Abstract
Mutations in the lamin A/C gene (LMNA) cause a variety of human diseases including Emery-Dreifuss muscular dystrophy, dilated cardiomyopathy, and Hutchinson-Gilford progeria syndrome. The tissue-specific effects of lamin mutations are unclear, in part because the function of lamin A/C is incompletely defined, but the many muscle-specific phenotypes suggest that defective lamin A/C could increase cellular mechanical sensitivity. To investigate the role of lamin A/C in mechanotransduction, we subjected lamin A/C–deficient mouse embryo fibroblasts to mechanical strain and measured nuclear mechanical properties and strain-induced signaling. We found that Lmna–/– cells have increased nuclear deformation, defective mechanotransduction, and impaired viability under mechanical strain. NF-κB–regulated transcription in response to mechanical or cytokine stimulation was attenuated in Lmna–/– cells despite increased transcription factor binding. Lamin A/C deficiency is thus associated with both defective nuclear mechanics and impaired mechanically activated gene transcription. These findings suggest that the tissue-specific effects of lamin A/C mutations observed in the laminopathies may arise from varying degrees of impaired nuclear mechanics and transcriptional activation.
Introduction
Lamins are structural components of the nuclear lamina, a protein network underlying the inner nuclear membrane that determines nuclear shape and size (1). In addition, lamins play an important role in organizing nuclear pore complexes (2) and recruiting other proteins such as emerin to the nuclear envelope (3, 4). Two types of lamins are found in mammalian cells: A-type lamins (lamin A, C, AΔ10, and C2) are encoded by a single gene (Lmna) and are developmentally regulated and expressed in differentiated cells. B-type lamins (B1 and B2/B3) are encoded by two distinct genes and are constitutively expressed in all cells (1, 5–10). Mutations in the gene encoding A-type lamins and their binding partners have been associated with a variety of human diseases, including Emery-Dreifuss muscular dystrophy, dilated cardiomyopathy, Dunnigan-type familial partial lipodystrophy, and Hutchinson-Gilford progeria syndrome (11–17).
The molecular mechanisms underlying the varied phenotypes are unknown, and two alternative hypotheses have been proposed to explain the tissue-specific effects observed in laminopathies. The “structural hypothesis” suggests that lamin mutations lead to increased nuclear fragility and eventual nuclear disruption in the mechanically stressed tissue, while the “gene regulation hypothesis” proposes a tissue-specific role of lamins in DNA transcription. Lmna–/– mice are indistinguishable from their littermates at birth but develop severe muscle wasting and contractures similar to Emery-Dreifuss muscular dystrophy by 3–4 weeks and die by 8 weeks (4). Cells derived from Lmna–/– mice have misshapen nuclei and obvious ultrastructural damage (4, 18). Distorted nuclear shape has also been demonstrated in fibroblasts from lipodystrophic patients with heterozygous R482Q/W mutations in the lamin A/C gene and in cells from Caenorhabditis elegans with reduced lamin levels (1, 19). Here we show that nuclear mechanics in cells from Lmna–/– mice are defective, with Lmna–/– nuclei displaying increased deformation and fragility under strain. In addition, we demonstrate that transcriptional activation in response to mechanical stimuli is attenuated in Lmna–/– cells, impairing viability of mechanically strained cells. These data suggest that the structural and gene regulation hypotheses of the laminopathies are in fact closely related, and different mutations may cause specific phenotypes by differentially affecting these processes.
Methods
Cells.
Lmna+/+ and Lmna–/– mouse embryo fibroblasts were maintained in DMEM (Invitrogen Corp., Carlsbad, California, USA) containing 10% FCS (HyClone Laboratories, Logan, Utah, USA) and penicillin/streptomycin (Invitrogen Corp.).
Nuclear strain experiments.
Cells were plated at 900 cells/cm2 on fibronectin-coated silicone membranes in DMEM supplemented with 10% FCS followed by serum starvation for 48 hours in DMEM containing insulin, transferrin, and selenium (ITS) supplement (Sigma-Aldrich, St. Louis, Missouri, USA). Preceding the strain experiments, cells were incubated with Hoechst 33342 nuclear stain (1 μg/ml; Molecular Probes Inc., Eugene, Oregon, USA) in DMEM plus ITS for 20 minutes. Membranes were placed on a custom-made strain device mounted on an Olympus IX-70 microscope (Olympus America Inc., Melville, New York, USA), and biaxial strain was applied in a stepwise fashion. Membrane and nuclear strain was computed on bright field and fluorescence images using a custom-written image-analysis algorithm. Normalized nuclear strain was defined as the ratio of nuclear strain to membrane strain to compensate for small variations in applied membrane strain (range 17.4–19.8%).
Magnetic bead microrheology.
Cells were plated on 35-mm polystyrene dishes (Corning-Costar Corp., Corning, New York, USA). The following day, cells were incubated with fibronectin-coated paramagnetic beads (Dynal Biotech Inc., Lake Success, New York, USA) for 30 minutes. To minimize nuclear effects, only beads attached more than 5 μm from the nucleus were selected for analysis. A sinusoidal force (amplitude 0.6 nanonewtons [nN], frequency 1 Hz, offset 0.6 nN) was applied through a magnetic trap, and bead displacement was monitored using a digital camera (Roper Scientific Inc., San Diego, California, USA). Displacement amplitudes were computed using custom-written MATLAB (The MathWorks Inc., Natick, Massachusetts, USA) algorithms. In separate experiments, smaller (2 μm) fibronectin-coated polystyrene beads (Bangs Laboratories Inc., Fishers, Indiana, USA) were incubated together with the magnetic beads for 1 hour to adhere to the cell surface. Cells containing single magnetic beads and several polystyrene beads were subjected to a brief force pulse (2.5 nN for 3 seconds). Using custom-written MATLAB algorithms, maximal induced magnetic and polystyrene bead displacements were computed and expressed in cylindrical coordinates (r, θ) with the magnetic bead at the origin and θ = 0 for the force direction. The induced strain field can be described by an analytical cell mechanics model proposed by Bausch et al. (20) expressing the radial component ur of the induced bead displacement as a function of the applied force F, cell stiffness μ*, the characteristic cut of radius κ−1, the distance from the magnetic bead center r, and the polar angle θ:
Equation
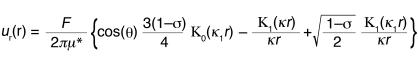 |
1 |
where Κ0 and Κ1 are modified Bessel functions of the second kind (order 0 and 1, respectively) and using κ1 = [(1 – ς)/2]1/2κ. The parameters μ* and κ were obtained by fitting equation 1 to the bead displacement data using the GraphPad Prism 4.0 robust curve fit function (GraphPad Software, San Diego, California, USA) and assuming ς = 0.5 for incompressible media and a magnetic bead contact radius of 2 μm.
