Abstract
Tandem mass spectrometry was applied to detect derangements in the pathways of amino acid and fatty acid metabolism in N-ethyl-N-nitrosourea–treated (ENU-treated) mice. We identified mice with marked elevation of blood branched-chain amino acids (BCAAs), ketoaciduria, and clinical features resembling human maple syrup urine disease (MSUD), a severe genetic metabolic disorder caused by the deficiency of branched-chain α-keto acid dehydrogenase (BCKD) complex. However, the BCKD genes and enzyme activity were normal. Sequencing of branched-chain aminotransferase genes (Bcat) showed no mutation in the cytoplasmic isoform (Bcat-1) but revealed a homozygous splice site mutation in the mitochondrial isoform (Bcat-2). The mutation caused a deletion of exon 2, a marked decrease in Bcat-2 mRNA, and a deficiency in both BCAT-2 protein and its enzyme activity. Affected mice responded to a BCAA-restricted diet with amelioration of the clinical symptoms and normalization of the amino acid pattern. We conclude that BCAT-2 deficiency in the mouse can cause a disease that mimics human MSUD. These mice provide an important animal model for study of BCAA metabolism and its toxicity. Metabolomics-guided screening, coupled with ENU mutagenesis, is a powerful approach in uncovering novel enzyme deficiencies and recognizing important pathways of genetic metabolic disorders.
Introduction
Although there is much current interest in the genome-wide analysis of organisms at the level of transcription (the transcriptome) and translation (the proteome), the third level of analysis, that of the metabolome, has been relatively unexplored to date (1). The term metabolome refers to the entire complement of all the small molecular weight metabolites inside an organism of interest.
Our laboratory has long pioneered the rapid analysis of metabolites at the whole-organism level, using methods such as tandem mass spectrometry, HPLC, and gas chromatography/mass spectrometry (2, 3). Tandem mass spectrometry is a branch of analytic chemistry in which compounds of interest (target analytes) can be detected and analyzed in complex mixture with little or no recourse to on-line chromatography separation. Acylcarnitines reflect the catabolism of fatty acids and certain amino acids, which take place primarily in the mitochondria, the organelle that provides the energy for the cell (2–4). Alpha amino acids share a common “backbone” to which different groups are attached, and they also are amenable to analysis by tandem mass spectrometry (5). Tandem mass spectrometry for amino acids and acylcarnitines determination has been fully automated and can reliably analyze more than 30 metabolites from a single blood spot (25 μl of whole blood) in one short-duration run and thereby provides a comprehensive assessment of the entire fatty acid and selected amino acid metabolic pathways. The method can detect more than 20 metabolic disorders in human (6, 7).
A systematic, genome-wide, phenotype-driven mouse mutagenesis program for gene function studies has been described (8, 9). Treatment with N-ethyl-N-nitrosourea (ENU) efficiently generates single-nucleotide mutations in mice, which can be screened for the disease phenotypes of interest. A systematic screening for metabolic diseases with tandem mass spectrometry has been proposed, but no results have been reported thus far (10).
Here we show how a metabolomics-guided screening of ENU mice can efficiently identify a mouse model of human metabolic disease. We applied the tandem mass spectrometry method to screen third-generation progeny of ENU-mutagenized mice (C57BL/6J) for abnormalities in the pathways of amino acid and fatty acid metabolism.
Methods
Generation of ENU-recessive mice.
ENU-treated mice were bred according to the three-generation breeding scheme as described (11). Briefly, C57BL/6J male mice were given three ENU intraperitoneal injections (100 mg/kg body weight) to generate G0 mice. G0 mice were then mated with normal B6 females to generate G1 male founder mice. Normal B6 females were mated with G1 male founders to generate G2 mice. G2 females were then backcrossed to G1 male mice to generate G3 offspring. Breeding and housing of the mice were conducted in the Mouse Mutagenesis Program Core facility of the Academia Sinica under specific pathogen-free conditions. Animal protocol was approved by the Institutional Animal Safety Committee. Mice were maintained on the regular rodent diet (PicoLab Mouse Diet 20 code5058, PMI LabDiet, St. Louis, Missouri, USA) or branched-chain amino acids–restricted (BCAAs-restricted) diet by mixing regular diet with BCAAs-free human formula (maple syrup urine disease [MSUD] diet powder from Mead Johnson Nutritionals, Evansville, Indiana, USA).
Tandem mass spectrometry screening of amino acids and acylcarnitines.
