence
Genetic map positions in cM from Rutgers combined linkage-physical map of the human genome Build 36 version
The linkage interval spans 17.53 cM region, which contains 11.67 Mb and 125 known genes. Nine hearing impairment loci involved in syndromic or nonsyndromic HI have been mapped to the short arm of chromosome 1 (1p). The syndromic loci include (a) STL2 (Stickler syndrome) at 1p21.1 which is due to mutations in the COL11A18 (MIM 120280) gene, (b) WS2B9 (Waardenburg syndrome type 2B; MIM 600193) at 1p21-p13.3, and (c) Bartter syndrome due to three genes, BSND10 (MIM 606412) at 1p32.3, and CLCNKA11 (MIM 602024) and CLCNKB11 (MIM 602023) both at 1p36.13. For autosomal dominant NSHI three loci have been mapped, (a) DFNA2A at 1p34.2 which is due to mutations in the KCNQ412 (MIM 603537) gene, (b) DFNA2B at 1p34.3 due to GJB313 (MIM 603324) mutations, and (c) DFNA3714 at 1p21 for which the gene is unknown. For ARNSHI only DFNB36 at 1p36.31 which is due to mutations in ESPN15 (MIM 606351) gene (Figure 1b) has been identified. Of these loci, the DFNB96 interval partially overlaps with the ESPN gene and also contains two Bartter syndrome genes, CLCNKA and CLCNKB. The genes ESPN, CLCNKA and CLCNKB were sequenced in hearing individual III-2 and two HI individuals IV-1 and IV-6 (Figure 1a). After sequencing using the BigDye Terminator v3.1 Cycle Sequencing Kit and Applied Biosystems 3730 DNA Analyzer, no potentially causal variants were found to segregate with HI in family 4514, thus excluding the three genes as the cause of HI in family 4514. The linkage region at 1p36.31-p36.13 was therefore assigned as the interval for the novel ARNSHI locus DFNB96. The identification of the gene for DFNB96 will provide us with additional insight into the genetic etiology of HI.
Acknowledgements
We are highly grateful to the family members who participated in this study. The study was supported by the Higher Education Commission, Islamabad, Pakistan (to W.A.) and the National Institutes of Health (NIH) – National Institute of Deafness and other Communication Disorders grant R01-DC03594 and R01-DC011651 (to S.M.L.). Genotyping services were provided by CIDR and were funded through a federal contract from the NIH to the Johns Hopkins University (Contract No N01-HG-65403).
Footnotes
Electronic Database Information The following URLs were accessed for data in this article: Hereditary Hearing Loss Homepage (http://hereditaryhearingloss.org) UCSC Genome Browser (http://genome.ucsc.edu) OMIM (http://www.omim.org)
References
- 1.O’Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am. J. Hum. Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin - rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 3.Cottingham RW, Jr, Idury RM, Schäffer AA. Faster sequential genetic linkage computations. Am. J. Hum. Genet. 1993;53:252–263. [PMC free article] [PubMed] [Google Scholar]
- 4.Matise TC, Chen F, Chen W, De La Vega FM, Hansen M, He C, et al. A second-generation combined linkage physical map of the human genome. Genome Res. 2007;17:1783–1786. doi: 10.1101/gr.7156307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gudbjartsson DF, Jonasson K, Frigge ML, Kong A. Allegro, a new computer program for multipoint linkage analysis. Nat. Genet. 2000;25:12–13. doi: 10.1038/75514. [DOI] [PubMed] [Google Scholar]
- 6.Lander E, Kruglyak L. Genetic dissection of complex traits: guidelines for interpreting and reporting linkage results. Nat. Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- 7.Sobel E, Lange K. Descent graphs in pedigree analysis: applications to haplotyping, location scores, and marker-sharing statistics. Am. J. Hum. Genet. 1996;58:1323–1337. [PMC free article] [PubMed] [Google Scholar]
- 8.Richards AJ, Yates JRW, Williams R, Payne SJ, Pope FM, Scott JD, et al. A family with Stickler syndrome type 2 has a mutation in the COL11A1 gene resulting in the substitution of glycine 97 by valine in alpha-1(XI) collagen. Hum. Mol. Genet. 1996;5:1339–1343. doi: 10.1093/hmg/5.9.1339. [DOI] [PubMed] [Google Scholar]
- 9.Lalwani AK, Baldwin CT, Morell R, Friedman TB, San Agustin TB, Milunsky A, et al. A locus for Waardenburg syndrome type II maps to chromosome 1p13.3-2.1. Am. J. Hum. Genet. 1994;55(suppl):A14. [Google Scholar]
- 10.Birkenhager R, Otto E, Schurmann MJ, Vollmer M, Ruf E-M, Maier-Lutz I, et al. Mutation of BSND causes Bartter syndrome with sensorineural deafness and kidney failure. Nat. Genet. 2001;29:310–314. doi: 10.1038/ng752. [DOI] [PubMed] [Google Scholar]
- 11.Schlingmann KP, Konrad M, Jeck N, Waldegger P, Reinalter SC, Holder M, et al. Salt wasting and deafness resulting from mutations in two chloride channels. New Eng. J. Med. 2004;350:1314–1319. doi: 10.1056/NEJMoa032843. [DOI] [PubMed] [Google Scholar]
- 12.Kubisch C, Schroeder BC, Friedrich T, Lutjohann B, El-Amraoui A, Marlin S, et al. KCNQ4, a novel potassium channel expressed in sensory outer hair cells, is mutated in dominant deafness. Cell. 1999;96:437–446. doi: 10.1016/s0092-8674(00)80556-5. [DOI] [PubMed] [Google Scholar]
- 13.Xia J, Liu C, Tang B, Pan Q, Huang L, Dai H, et al. Mutations in the gene encoding gap junction protein beta-3 associated with autosomal dominant hearing impairment. Nat. Genet. 1998;20:370–373. doi: 10.1038/3845. [DOI] [PubMed] [Google Scholar]
- 14.Talebizadeh Z, Kenyon JB, Askew JW, Smith SD. A new locus for dominant progressive hearing loss DFNA37 mapped to chromosome 1p21. Am. J. Hum. Genet. 2000;67(suppl 2):314. [Google Scholar]
- 15.Naz S, Griffith AJ, Riazuddin S, Hampton LL, Battey JF, Jr., Khan SN, et al. Mutations of ESPN cause autosomal recessive deafness and vestibular dysfunction. J. Med. Genet. 2004;41:591–595. doi: 10.1136/jmg.2004.018523. [DOI] [PMC free article] [PubMed] [Google Scholar]