The magnetic trap calibration was performed as described previously (21). In brief, magnetic beads suspended in viscous solution were monitored while being attracted to the magnetic trap operated at various currents. The applied force as a function of current and distance from the magnetic trap was then computed based on Stokes’ law.
Microinjection.
Cells were plated on fibronectin-coated glass dishes (WillCo Wells BV, Amsterdam, The Netherlands) or silicone dishes and incubated overnight. Microinjections were performed using an Eppendorf microinjector with Eppendorf Femtotips (Eppendorf AG, Hamburg, Germany). In each dish, 20–50 cells were injected with Texas Red–labeled 70-kDa dextran (Molecular Probes Inc.) dissolved at 10 mg/ml in PBS (Invitrogen Corp.), either into the cytoplasm (injection pressure 500 hectopascals [hPa], injection time 0.6 seconds) or into the nucleus (injection pressure 10, 100, 500, and 1,500 hPa, injection time 0.6 seconds). Following microinjection, cells were washed in HBSS (Invitrogen Corp.), and intracellular localization of dextran–Texas Red was recorded under a fluorescent microscope. Selected silicone membranes were subjected to constant biaxial strain (about 32% for 30 minutes) or cyclic biaxial strain (10% at 1 Hz for 24 hours), and localization of Texas Red–labeled dextran in strained and control cells was analyzed on a fluorescence microscope (Olympus America Inc.).
Strain experiments.
Strain stimulation was carried out as previously described (22). In brief, cells were plated on fibronectin-coated silicone membranes (2,500–3,300 cells/cm2). After 72 hours of serum starvation, cells were subjected to biaxial cyclic strain (4% or 10% at 1 Hz). For chemical stimulation, cells were incubated with IL-1β (25 ng/ml; R&D Systems Inc., Minneapolis, Minnesota, USA) or PMA (200 ng/ml, Sigma-Aldrich) in DMEM plus ITS.
DsRed/peroxiredoxin-2 localization.
Cells were plated on fibronectin-coated glass dishes or fibronectin-coated silicone membranes. Following overnight incubation, cells were transfected with a CMV promoter–driven DsRed/peroxiredoxin-2 fusion construct (CLONTECH Inc., Palo Alto, California, USA) using FuGENE 6 (F. Hoffman–La Roche Ltd., Basel, Switzerland) and incubated for 24 hours. Selected silicone membranes were subjected to constant (about 19% for 60 minutes) or cyclic biaxial strain (10% at 1 Hz for 3 hours), and localization of DsRed-labeled peroxiredoxin-2 in strained and control cells was analyzed on a fluorescence microscope (Olympus America Inc.).
Flow cytometry.
For cell viability assays, propidium iodide (2 μg/ml, Sigma-Aldrich) was added to the dishes after 24 hours of strain application. Cells were collected and analyzed using a Cytomics FC 500 flow cytometer (Beckman Coulter Inc., Fullerton, California, USA), counting 10,000–30,000 events in each group. Thresholds for propidium iodide incorporation were determined based on negative (no propidium iodide staining) and positive (cells permeabilized by 50% ethanol) controls. Apoptotic and necrotic cell fractions were measured in similar experiments using the Vybrant Apoptosis Assay Kit no. 3 (Molecular Probes Inc.).
Northern and Western analyses.
Expression of iex-1 and egr-1 mRNA was assessed by Northern analysis as described previously (23). Protein expression was analyzed by Western analysis of nuclear and cytoplasmic cell fractions using antibodies against NF-κB p65 (antibody does not recognize p50 or p105), IκBα (both from Santa Cruz Biotechnology Inc., Santa Cruz, California, USA), and actin (Sigma-Aldrich). Additional immunoblotting was performed on whole cell lysates using specific antibodies against total ERK1/2 (Santa Cruz Biotechnology Inc.) and phospho-p44/p42 MAP kinase (Cell Signaling Technology, Beverly, Massachusetts, USA). After incubation with HRP-conjugated secondary antibody (Bio-Rad Laboratories Inc., Hercules, California, USA), specific bands were visualized by enhanced chemiluminescence (PerkinElmer Inc., Boston, Massachusetts, USA).
Luciferase experiments.
Cells were transfected with plasmids for NF-κB–controlled luciferase expression and SV40-regulated β-gal (Promega Corp., Madison, Wisconsin, USA) using FuGENE 6 (F. Hoffman–La Roche Ltd.). Following transfection, cells were serum starved in DMEM plus ITS medium for 48 hours, followed by overnight stimulation with PMA (200 ng/ml) or IL-1β (25 ng/ml). Luciferase assays were quantified in a Victor2 Multilabel Counter (Perkin Elmer Inc.). Results were normalized for β-gal activity and expressed as percent WT control.
Immunohistochemistry.
Cells were plated on untreated or fibronectin-coated chamber slides and serum-starved for 24–72 hours followed by stimulation with IL-1β. Cells were fixed in 4% paraformaldehyde or methanol, washed in PBS, and permeabilized with 0.1% Triton X-100. After blocking, cells were incubated overnight with primary rabbit antibody anti–NF-κB (p65; Santa Cruz Biotechnology Inc.) at 4°C or Alexa Fluor 568 phalloidin (A-12380; Molecular Probes Inc.) for 1 hour at 25°C, followed by 1 hour of incubation with secondary FITC- or TRITC-conjugated antibodies.
Cellular protein fractions and electrophoretic mobility shift assay.
Nuclear extracts were prepared as described previously (24) with the following modifications. Cells were washed with ice-cold PBS and lysed in buffer A, which consisted of 0.1% Triton X-100, 10 mM EDTA, 10 mM EGTA, 10 mM KCl, 10 mM HEPES, 1 mM DTT, 0.5 mM PMSF, and protease inhibitor cocktail (P-8340; Sigma-Aldrich). After centrifugation at 1,200 g for 10 minutes, the supernatant was stored as the cytoplasmic cell fraction, while the nuclear pellet was washed once in PBS, resuspended in buffer C (1 mM EDTA, 1 mM EGTA, 0.4 M NaCl, 20 mM HEPES, 5 mM MgCl2, 25% glycerol, 1 mM DTT, 0.5 mM PMSF, and protease inhibitor cocktail), and incubated at 4°C for 10 minutes. The nuclear extract was centrifuged for 10 minutes at 4°C at 12,000 g, and the supernatant was used for Western analysis and electrophoretic mobility shift assay. NF-κB–specific oligonucleotides (Promega Corp.) were end-labeled using T4 polynucleotide kinase and [γ-32P]ATP (DuPont NEN Research Products, Boston, Massachusetts, USA). Nuclear extracts were preincubated for 10 minutes in binding buffer followed by 20 minutes of incubation at room temperature with labeled oligonucleotide. Samples were separated on a 4% native polyacrylamide gel. For competition studies, an excess (50 times) of unlabeled oligonucleotide was used, and for the supershift assay, the nuclear extracts were incubated with 2 μg of anti-p50 or anti-p65 antibody (Santa Cruz Biotechnology Inc.).