Standardized filter papers (S&S 903; Schleicher & Shuell, Dassel, Germany) were impregnated with 25 μl whole blood from the tail vein of 2- to 3-month- old G3 mice. Blood was eluted from blood spots, and amino acids and acylcarnitines were derivatized as previously described (6, 7). We used a tandem mass spectrometer (Quattro Micro; Micromass, Beverly, Massachusetts, USA) to analyze acylcarnitines and amino acids. Acylcarnitines were detected with a positive precursor ion scan of 85 Da, and amino acids were detected by positive neutral loss scans of 102, 119, and 161 Da, respectively. The ratios of molecular signals to their respective internal standards were used to quantify the analytes. Amino acids were also determined by ion exchange chromatography on an amino acid analyzer (Beckman Instruments, Palo Alto, California, USA), which enabled the complete separation and quantification of leucine, isoleucine, and allo-isoleucine.
Urinary organic acids.
We collected 100 μl urine from each mouse. The urine samples were diluted to 1 ml, derivatized with ethoxyamine hydrochloride at pH 10, acidified to pH 1, and then extracted four times with 2 ml of ethyl acetate. After evaporation of the combined extracts, the residue was silylated with 100 μl of N,O-bis(trimethylsilyl) trifluoroacetamide with 1% trimethylchlorosilane plus 10 μl pyridine and analyzed by capillary column gas chromatography/mass spectrometry, using the analytical conditions described previously (12).
Enzyme assays.
Branched-chain α-keto acid dehydrogenase (BCKD) activity was assayed spectrophotometrically using α-ketoisocaproate (α-KIC) as the substrate as described (13). The assay reaction mixture consisted of 1 mM α-KIC, 30 mM potassium phosphate, 2 mM MgCl2, 0.4 mM thiamine pyrophosphate, 0.4 mM CoA, 1 mM NAD+, 0.1% (w/v) Triton X-100, 2 mM DTT, pig heart dihydrolipoamide dehydrogenase (5 units/ml), and 2 μl of tissue extract. Absorbance at 340 nm was recorded. One unit of BCKD enzyme activity was defined as the formation of 1 μmol of NADH per minute at 30°C.
For determination of branched-chain aminotransferase (BCAT) activity, frozen tissue (50–100 mg) was pulverized with a mortar and pestle cooled in liquid nitrogen. The powder was transferred to a cooled centrifuge tube and suspended in 10 volumes of ice-cold homogenization buffer consisting of 20 mM EDTA, 20 mM EGTA, 0.4% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]-2-hydroxy-1-propanesulfonate, 5 mM DTT, 10 μl/ml protease inhibitor cocktail, and 25 mM HEPES (pH 7.4). The tissue homogenates were then subjected to three rounds of freeze-thaw sonication before centrifugation at 15,000 g for 30 minutes at 4°C. The supernatant was used as a source for total BCAT activity. The BCAT-1 (BCAT in cytoplasm) and BCAT-2 (BCAT in mitochondria) isozyme activities were also measured individually after separating mitochondrial and cytosolic fractions using a mitochondria isolation kit, MITO-ISO1 (Sigma-Aldrich, St. Louis, Missouri, USA). All BCAT enzyme activities were measured by a continuous 96-well plate spectrophotometric assay method (14) using MultiSkan Spectrum (Thermo Labsystems, Stockholm, Sweden). One unit of BCAT enzyme activity was defined as 1 μmol NADH reduced per minute at 37°C.
Western blot analysis.
Anti-BCAT-2 antibody and recombinant human BCAT-2 were a generous gift of Susan Hutson of Wake Forest University (Winston-Salem, North Carolina, USA). The antibody was a polyclonal antibody raised in rabbit against purified human recombinant BCAT-2. Tissue samples were subjected to SDS/PAGE using a NuPAGE Novex 4–12% Bis-Tris Gel (Invitrogen, Carlsbad, California, USA). Ten micrograms of total protein were loaded onto each lane. Recombinant human BCAT-2 (0.7 μg) was included as a standard. Resolved proteins were transferred to Immobilon P membranes. Membranes were blocked with 1% BSA/PBS and incubated with rabbit anti-human BCAT-2 (0.45 μg/ml) antixbodies. Bound antibodies were detected with anti-rabbit IgG conjugated with alkaline phosphatase (Promega, Madison, Wisconsin, USA). The immunoreactive protein bands were visualized using the bromochloroindolyl phosphate/nitro blue tetrazolium alkaline phosphatase substrate system according to the manufacturer’s instruction (Sigma-Aldrich).
PCR and DNA sequencing.
Genomic DNA was purified from 300 μl whole blood using the Puregene DNA purification kit (Gentra Systems, Minneapolis, Minnesota, USA). All exons of candidate genes (Bcat1, Bcat2, and genes of the BCKD complex, including Bckdha, Bckdhb, Dbt, and Dld) were amplified and sequenced. Primers were designed by the Primer3 PCR primer-picking program (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi). The primers used to detect exon 2 mutations in the Bcat2 gene were: 5′-GGGCTGGAGCTGACTTTAGTT, sense strand in intron 1 and 5′-CGCCAACCTGAGAGACTACA, anti-sense strand in intron 2. The PCR reactions were performed in a final volume of 50 μl, containing 0.2 μM each primer, 10 mM Tris-HCl (pH 8.3), 50 mM KCl,1.5 mM MgCl2, 0.2 mM deoxyribonucleoside triphosphate (dNTP), and 1 unit TaKaRa Taq (Takara, Shiga, Japan). Amplification conditions consisted of an initial denaturation of 3 min at 94°C, followed by 20 cycles of touchdown PCR in 30 seconds at 94°C, 30 seconds at 65°C (decrease 0.5°C per cycle), 40 seconds at 72°C, and a final 20 cycles in 30 seconds at 94°C, 30 seconds at 55°C, and 40 seconds at 72°C.
Total RNA was extracted from tissues using REzol C&T (PROtech Technologies, Taiwan). First-strand cDNA was synthesized using oligo-dT15 primer and Moloney murine leukemia virus reverse transcriptase RNase H minus (Promega) in 25 μl of 0.5 mM dNTPs, 10 mM Tris-HCl (pH 8.3), 2.5 mM KCl, 0.6 mM MgCl2, 20 U RNase inhibitor, and 2 mM DTT. RT-PCR amplification of Bcat2 was conducted by using the following primer set: 5′-CACACACCGGATCATGGCTG, sense strand in Bcat2 exon1 and 5′-CGCCTAGCAGAACGTAGCAT, anti-sense strand in Bcat2 exon4.
All amplified PCR fragments were purified using NucleoFast 96 PCR plate (Macherey-Nagel, Easton, Pennsylvania, USA), according to the manufacturer’s instructions, to remove unincorporated primers and dNTPs. The purified PCR products were sequenced using the BigDye Terminator Cycle Sequencing Kit v1.1/3.1 (Applied Biosystems, Foster City, California, USA) following the manufacturer’s instructions. Sequencing products were separated on either an ABI PRISM 3100 Genetic Analyzer or an ABI PRISM 3700 DNA Analyzer (Applied Biosystems). Raw sequencing data was analyzed by the DNA Sequencing Analysis Software v3.7 (Applied Biosystems).
Real-time quantitative RT-PCR.
Real-time quantitative RT-PCR analysis was performed using the ABI PRISM 7700 Sequence Detection System (Applied Biosystems). PCR primers and TaqMan MGB probe for 18S RNA quantitation were purchased as an Assay-on-Demand kit from Applied Biosystems. The TaqMan MGB probe was linked to the reporter dye (6-FAM) at the 5′ end and to the nonfluorescent quencher (NFQ) at the 3′ end. For Bcat-2 mRNA quantitation, the primers and probes were designed using PrimerExpress (Applied Biosystems). EX-1,3 was designed to amplify specifically the alternative splicing transcript that has exon 2 deleted. EX-3,4 was designed to amplify the total Bcat-2 transcripts. EX-1,3 primers were: 5′-primer, 5′-CGCACACACCGGATCATG-3′; 3′-primer, 5′-CTTCTGTGGTTCTTTGGTCATCTGA-3′; and the probe was 5′-FAM-TCTGCAGCCTGTCCTAGTG-NFQ-MGB-3′. EX-3,4 primers were: 5′-primer, 5′-CCTGCTCTGGTCTGCACT-3′; 3′-primer, 5′-GTCTCCACCTTTGTATGCTTTCAAG-3′; and the probe was 5′-FAM-ACTCTCTGCAGCTCTTTG –NFQ-MGB-3′.
The cDNA corresponding to 75 ng of reversed transcribed total RNA was amplified in a final volume of 20 μl using TaqMan Universal PCR Mastermix (Applied Biosystems) in duplicate assays for Bcat-2 EX-1,3, Bcat-2 EX-3,4, and the endogenous 18S RNA. Final concentrations of PCR primers and TaqMan MGB probes were 900 nM and 250 nM, respectively.
An analysis of the results was based on the Ct calculation, where Ct represents the cycle number at which fluorescence of the PCR samples crossed a given threshold. The expression level of the 18S RNA was taken as the first “calibrator” to normalize the total Bcat-2 mRNA in each tissue (ΔCt). The expression of the Bcat-2 in each of the control mouse tissue was then taken as the second “calibrator” to normalize the expression of Bcat-2 in the affected tissue accordingly (ΔΔCt). Final results were given as the relative amounts of Bcat-2 mRNA in the affected mouse tissues as compared to the control.
Results
Mutant mice with an abnormal amino acid pattern.