Statistical analysis.
All experiments were performed at least three independent times. Data are expressed as mean ± SEM. Statistical analysis was performed using PRISM 4.0 and INSTAT software (GraphPad Software Inc., San Diego, California, USA). The data were analyzed by unpaired t test (allowing different SDs),one-way ANOVA, or the Mann-Whitney test in case of nonparametric distribution. A two-tailed P value below 0.05 was considered significant.
Results
Nuclear mechanics.
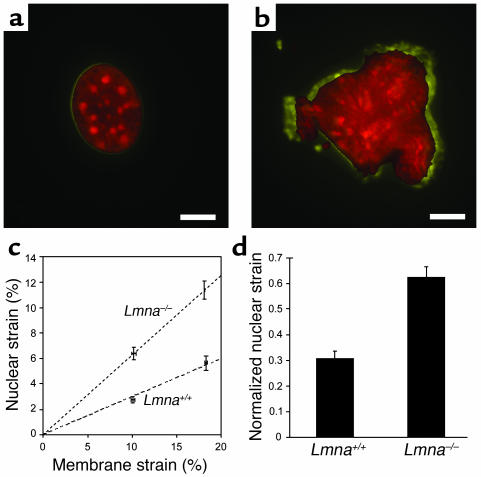
To explore the role of lamin A/C in nuclear mechanics, mouse embryo fibroblasts derived from Lmna–/– and Lmna+/+ mice were cultured on transparent membranes and subjected to stepwise increasing biaxial strain (first step: 10.1% ± 0.18 %; second step: 18.2% ± 0.07%). The cytoskeleton, attached to the membrane through integrin receptors, is exposed to the same biaxial strain as the membrane, while the stiffer nucleus exhibits only small deformations in WT cells (25). The induced nuclear deformations were calculated by tracking the fluorescently labeled nucleus (Figure 1, a and b) and normalized to membrane strain to compensate for the small variation in applied membrane strain. For each cell type, nuclear deformation increased approximately linearly with applied membrane strain (Figure 1c), but Lmna–/– nuclei showed significantly larger deformations than did WT cells (Figure 1, c and d). Fitting a linear regression to the nuclear deformation data revealed a significantly larger slope for the Lmna–/– nuclei, and the maximal normalized nuclear deformation was significantly larger for Lmna–/– cells, indicating impaired nuclear mechanics in lamin A/C–deficient nuclei.
Figure 1.
Nuclear mechanics is impaired in lamin A/C–deficient cells. (a) Nucleus of WT fibroblast before strain (red) and at 22% strain (yellow). Scale bar: 10 μm. (b) Lmna–/– nucleus before strain (red) and at 19% strain (yellow). Scale bar: 10 μm. (c) Nuclear strain as a function of applied membrane strain. Dashed lines represent linear regression of the data for each cell type forced through the origin (Lmna+/+: y = 0.299x, Lmna–/–: y = 0.626x). (d) Maximal normalized nuclear strain was significantly increased in Lmna–/– fibroblasts (0.306 ± 0.029 vs. 0.626 ± 0.039; P < 0.0001, n = 21).
Cytoskeletal mechanics.
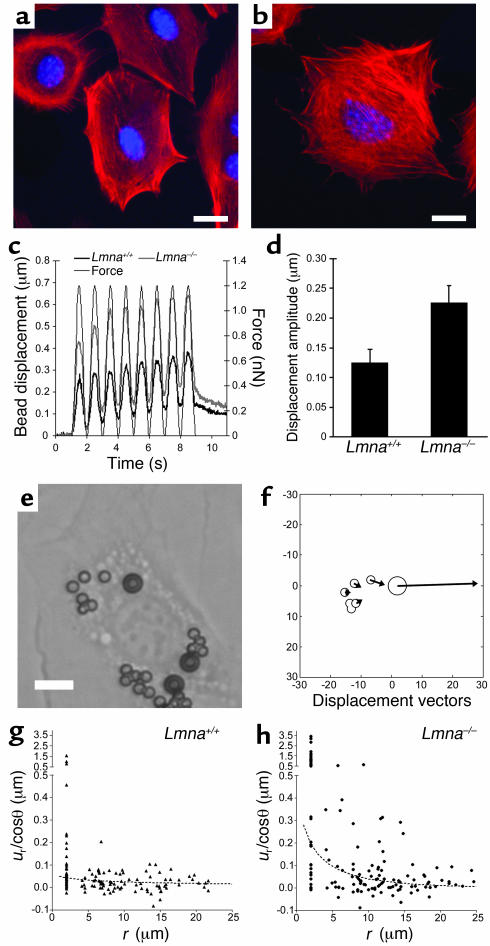
To evaluate the possibility that the observed increase in nuclear deformation was caused by more direct force transmission to the nucleus, we examined cytoskeletal organization by staining actin stress fibers with phalloidin. No apparent differences in cytoskeletal architecture were found between WT (Figure 2a) and Lmna–/– cells (Figure 2b). Magnetic bead microrheology (20) was used for quantitative evaluation of cytoskeletal stiffness. Small (4.5 μm), fibronectin-coated paramagnetic beads were attached to the cell, and the bead displacement in response to an applied magnetic force was used as an indicator of cytoskeletal stiffness (Figure 2c). The induced bead displacement amplitude was significantly increased in Lmna–/– fibroblasts (Figure 2d), indicating decreased cytoskeletal stiffness. Since magnetic bead displacement depends not only on cytoskeletal stiffness but also on the binding characteristics of the cell to the bead, we applied a second microrheology method that measures the induced displacement in polystyrene beads located on the cell surface close to the magnetic bead (Figure 2, e and f). Lamin A/C–deficient cells exhibited significantly larger induced polystyrene bead displacements that decreased quickly with increasing distance from the magnetic bead, whereas bead displacements in WT cells were smaller and hardly exceeded the detection limit of less than 0.1 μm (Figure 2, g and h). Based on an analytical model proposed by Bausch et al. (20), the induced polystyrene bead displacement is a function of the applied force, the position relative to the magnetic bead, and two cellular mechanics parameters μ* and κ that describe the cell stiffness and intracellular strain dissipation, respectively. By separately fitting the theoretical displacement field to the observed bead data for Lmna–/– and WT fibroblasts, one can estimate the parameters for cellular stiffness and dissipation (Figure 2, g and h), confirming that Lmna–/– cells exhibit significantly decreased cytoskeletal stiffness.