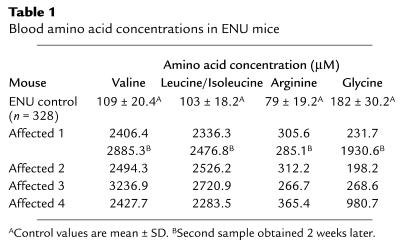
In screening 614 G3 mice from 39 families, we identified one mutant mouse having striking elevation of blood BCAA (valine, leucine/isoleucine, 20–30 fold) and moderate elevation of arginine (4-fold) (Table 1). Leucine and isoleucine were grouped together because they have the same molecular mass and are not differentiated by the tandem mass spectrometry method used in our laboratory. Using ion exchange chromatography (Beckman amino acid analyzer), the elevations of valine, leucine, and isoleucine were confirmed. In addition, it was found that leucine and isoleucine were present in a 60:40 ratio. The abnormal amino acid profile was confirmed in the second sample of the same mouse obtained 2 weeks later. Subsequently, we identified three more mutant mice in the same family having valine, leucine/isoleucine, and arginine elevations (Table 1). The finding of the same abnormal amino acid profile in multiply affected mice in independent litters of the same family suggested that this phenotype was heritable. In addition to the elevation of valine, leucine/isoleucine, and arginine, glycine was also elevated in two of the four mutant mice, although its elevation was not invariably seen even in the same mouse (Table 1). All mice had normal acylcarnitine profiles.
Table 1.
Blood amino acid concentrations in ENU mice
Other clinical phenotypes.
In addition to the abnormal amino acid profile, these mutant mice (three males, one female) displayed failure to thrive (body weight 30–40% less than the unaffected siblings), weakness with decreased spontaneous movement, and hairs that were thin and scanty and with decreased luster. Family history revealed that many of the mutant mice expired prior to weaning and prior to metabolic screening at 2 months of age.
Why are these amino acids elevated?
Since the most striking amino acid elevations were in valine, leucine, and isoleucine, all of which are BCAAs that share common metabolic pathways, we focused our investigations on these analytes. Conditions with elevated blood levels of BCAAs include starvation, diabetes mellitus, obesity, and inborn errors of metabolism (15). Starvation, diabetes, and obesity were ruled out in our mice; therefore, we considered the likelihood that the fundamental defect was an inborn error of BCAA catabolism.
BCAA catabolism.
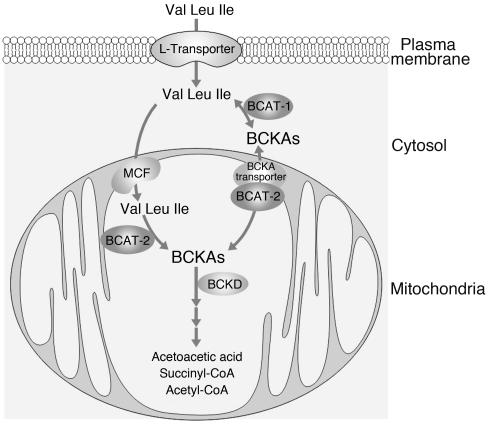
BCAA oxidation begins with the transport of these amino acids into cells, where they undergo transamination, oxidative decarboxylation, and dehydrogenation (Figure 1). Transamination is catalyzed by BCATs that are either cytosolic or mitochondrial. Oxidative decarboxylation and dehydrogenase are catalyzed by a single BCKD complex, which is comprised of three catalytic components and two regulatory units (15).
Figure 1.
Schematic catabolic pathway of branched-chain amino acids valine (Val), leucine (Leu), and isoleucine (Ile). MCF, mitochondrial carrier family.
Mammalian amino acid transporters have been classified into a distinct “system,” depending upon substrate specificity, transport mechanism, and regulatory properties (16) There are more than 20 different transport systems known, which can fall into five superfamilies: two are preferentially Na+-coupled transporters and the other three often function as H+-coupled systems. BCAAs, along with other neutral amino acids, such as phenylalanine, tyrosine, and tryptophan, enter the cells through the sodium independent “L” system transporter (17). However, the unique amino acid pattern in our mutant mice did not support a defect in either the L system transporter or any of the known amino acid transporters.
To further investigate the abnormalities in the BCAA catabolism, we examined urine organic acids. Branched-chain α-keto acids (BCKAs), 2-oxoisovaleric acid, 2-oxoisocaproic acid, and 2-oxo-3-methylvaleric acid were present in the affected mice but not in the normal mice (data not shown); these keto acids are transamination products of valine, leucine, and isoleucine, respectively. The elevation of BCAAs as well the corresponding BCKAs suggest that the block may be at the BCKD complex (Figure 1): the enzyme complex catalyzes the decarboxylation and dehydrogenation steps of BCKAs, and, when deficient, causes human MSUD. Clinically, MSUD patients present in early infancy with failure to thrive, poor feeding, lethargy, ketoacidosis, and, if left untreated, early death (15). These phenotypic features are similar to what we observed in our mice. The BCKD complex activity in both liver and muscle of our mutant mice, however, were normal (Table 1). BCKD comprises three catalytic components: a thiamine pyrophosphate-dependent decarboxylase (E1α, E1β), a transacyclase (E2), and a dehydrogenase (E3). Sequencing the genes encoding these components revealed no mutation. Furthermore, no alloisoleucine, a pathognomonic marker for human MSUD (18), was detected in the urine of the affected mice.
Defect in BCAT.
The possibility that a defect in the BCAT is responsible for the MSUD-like mice was investigated. In mammals, there are two BCAT isozymes, a cytosolic (BCATc or BCAT-1) and a mitochondrial (BCATm or BCAT-2) form (19) (Figure 1); each is encoded by a single gene. Interestingly, BCAT-2 may also play a role in the transport of BCKAs, either it functions as a transport protein that transports BCKAs across the mitochondrial membrane, or it interacts with the inner mitochondrial membrane for BCKAs transport (20, 21).
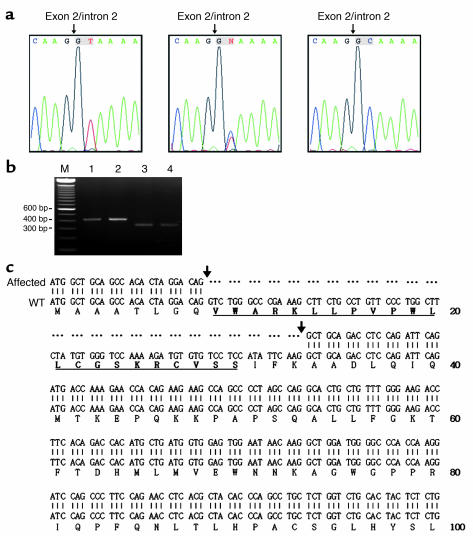
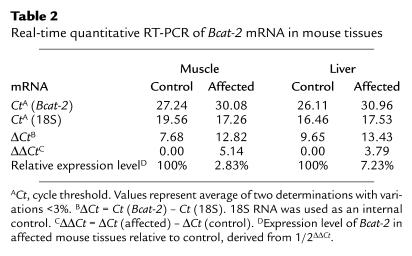
Sequencing the Bcat genes revealed no mutation in the Bcat1 gene. However, a homozygous T to C nucleotide change in the 5′ splicing site consensus sequence of the exon 2 and intron 2 (IVS2 + 2T > C) of the Bcat2 gene was identified (Figure 2a). Both parents were heterozygous for the mutation. RT-PCR amplification of liver and muscle Bcat2 mRNA with primers located in exon 1 and exon 4 resulted in DNA fragments of 414 bp in WT and 339 bp fragments in the affected mouse tissues, respectively (Figure 2b). Sequencing the RT-PCR products of the affected mouse tissues revealed that the entire exon 2 of Bcat2 mRNA was deleted (Figure 2c). Real-time quantitative RT-PCR showed that the amounts of Bcat-2 mRNA were markedly reduced in the affected mouse tissues (2.83% in muscle and 7.23% in liver) as compared with the control (Table 2). The Bcat-2 mRNA detected in the affected mouse tissues was predominantly the spliced variant (exon 2 deleted), and this spliced variant was not detected in the control mouse tissues (data not shown), which was consistent with data obtained from the qualitative RT-PCR experiments (Figure 2b). These data indicated that the splicing site mutation resulted in a mutant form of Bcat-2 mRNA (lacking exon 2), which had a decreased stability.
Figure 2.
Sequence analysis of mouse Bcat2 gene. (a) Nucleotide sequences showing T/T (WT, left panel), T/C (G1 father, middle panel) and C/C (affected, right panel) at second nucleotide position of intron 2. (b) RT-PCR results of mRNA amplified with primers located in exon 1 and exon 4 as described in Methods. Lane 1, WT liver; lane 2, WT muscle; lane 3 affected liver; lane 4, affected muscle; M, molecular weight markers. (c) Alignment of nucleotide and deduced amino acid sequences derived from RT-PCR results in b. Mitochondrial targeting leader sequences are underlined. Dots represent gaps. Sequences marked between two arrows represent exon 2.
Table 2.