Figure 2.
Cytoskeletal stiffness is reduced in lamin A/C–deficient cells. (a) Phalloidin staining for actin stress fibers in WT (Lmna+/+) fibroblasts. Scale bar: 20 μm. (b) Phalloidin staining for actin stress fibers in Lmna–/– cells. Scale bar: 20 μm. (c) Magnetic bead microrheology. Representative examples of magnetic bead displacement in response to applied sinusoidal force (thin black line) for WT (thick black line) and Lmna–/– (thick gray line) fibroblasts. (d) Bead displacement amplitude in response to applied magnetic forces was significantly increased in Lmna–/– fibroblasts, indicating reduced cytoskeletal stiffness in lamin A/C–deficient cells (0.124 ± 0.024 μm vs. 0.226 ± 0.029 μm; P < 0.01, n = 60). (e) Fibroblast with magnetic (diameter 4.5 μm) and polystyrene beads (diameter 2 μm) attached to the cell membrane. Scale bar: 10 μm. (f) Graphic representation of the displacement field after a brief force pulse (2.5 nN for 3 seconds). Bead sizes and positions are drawn to scale, while bead deflections are enlarged by a factor of 10. (g and h) Distance dependence of the angle-corrected radial bead displacement component ur/cosθ as defined in equation 1. The dotted line is an optimal fit to equation 1, yielding estimates for cellular stiffness μ* and dissipation κ for WT (g) and Lmna–/– cells (h), respectively (μ*: 27,537 ± 8,458 pN/μm vs. 2,417 ± 734.7 pN/μm; P < 0.01, n = 128 [WT], 153 [Lmna–/–]; κ: 0.020 ± 0.017 μm–1 vs. 0.201 ± 0.072 μm–1; P < 0.05, n = 128 [WT], 153 [Lmna–/–]). pN, piconewton.
Therefore, the observed increased nuclear deformation in lamin-deficient cells is unlikely to be due to altered force transmission to the nucleus. In contrast, the softer cytoskeleton would result in an underestimation of the nuclear stiffness. Based on these findings we conclude that lamin A/C–deficient cells have decreased nuclear stiffness and altered nuclear mechanics.
Cellular response to mechanical strain.
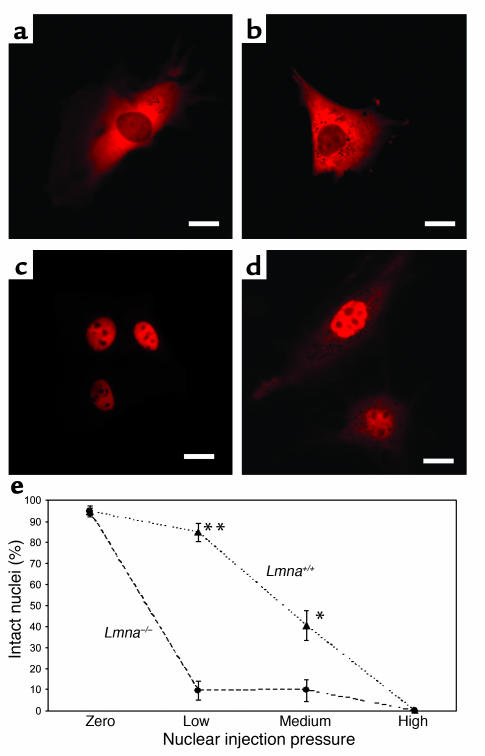
Strain-induced damage to the more fragile nucleus could provide one explanation for tissue-specific effects of lamin A/C mutations, for example, in mechanically active tissues like myocardium and skeletal muscle. To examine nuclear envelope integrity, we monitored the subcellular location of fluorescently labeled 70-kDa dextran microinjected into either the cytoplasm or nucleus of adherent fibroblasts. Cytoplasmic injection revealed that the high-molecular-weight dextran was excluded from the nucleus in both WT and lamin-deficient cells (Figure 3, a and b), indicating that nuclear integrity is not significantly impaired in Lmna–/– cells under resting conditions. These results were confirmed by transfecting the cells with a DsRed/peroxiredoxin-2 fusion protein. The protein lacks a nuclear localization sequence and is too large (mol wt 57 kDa) to passively diffuse into the intact nucleus. Consequently, the fusion protein was excluded from the nucleus of WT and Lmna–/– cells.
Figure 3.
Nuclear fragility is increased in lamin A/C–deficient cells. (a and b) Fluorescently labeled 70-kDa dextran is excluded from the nucleus following cytoplasmic injection, indicating an intact nuclear membrane in WT (a) and Lmna–/– (b) cells under resting conditions. Scale bar: 20 μm. (c) Nuclear injection at low pressure results in fluorescently labeled dextran contained in the nucleus of WT cells, indicating that the nuclear integrity is preserved during injection. Scale bar: 20 μm. (d) In contrast, nuclear integrity is disrupted in nuclei of Lmna–/– cells even at low pressure, leading to fluorescently labeled dextran escaping into the cytoplasm during injection. Scale bar: 20 μm. (e) Nuclear rupture as a function of increasing injection pressure of dextran microinjection into the nucleus. Zero pressure: cytoplasmic injection at 500 hPa. Low pressure: nuclear injection at 10–20 hPa (**84.7% ± 4.24% vs. 9.5% ± 4.53% intact nuclei for WT and Lmna–/– cells, respectively; P < 0.0001, n = 72 [WT], 42 [Lmna–/–]). Medium pressure: nuclear injection at 100–500 hPa (*40.4% ± 7.16% vs. 9.5% ± 4.53% intact nuclei for WT and Lmna–/– cells respectively; P < 0.01, n = 47 [WT], 31 [Lmna–/–]). High pressure: nuclear injection at 1,500 hPa; all cells showed nuclear rupture.
However, when dextran was injected directly into the nucleus at low and medium injection pressures (10–500 hPa), nuclear integrity in lamin A/C–deficient cells was at least temporarily compromised, resulting in fluorescently labeled dextran escaping into the cytoplasm (Figure 3d). In contrast, dextran was confined to the nucleus in most WT cells when injected at low pressure (Figure 3c), and even at medium pressure significantly more WT cells than Lmna–/– cells maintained their nuclear integrity. Using sufficiently high pressure (1,500 hPa), both WT and lamin-deficient nuclei could be ruptured (Figure 3e).