Real-time quantitative RT-PCR of Bcat-2 mRNA in mouse tissues
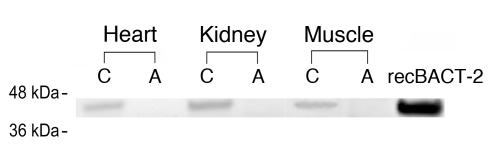
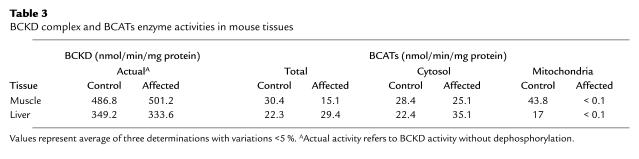
Western blot analysis using anti-human BCAT-2 antibody detected mouse BCAT-2 protein in the WT mouse heart, kidney, and muscle but not in the affected mouse tissues (Figure 3). Enzyme activity measurements showed that total BCAT activity was decreased 50% in the affected mouse muscle (Table 3). However, when BCAT-1 and BCAT-2 were measured separately, only mitochondrial BCAT activities were markedly reduced in the affected mouse tissues, whereas cytosolic BCAT activities were retained. These data established that the fundamental defect in our MSUD-like mice lies in the Bcat2 gene. The fact that BCAT-2 has a role in BCKA transport (20, 21) may explain why BCKAs (presumably derived from intact BCAT-1 activity) were elevated in our mice.
Figure 3.
Western blot analysis of BCAT-2 in control (C) and affected (A) mouse tissues. Each lane contains 10 0g of total protein from tissue extracts. Recombinant human BCAT-2 (recBCAT-2; 0.7 μg) was used as a standard.
Table 3.
BCKD complex and BCATs enzyme activities in mouse tissues
The spliced site mutation identified in our mutant mice caused a deletion of entire exon 2 of the Bcat-2 mRNA. Because exon 2 contains the mitochondrial targeting leader sequence (Figure 2c), the deletion might be expected to have an effect on the BCAT-2 targeting, as in the case of an alternatively spliced Bcat2 variant mRNA detected in human tissues (22). This human spliced variant lacks 276 nucleotides in the 5′ end, including the entire mitochondrial targeting sequence. The spliced variant maintains the same reading frame and produces a truncated protein at a very low level in the cytosol (22). Whether the human spliced variant also has a decreased stability is not known. The pathogenesis of the mutation in our mutant mice, however, was primarily an unstable, mutant form of Bcat-2 mRNA (lacking exon 2) that resulted in the loss of BCAT-2 protein and enzyme activity.
Response to a diet low in BCAAs.
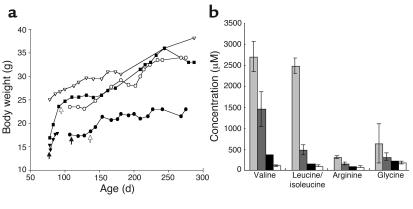
Our BCAT-deficient mice exhibited failure to thrive, weakness with decreased spontaneous movement, hair loss, and decreased luster. Without treatment, they died before 4 months of age. In an attempt to rescue these mice, we placed them on a low-BCAA diet by mixing human MSUD diet with regular rodent diet at a proportion of 2:1 or 3:1 (Figure 4). Human MSUD diet is an infant formula completely devoid of BCAAs that has been used successfully to treat human MSUD patients (23). Significant weight gains were seen on the low-BCAA diet (Figure 4a), along with improvements in hair quality and spontaneous activity. Concomitant with amelioration of clinical symptoms, BCAA levels decreased (>85%) and arginine and glycine normalized (Figure 4b). Importantly, the mice became fertile and could be bred with the diet. A breeding colony had been established. All affected mice (now a total of 24) responded to the diet with an average of 44% weight gains and reversal of the clinical symptoms in 5 weeks.
Figure 4.
Body weight and amino acid changes with diet restricted in BCAAs. (a) Body weight changes of five mice in two litters: affected males (filled squares and filled triangles); affected female (filled circles); unaffected siblings (male, open triangles; female, open circles). Black arrows indicate when the restricted diet (MSUD diet/regular diet, 2:1) was initiated. Open arrows denote that diet was further changed to MSUD diet/regular diet, 3:1. One mouse (filled triangle) died accidentally at day 89. (b) Changes in amino acid concentrations. Light gray bars, affected mice with regular diet; dark gray bars, MSUD diet/regular diet, 2:1; black bars, MSUD diet/regular diet, 3:1; white bars, control ENU mice with regular diet.
Discussion
The BCAT-deficient mice had clinical and laboratory findings resembling human MSUD and, like human MSUD, responded to diet low in BCAAs. Compared with human patients, however, there were subtle differences, including elevations of arginine and glycine and higher levels of valine than leucine or isoleucine, whereas in the human, leucine is the highest among three elevated BCAAs. We hypothesize that arginine and glycine elevations are secondary to the increase in BCAAs, because BCAA restricted diet has resulted in normalization of these two amino acids in our mice. The difference between mouse and human with regard to individual BCAA elevations cannot be due to the diet, as both rodent and human diets have similar individual BCAA compositions. We reason that this difference is likely to result from the fundamental genetic defect, which is BCAT-2 deficiency in our mice. BCAT-2 might have a higher affinity for valine than leucine or isoleucine. It would be interesting to know whether there exist any unclassified human MSUD patients that might be due to BCAT-2 deficiency. They might be expected to have elevated BCAAs, especially valine, with no allo-isoleucine. Several early reports of patients with hypervalinemia, vomiting, failure to thrive, mental retardation, hyperkinesias, and nystagmus (24–26) suggest such patients may well exist and further study will be needed to confirm the presence of human BCAT-2 deficiency.