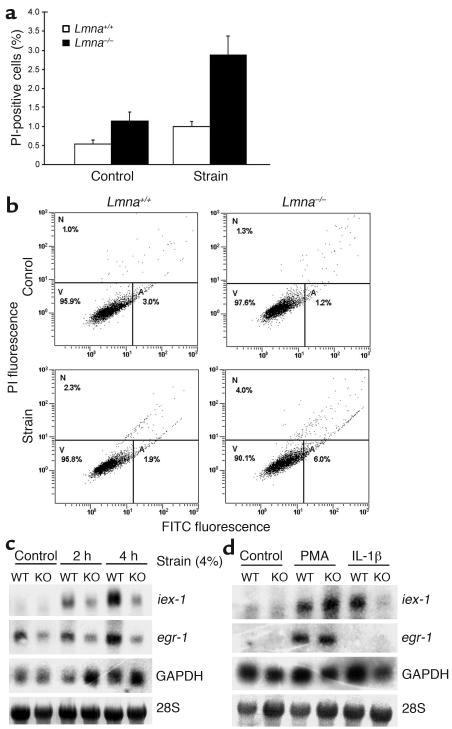
To determine whether Lmna–/– fibroblasts are thus more susceptible to mechanical strain, cells were subjected to cyclic biaxial strain (10% strain at 1 Hz). After 24 hours of strain, Lmna–/– fibroblasts had a significantly increased fraction of dead (i.e., propidium iodide–positive) cells compared with controls not subjected to strain (Figure 4a). Differences within controls (WT and Lmna–/–) and between WT control and strained cells were not significant. Dual labeling with propidium iodide and an FITC-conjugated annexin V antibody revealed that the decrease in viability was due to an increase in both necrotic and apoptotic cell fractions compared with WT cells (Figure 4b).
Figure 4.
Impaired mechanotransduction in lamin A/C–deficient cells. (a) Lmna–/– fibroblasts exhibited a significantly higher percentage of propidium iodide–positive cells than did WT cells (2.88% ± 0.49% vs. 1.14% ± 0.14%; P < 0.01, n = 7 [WT], 8 [Lmna–/–]) after 24 hours of strain application (10% at 1 Hz). Differences in unstrained cells were not significant (n = 9 [WT], 10 [Lmna–/–]). (b) Dual labeling with FITC-conjugated annexin V and propidium iodide uptake indicated that apoptotic (A) and necrotic (N) cell fractions are increased in Lmna–/– cells following prolonged strain. Viable cells (V) are propidium iodide–negative and FITC-negative. Top left: WT unstrained control; top right: Lmna–/– unstrained control; bottom left: WT cells after 10% strain for 24 hours; bottom right: Lmna–/– cells after 10% strain for 24 hours. PI, propidium iodide. (c) Lmna–/– (KO) fibroblasts exhibited attenuated mechanical induction of egr-1 and iex-1 after 2 hours and 4 hours of strain (4%) compared with Lmna+/+ cells (WT). Expression of GAPDH was not negatively affected. (d) Cytokine–induced expression of iex-1 was impaired in Lmna–/– cells, while PMA responsiveness remained intact in Lmna–/– cells.
Interestingly, when cells were injected with the fluorescently labeled 70-kDa dextran into the cytoplasm and subjected to either constant (30% for 30 minutes) or cyclic (10% at 1 Hz for 24 hours) biaxial strain, strain application did not significantly increase the number of dextran-positive nuclei in either WT or Lmna–/– fibroblasts (data not shown), suggesting that the extreme event of nuclear rupture under mechanical strain occurs in only a small fraction of cells that cannot be detected based on a small number (n = 20–30) of single cell observations.
The increased fraction of apoptotic cells in mechanically strained Lmna–/– fibroblasts indicates that necrosis through nuclear rupture can only partly explain the increased sensitivity to mechanical stimulation. Therefore, we investigated the cellular response to mechanical stimulation in more detail. Interestingly, expression of the mechanosensitive genes egr-1 and iex-1 in response to mechanical stimulation was impaired in Lmna–/– cells (Figure 4c), whereas expression of the mechanically unresponsive genes thioredoxin-1 and GAPDH was unaltered (data not shown), indicating that transcription is not impaired in a nonspecific manner. To test whether the observed changes in Lmna–/– cells were specific to mechanical stimulation or represented a more general deficiency in signal transduction, we measured the expression levels of iex-1 and egr-1 in response to stimulation with PMA or the cytokine IL-1β. Cytokine stimulation led to an attenuated response in iex-1 but not egr-1 expression in Lmna–/– cells, while PMA responsiveness remained intact for both genes in Lmna–/– cells (Figure 4d).
NF-κB signaling.
Because iex-1 is an NF-κB–dependent survival gene (23), and because NF-κB can be biomechanically activated (26), we examined whether biomechanical signaling through NF-κB is disturbed in Lmna–/– cells. The MAPK ERK1/2 is an important regulator of mechanically induced gene expression and has been linked to NF-κB activation (27). Analysis of MAPK phosphorylation after chemical (PMA) or mechanical stimulation revealed no differences between Lmna–/– and WT cells (data not shown), indicating that the observed changes are caused by alterations in signal transduction other than impaired cytoplasmic MAPK activation.
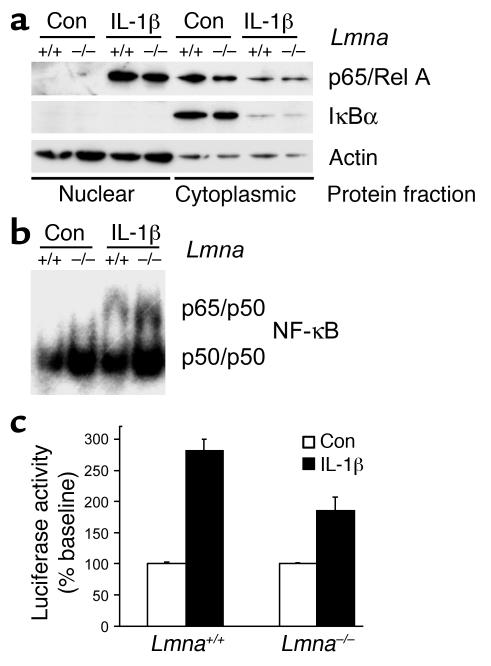
In resting cells, NF-κB is sequestered in the cytoplasm by the inhibitor IκB. Upon stimulation, IκB is ubiquinated and degraded, allowing NF-κB to translocate into the nucleus and activate target genes. Figure 5a shows that cytokine-induced cytoplasmic degradation of IκBα and translocation of the NF-κB subunit p65/RelA into the nucleus was not impaired in Lmna–/– cells. This finding was confirmed in immunofluorescence directed against p65/RelA (data not shown). Total levels of NF-κB subunits p50, p65, and p105 in whole cell extracts from Lmna–/– cells were indistinguishable from those in WT cells (data not shown). Interestingly, binding of nuclear NF-κB to its transcription factor binding site was increased in lamin A/C–deficient cells compared with WT cells (Figure 5b). Surprisingly, despite the increased levels of transcription factor binding, IL-1β–induced activity of NF-κB–dependent luciferase was significantly impaired in Lmna–/– fibroblasts (Figure 5c). These results indicate that the deficient response of Lmna–/– cells to mechanical or cytokine stimulation is based on a role for lamin A/C in transcriptional activation following transcription factor binding.
Figure 5.
Defective NF-κB signaling in lamin A/C–deficient cells. (a) Western blot analysis of nuclear and cytoplasmic protein fractions. Cytokine-induced nuclear translocation of p65/RelA and degradation of cytoplasmic IκBα was indistinguishable between WT and Lmna–/– fibroblasts. Cytoplasmic proteins were loaded at a lower concentration. Con, control. (b) Electrophoretic mobility shift assay for NF-κB target sequence using protein from the same nuclear fractions as in a. Probe specificity was confirmed using unlabeled competitive and noncompetitive probes. Identity of NF-κB subunits p50 and p65 was confirmed by supershift assay. (c) Cytokine-induced NF-κB–regulated luciferase activity was significantly impaired in Lmna–/– cells (percent of baseline: 282% ± 18.5% vs. 185% ± 22.2%; P < 0.001, n = 9). Baseline activity was not significantly different between WT and Lmna–/– cells.
Discussion
Our results establish the importance of lamin A/C for nuclear stability and highlight its role in transcriptional regulation in response to mechanical or chemical stimulation. Measurements of nuclear mechanics were obtained using a novel technique to measure nuclear deformation with biaxial strain applied to the cells. This method yields quantitative measurements of nuclear stiffness compared with cytoskeletal stiffness in living cells without having to isolate the nuclei. Our results for nuclear deformation in strained Lmna–/– cells are in excellent agreement with measurements obtained by Caille et al. (25) in endothelial cells under uniaxial strain and on isolated endothelial cell nuclei (28). Increased nuclear fragility in Lmna–/– cells was further confirmed by nuclear microinjection experiments, which demonstrated increased nuclear rupture at low and moderate pressures compared with that in WT cells. The markedly increased nuclear deformability and fragility in Lmna–/– cells compared with WT cells has important implications for the cellular response to mechanical strain. Impaired nuclear stability can lead to rupture of the nucleus, resulting directly in cell death. Evidence of fragmented nuclei has been reported in skeletal muscle fibers from emerin-deficient Emery-Dreifuss muscular dystrophy patients and in fibroblasts from Dunnigan-type familial partial lipodystrophy patients following heat shock treatment (19, 29). This direct effect of impaired nuclear stability is also consistent with the impaired cell viability after cyclic strain observed in our experiments.
In addition to the direct effect of nuclear rupture, altered nuclear mechanics can affect cells through impaired nuclear mechanotransduction. Recent studies have reported desmin intermediate filament–mediated changes in chromatin in response to mechanical strain, and hypothesized that stretch-induced changes in chromatin can lead to activation of hypertrophy-associated genes (30). Mechanical connections between integrins, cytoskeletal filaments, and the nucleus have also been demonstrated by micromanipulation with microbeads and micropipettes in endothelial cells (31). Our nuclear strain experiments demonstrate that external strain application results in increased nuclear strain, indicating mechanical coupling between the extracellular matrix and the nucleus mediated through focal adhesion sites and the cytoskeleton. Mutations in nuclear envelope proteins such as lamin or emerin could interrupt some of these connections and impair nuclear mechanotransduction pathways. In addition, the impaired nuclear and cytoskeletal mechanics observed in Lmna–/– cells lead to significantly increased nuclear strain, which could further result in altered nuclear mechanosensing.
The observation of decreased cytoskeletal stiffness in Lmna–/– cells raises additional questions. Alterations in cytoskeletal stiffness could arise as a compensatory mechanism to protect a more fragile nucleus, but could also play a pivotal role in the pathophysiology of the disease. Altered cytoskeletal mechanics not only affect the transmitted force to the nucleus under applied strain, but can also play an important role in cell shape, migration, and other critical functions with direct consequences to the affected tissue.
However, increased nuclear fragility can only partially explain the phenotypes observed in several laminopathies. Even though nuclear mechanics are clearly impaired in the vast majority of Lmna–/– cells, only a very small fraction of cells (about 3–5%) exhibited nuclear rupture under strain in our experiments, and only a small number of ruptured nuclei are found in affected muscle tissue in patients suffering from Emery-Dreifuss muscular dystrophy (29). In addition, fibroblasts obtained from lipodystrophic patients with the lamin R482Q/W mutation also exhibit defective nuclear mechanics, even though these patients lack a muscular phenotype (19). Therefore, most cells appear to be functional despite distorted nuclear shape and altered nuclear mechanics, and additional events might be necessary to trigger nuclear failure. Therefore, defective nuclear mechanics could play a more important role in muscle tissue that is subjected to large mechanical strain and stress compared with (for example) adipose tissue, with cumulative cellular damage through both direct nuclear rupture and impaired mechanotransduction signaling, eventually leading to the muscular dystrophy observed in several laminopathies.
In addition to their role as nuclear envelope proteins, lamins form stable structures within the nucleoplasm as shown by fluorescence recovery after photobleaching (FRAP) (32). The impaired response of iex-1 and egr-1 expression to mechanical and cytokine stimulation, as well as the attenuated response of NF-κB–regulated luciferase activity in Lmna–/– fibroblasts despite increased transcription factor binding, indicate an important role of lamin A/C in transcriptional activation. Interactions between lamin and nuclear transcription factors have been previously demonstrated in vivo and in vitro (33, 34), and lamin A/C speckles can mediate spatial organization of splicing factor compartments and RNA polymerase II transcription (35). Disrupting the normal organization of nuclear lamins by expression of a dominant-negative lamin mutant has been shown to inhibit RNA polymerase II–dependent transcription in mammalian cells and active embryonic nuclei from Xenopus laevis (36). The impaired transcriptional activation observed in our experiments indicates a yet-unknown role of lamin A/C in the assembly of enhancesomes or as a scaffolding protein for transcription factors and coactivators.