ENU mutagenesis is a powerful way to produce and screen genetic variants for gene function studies in the whole organism. A potential problem is the presence of other mutations induced by ENU, which may confound the observed phenotypes. In our study, we applied a three-generation strategy to detect recessive mutations. We have further bred the affected mice with WT mice and carried out outcross breeding. All mice, now through the sixth generation, continued to show 100% phenotype and genotype correlations due to the Bcat2 gene mutation. Other mutations, if present, are likely to be diluted out and contribute little, if any, to the observed phenotypes.
In conclusion, we have identified a mouse model with a novel enzyme deficiency resembling human MSUD. These mice could provide an important animal model for study of BCAA metabolism and its toxicity. Our data further suggest that metabolomics-guided screening, coupled with ENU mutagenesis, is a powerful approach to uncover novel enzyme deficiencies and recognize important pathways for genetic metabolic disorders.
Acknowledgments
We thank Yi-Jung Lin, Cheng-Chih Huang, and the National Genotyping Center for technical support and Mouse Mutagenesis Core for providing ENU mice. We thank Susan Hutson for providing the anti-BCAT-2 antibody and helpful discussion on the project. This work was supported by the National Science & Technology Program for Genomic Medicine from the National Science Council, Taiwan, and the Genomics and Proteomics Program from the Academia Sinica, Taiwan.
Footnotes
See the related Commentary beginning on page 354.
Jer-Yuarn Wu and Hsiao-Jung Kao contributed equally to this work.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: N-ethyl-N-nitrosourea (ENU); branched-chain amino acid (BCAA); maple syrup urine disease (MSUD); branched-chain α-keto acid dehydrogenase (BCKD); branched-chain aminotransferase (BCAT); nonfluorescent quencher (NFQ); deoxyribonucleoside triphosphate (dNTP); branched-chain α-keto acid (BCKA).
References
- 1.Fields S. The future is function. Nat. Genet. 1997;15:325–327. doi: 10.1038/ng0497-325. [DOI] [PubMed] [Google Scholar]
- 2.Millington DS, Kodo N, Terada N, Roe D, Chace DH. The analysis of diagnostic markers of genetic disorders in human blood and urine using tandem mass spectrometry with liquid secondary ion mass spectrometry. Int. J. Mass Spectrom. Ion Process. 1991;111:211–228. [Google Scholar]
- 3.Millington DS, Norwood DL, Kodo N, Roe CR, Inoue F. Application of fast atom bombardment with tandem mass spectrometry and liquid chromatography/mass spectrometry to the analysis of acylcarnitines in human urine, blood, and tissue. Anal. Biochem. 1989;180:331–339. doi: 10.1016/0003-2697(89)90441-7. [DOI] [PubMed] [Google Scholar]
- 4.Millington DS, Kodo N, Norwood DL, Roe CR. Tandem mass spectrometry: a new method for acylcarnitine profiling with potential for neonatal screening for inborn errors of metabolism. J. Inherit. Metab. Dis. 1990;13:321–324. doi: 10.1007/BF01799385. [DOI] [PubMed] [Google Scholar]
- 5.Chace DH, et al. Rapid diagnosis of maple syrup urine disease in blood spots from newborns by tandem mass spectrometry. Clin. Chem. 1995;41:62–68. [PubMed] [Google Scholar]
- 6.Van Hove JL, et al. Medium-chain acyl-CoA dehydrogenase (MCAD) deficiency: diagnosis by acylcarnitine analysis in blood. Am. J. Hum. Genet. 1993;52:958–966. [PMC free article] [PubMed] [Google Scholar]
- 7.Rashed MS, Ozand PT, Bucknall MP, Little D. Diagnosis of inborn errors of metabolism from blood spots by acylcarnitines and amino acids profiling using automated electrospray tandem mass spectrometry. Pediatr. Res. 1995;38:324-–331. doi: 10.1203/00006450-199509000-00009. [DOI] [PubMed] [Google Scholar]
- 8.Hrabe de Angelis MH, et al. Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat. Genet. 2000;25:444–447. doi: 10.1038/78146. [DOI] [PubMed] [Google Scholar]