NF-κB is a mechanical stress–responsive transcription factor that can function as an antiapoptotic signal. Impaired transcriptional activation can therefore lead to increased apoptosis in mechanically strained tissue. Furthermore, direct evidence for the role of lamin in apoptosis has previously been demonstrated in cultured cells that expressed an uncleavable mutant form of lamin (37). In these cells, chromatin failed to condense and DNA cleavage was delayed despite the activation of caspases. Kumar et al. demonstrated in ex vivo experiments that mechanical stress activated NF-κB in skeletal muscle fibers and that this response is altered in mdx mice, a model for Duchenne muscular dystrophy, stressing the importance of NF-κB signaling in muscle tissue that is affected most often in laminopathies (38).
Interestingly, several cells such as lymphoblasts, basal skin cells, and early embryonic cells do not express lamin A/C. We speculate that nuclear mechanics and mechanotransduction in these cells would be normal, since these cells are known to be capable of normal function. In these cells, the role of lamin A/C might be taken over by a different protein or complex of proteins. However, it is important to note that our experiments used only fibroblasts, and future studies of cells from different tissues are essential.
While all experiments were performed in fibroblasts that completely lack lamin A and lamin C, most human diseases arise from heterozygous lamin mutations. In these situations mutant lamin often appears as stable as WT lamin and is expressed at similar levels (39). In many cases, especially in phenotypes involving striated muscle, the mutation may lead to a structurally impaired form of lamin A/C that could act as a dominant negative and lead to cellular mechanical deficiencies as observed in the lamin A/C–null cells. Other mutations, such as those resulting in lipodystrophy, could affect the binding of lamin to other proteins or chromatin with fewer effects on the structural role of lamin itself, resulting in a partially functional protein that might affect only specific signaling pathways. Furthermore, combinations of mechanical and transcriptional regulation defects could result in complex phenotypes affecting several tissue types in diseases such as progeria. By providing independent tests for measuring structural and gene-regulatory functions of lamin A/C, our experiments could help distinguish the effects of individual mutations on the function of lamin.
Tissue-specific effects observed in several laminopathies may thus arise from two mechanisms: the impaired nuclear stability renders mechanically strained tissue more susceptible to cellular damage, and abnormal transcriptional activation impairs adaptive and protective pathways. Individual mutations in the lamin A/C gene could potentially selectively interfere with any of these functions, explaining the diversity of observed phenotypes.
Footnotes
See the related Commentary beginning on page 349.
Conflict of interest: The authors have declared that no conflict of interest exists.
Nonstandard abbreviations used: insulin, transferrin, and selenium (ITS); nanonewton (nN); hectopascal (hPa).
References
- 1.Liu J, et al. Essential roles for Caenorhabditis elegans lamin gene in nuclear organization, cell cycle progression, and spatial organization of nuclear pore complexes. Mol. Biol. Cell. 2000;11:3937–3947. doi: 10.1091/mbc.11.11.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hutchison CJ. Lamins: building blocks or regulators of gene expression? Nat. Rev. Mol. Cell Biol. 2002;3:848–858. doi: 10.1038/nrm950. [DOI] [PubMed] [Google Scholar]
- 3.Holt I, et al. Effect of pathogenic mis-sense mutations in lamin A on its interaction with emerin in vivo. J. Cell Sci. 2003;116:3027–3035. doi: 10.1242/jcs.00599. [DOI] [PubMed] [Google Scholar]
- 4.Sullivan T, et al. Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 1999;147:913–920. doi: 10.1083/jcb.147.5.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Biamonti G, et al. The gene for a novel human lamin maps at a highly transcribed locus of chromosome 19 which replicates at the onset of S-phase. Mol. Cell. Biol. 1992;12:3499–3506. doi: 10.1128/mcb.12.8.3499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin F, Worman HJ. Structural organization of the human gene encoding nuclear lamin A and nuclear lamin C. J. Biol. Chem. 1993;268:16321–16326. [PubMed] [Google Scholar]
- 7.Furukawa K, Inagaki H, Hotta Y. Identification and cloning of an mRNA coding for a germ cell-specific A-type lamin in mice. Exp. Cell Res. 1994;212:426–430. doi: 10.1006/excr.1994.1164. [DOI] [PubMed] [Google Scholar]
- 8.Lin F, Worman HJ. Structural organization of the human gene (LMNB1) encoding nuclear lamin B1. Genomics. 1995;27:230–236. doi: 10.1006/geno.1995.1036. [DOI] [PubMed] [Google Scholar]
- 9.Machiels BM, et al. An alternative splicing product of the lamin A/C gene lacks exon 10. J. Biol. Chem. 1996;271:9249–9253. doi: 10.1074/jbc.271.16.9249. [DOI] [PubMed] [Google Scholar]
- 10.Harborth J, Elbashir SM, Bechert K, Tuschl T, Weber K. Identification of essential genes in cultured mammalian cells using small interfering RNAs. J. Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 11.Bonne G, et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 1999;21:285–288. doi: 10.1038/6799. [DOI] [PubMed] [Google Scholar]
- 12.Fatkin D, et al. Missense mutations in the rod domain of the lamin A/C gene as causes of dilated cardiomyopathy and conduction-system disease. N. Engl. J. Med. 1999;341:1715–1724. doi: 10.1056/NEJM199912023412302. [DOI] [PubMed] [Google Scholar]
- 13.Cao H, Hegele RA. Nuclear lamin A/C R482Q mutation in canadian kindreds with Dunnigan-type familial partial lipodystrophy. Hum. Mol. Genet. 2000;9:109–112. doi: 10.1093/hmg/9.1.109. [DOI] [PubMed] [Google Scholar]
- 14.Shackleton S, et al. LMNA, encoding lamin A/C, is mutated in partial lipodystrophy. Nat. Genet. 2000;24:153–156. doi: 10.1038/72807. [DOI] [PubMed] [Google Scholar]
- 15.Burke B, Stewart CL. Life at the edge: the nuclear envelope and human disease. Nat. Rev. Mol. Cell Biol. 2002;3:575–585. doi: 10.1038/nrm879. [DOI] [PubMed] [Google Scholar]
- 16.De Sandre-Giovannoli A, et al. Lamin A truncation in Hutchinson-Gilford progeria. Science. 2003;300:2055. doi: 10.1126/science.1084125. [DOI] [PubMed] [Google Scholar]
- 17.Eriksson M, et al. Recurrent de novo point mutations in lamin A cause Hutchinson-Gilford progeria syndrome. Nature. 2003;423:293–298. doi: 10.1038/nature01629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raharjo WH, Enarson P, Sullivan T, Stewart CL, Burke B. Nuclear envelope defects associated with LMNA mutations cause dilated cardiomyopathy and Emery-Dreifuss muscular dystrophy. J. Cell Sci. 2001;114:4447–4457. doi: 10.1242/jcs.114.24.4447. [DOI] [PubMed] [Google Scholar]
- 19.Vigouroux C, et al. Nuclear envelope disorganization in fibroblasts from lipodystrophic patients with heterozygous R482Q/W mutations in the lamin A/C gene. J. Cell Sci. 2001;114:4459–4468. doi: 10.1242/jcs.114.24.4459. [DOI] [PubMed] [Google Scholar]
- 20.Bausch AR, Ziemann F, Boulbitch AA, Jacobson K, Sackmann E. Local measurements of viscoelastic parameters of adherent cell surfaces by magnetic bead microrheometry. Biophys. J. 1998;75:2038–2049. doi: 10.1016/S0006-3495(98)77646-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lammerding J, Kazarov AR, Huang H, Lee RT, Hemler ME. Tetraspanin CD151 regulates alpha6beta1 integrin adhesion strengthening. Proc. Natl. Acad. Sci. U. S. A. 2003;100:7616–7621. doi: 10.1073/pnas.1337546100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng GC, et al. Mechanical strain tightly controls fibroblast growth factor-2 release from cultured human vascular smooth muscle cells. Circ. Res. 1997;80:28–36. doi: 10.1161/01.res.80.1.28. [DOI] [PubMed] [Google Scholar]
- 23.De Keulenaer GW, et al. Identification of IEX-1 as a biomechanically controlled nuclear factor-kappaB target gene that inhibits cardiomyocyte hypertrophy. Circ. Res. 2002;90:690–696. doi: 10.1161/01.res.0000012922.40318.05. [DOI] [PubMed] [Google Scholar]
- 24.Remoli ME, et al. Selective expression of type I IFN genes in human dendritic cells infected with Mycobacterium tuberculosis. J. Immunol. 2002;169:366–374. doi: 10.4049/jimmunol.169.1.366. [DOI] [PubMed] [Google Scholar]
- 25.Caille N, Tardy Y, Meister JJ. Assessment of strain field in endothelial cells subjected to uniaxial deformation of their substrate. Ann. Biomed. Eng. 1998;26:409–416. doi: 10.1114/1.132. [DOI] [PubMed] [Google Scholar]
- 26.Inoh H, et al. Uni-axial cyclic stretch induces the activation of transcription factor nuclear factor kappaB in human fibroblast cells. FASEB J. 2002;16:405–407. doi: 10.1096/fj.01-0354fje. [DOI] [PubMed] [Google Scholar]
- 27.Granet C, Boutahar N, Vico L, Alexandre C, Lafage-Proust MH. MAPK and SRC-kinases control EGR-1 and NF-kappa B inductions by changes in mechanical environment in osteoblasts. Biochem. Biophys. Res. Commun. 2001;284:622–631. doi: 10.1006/bbrc.2001.5023. [DOI] [PubMed] [Google Scholar]
- 28.Caille N, Thoumine O, Tardy Y, Meister JJ. Contribution of the nucleus to the mechanical properties of endothelial cells. J. Biomech. 2002;35:177–187. doi: 10.1016/s0021-9290(01)00201-9. [DOI] [PubMed] [Google Scholar]
- 29.Markiewicz E, et al. Increased solubility of lamins and redistribution of lamin C in X-linked Emery-Dreifuss muscular dystrophy fibroblasts. J. Struct. Biol. 2002;140:241–253. doi: 10.1016/s1047-8477(02)00573-7. [DOI] [PubMed] [Google Scholar]
- 30.Bloom S, Lockard VG, Bloom M. Intermediate filament-mediated stretch-induced changes in chromatin: a hypothesis for growth initiation in cardiac myocytes. J. Mol. Cell. Cardiol. 1996;28:2123–2127. doi: 10.1006/jmcc.1996.0204. [DOI] [PubMed] [Google Scholar]
- 31.Maniotis AJ, Chen CS, Ingber DE. Demonstration of mechanical connections between integrins, cytoskeletal filaments, and nucleoplasm that stabilize nuclear structure. Proc. Natl. Acad. Sci. U. S. A. 1997;94:849–854. doi: 10.1073/pnas.94.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moir RD, Yoon M, Khuon S, Goldman RD. Nuclear lamins A and B1: different pathways of assembly during nuclear envelope formation in living cells. J. Cell Biol. 2000;151:1155–1168. doi: 10.1083/jcb.151.6.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dreuillet C, Tillit J, Kress M, Ernoult-Lange M. In vivo and in vitro interaction between human transcription factor MOK2 and nuclear lamin A/C. Nucleic Acids Res. 2002;30:4634–4642. doi: 10.1093/nar/gkf587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lloyd DJ, Trembath RC, Shackleton S. A novel interaction between lamin A and SREBP1: implications for partial lipodystrophy and other laminopathies. Hum. Mol. Genet. 2002;11:769–777. doi: 10.1093/hmg/11.7.769. [DOI] [PubMed] [Google Scholar]
- 35.Kumaran RI, Muralikrishna B, Parnaik VK. Lamin A/C speckles mediate spatial organization of splicing factor compartments and RNA polymerase II transcription. J. Cell Biol. 2002;159:783–793. doi: 10.1083/jcb.200204149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Spann TP, Goldman AE, Wang C, Huang S, Goldman RD. Alteration of nuclear lamin organization inhibits RNA polymerase II-dependent transcription. J. Cell Biol. 2002;156:603–608. doi: 10.1083/jcb.200112047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rao L, Perez D, White E. Lamin proteolysis facilitates nuclear events during apoptosis. J. Cell Biol. 1996;135:1441–1455. doi: 10.1083/jcb.135.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kumar A, Boriek AM. Mechanical stress activates the nuclear factor-kappaB pathway in skeletal muscle fibers: a possible role in Duchenne muscular dystrophy. FASEB J. 2003;17:386–396. doi: 10.1096/fj.02-0542com. [DOI] [PubMed] [Google Scholar]
- 39.Östlund C, Bonne G, Schwartz K, Worman HJ. Properties of lamin A mutants found in Emery-Dreifuss muscular dystrophy, cardiomyopathy and Dunnigan-type partial lipodystrophy. J. Cell Sci. 2001;114:4435–4445. doi: 10.1242/jcs.114.24.4435. [DOI] [PubMed] [Google Scholar]