- 9.Nolan PM, et al. A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat. Genet. 2000;25:440–443. doi: 10.1038/78140. [DOI] [PubMed] [Google Scholar]
- 10.Rolinski B, et al. The biochemical metabolite screen in the Munich ENU Mouse Mutagenesis Project: determination of amino acids and acylcarnitines by tandem mass spectrometry. Mamm. Genome. 2000;11:547–551. doi: 10.1007/s003350010105. [DOI] [PubMed] [Google Scholar]
- 11.Weber JS, Salinger A, Justice MJ. Optimal N-ethyl-N-nitrosourea (ENU) doses for inbred mouse strains. Genesis. 2000;26:230–233. [PubMed] [Google Scholar]
- 12.Roe CR, et al. Diagnostic and therapeutic implications of medium-chain acylcarnitines in the medium-chain acyl-coA dehydrogenase deficiency. Pediatr. Res. 1985;19:459–466. doi: 10.1203/00006450-198505000-00011. [DOI] [PubMed] [Google Scholar]
- 13.Nakai N, Kobayashi R, Popov KM, Harris RA, Shimomura Y. Determination of branched-chain alpha-keto acid dehydrogenase activity state and branched-chain alpha-keto acid dehydrogenase kinase activity and protein in mammalian tissues. Methods Enzymol. 2000;324:48–62. doi: 10.1016/s0076-6879(00)24218-3. [DOI] [PubMed] [Google Scholar]
- 14.Cooper AJ, Conway M, Hutson SM. A continuous 96-well plate spectrophotometric assay for branched-chain amino acid aminotransferases. Anal. Biochem. 2002;308:100–105. doi: 10.1016/s0003-2697(02)00243-9. [DOI] [PubMed] [Google Scholar]
- 15.Chuang, D.T., and Shih, V.E. 2001. Maple syrup urine disease (branched-chain ketoaciduria). In The metabolic and molecular bases of inherited disease. C.R. Scriver, et al., editors. McGraw-Hill. New York, New York, USA. 1971–2005.
- 16.Hyde R, Taylor PM, Hundal HS. Amino acid transporters: roles in amino acid sensing and signalling in animal cells. Biochem. J. 2003;373:1–18. doi: 10.1042/bj20030405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Cam A, Freychet P. Neutral amino acid transport. Characterization of the A and L systems in isolated rat hepatocytes. J. Biol. Chem. 1977;252:148–156. [PubMed] [Google Scholar]
- 18.Matthews DE, Ben-Galim E, Haymond MW, Bier DM. Alloisoleucine formation in maple syrup urine disease: isotopic evidence for the mechanism. Pediatr. Res. 1980;14:854–857. doi: 10.1203/00006450-198007000-00002. [DOI] [PubMed] [Google Scholar]
- 19.Davoodi J, et al. Overexpression and characterization of the human mitochondrial and cytosolic branched-chain aminotransferases. J. Biol. Chem. 1998;273:4982–4989. doi: 10.1074/jbc.273.9.4982. [DOI] [PubMed] [Google Scholar]
- 20.Hutson SM, Hall TR. Identification of the mitochondrial branched chain aminotransferase as a branched chain alpha-keto acid transport protein. J. Biol. Chem. 1993;268:3084–3091. [PubMed] [Google Scholar]
- 21.Drown PM, Torres N, Tovar AR, Davoodi J, Hutson SM. Use of sulfhydryl reagents to investigate branched chain alpha-keto acid transport in mitochondria. . Biochim. Biophys. Acta. 2000; 1468:273–284. doi: 10.1016/s0005-2736(00)00266-2. [DOI] [PubMed] [Google Scholar]
- 22.Than NG, Sumegi B, Than GN, Bellyei S, Bohn H. Molecular cloning and characterization of placental tissue protein 18 (PP18a)/human mitochondrial branched-chain aminotransferase (BCATm) and its novel alternatively spliced PP18b variant. Placenta. 2001;22:235–243. doi: 10.1053/plac.2000.0603. [DOI] [PubMed] [Google Scholar]
- 23.Snyderman SE, Goldstein F, Sansaricq C, Norton PM. The relationship between the branched chain amino acids and their alpha-ketoacids in maple syrup urine disease. Pediatr. Res. 1984;18:851–853. doi: 10.1203/00006450-198409000-00009. [DOI] [PubMed] [Google Scholar]
- 24.Wada Y, et al. Ideopathic hypervalinemia-probably new entity of inborn error of valine metabolism. Tohoku J. Exp. Med. 1963;81:46–55. [PubMed] [Google Scholar]
- 25.Dancis J, et al. Hypervalinemia. A defect in valine transamination. Pediatrics. 1967;39:813–817. [PubMed] [Google Scholar]
- 26.Reddi OS, Reddy SV, Reddy KR. A sibship with hypervalinemia. Hum. Genet. 1977;39:139–142. doi: 10.1007/BF00273165. [DOI] [PubMed] [Google Scholar]