Abstract
About one-third of the world’s rice area is in rain-fed lowlands and most are prone to water shortage. The identification of genes imparting tolerance to drought in the model cereal plant, rice, is an attractive strategy to engineer improved drought tolerance not only rice but other cereals as well. It is demonstrated that RNAi-mediated disruption of a rice farnesyltransferase/squalene synthase (SQS) by maize squalene synthase improves drought tolerance at both the vegetative and reproductive stages. Twenty-day-old seedlings of wild type (Nipponbare) and seven independent events of transgenic RNAi lines showed no difference in morphology. When subjected to water stress for a period of 32 d under growth chamber conditions, transgenic positives showed delayed wilting, conserved more soil water, and improved recovery. When five independent events along with wild-type plants were subjected to drought at the reproductive stage under greenhouse conditions, the transgenic plants lost water more slowly compared with the wild type, through reduced stomatal conductance and the retention of high leaf relative water content (RWC). After 28 d of slow progressive soil drying, transgenic plants recovered better and flowered earlier than wild-type plants. The yield of water-stressed transgenic positive plants ranged from 14–39% higher than wild-type plants. When grown in plates with Yoshida’s nutrient solution with 1.2% agar, transgenic positives from three independent events showed increased root length and an enhanced number of lateral roots. The RNAi-mediated inactivation produced reduced stomatal conductance and subsequent drought tolerance.
Keywords: ABA, filled grain weight, maize, relative water content, rice, RNAi, root number stomatal conductance, squalene synthase, water deficit
Introduction
Rice, wheat, and corn, the three major food crops of the world, have essentially the same set of genes with a large difference in genome size. Rice (Oryza sativa L.), with a genome size of ∼390 Mb (Lin et al., 2008) is a model genome, an experimental model plant for understanding the biology of all cereals and, above all, it is the most important crop for human consumption, providing staple food for more than half of the world’s population (Praba et al., 2009). Global water deficit is a more common scenario for cultivated rice which is one of the world’s most important foods, a labour-intensive crop that also requires plenty of water. It has been reported that, the global reduction in rice production due to drought averages 18 million tonnes annually (O’Toole, 2004). Even for deep water rice, accounting for 11% of the world’s rice area, and upland rice which is grown under aerobic conditions and rain-fed fields without standing water and accounts for 13% of the world’s rice area, the productivity is threatened by the scarcity of water at the seedling stage before the floods (Evenson et al., 1996) and a lack of standing water, respectively (Wang et al., 2009a).
The impact of drought in rice is influenced by cultivar, stage of drought imposition, and duration of stress. Rice’s susceptibility to water stress is more pronounced at the reproductive stage and causes the greatest reduction in grain yield when stress coincides with the irreversible reproductive processes (Cruz and O’Toole, 1984). Drought tolerance has been considered as an important breeding target to partially compensate for the loss in yield. Selecting rice for drought tolerance based on unknown genetic mechanisms will result in inefficient genetic improvement. Serraj et al. (2009) suggested that development of rice cultivars by combining improved drought resistance with yield potential under favourable conditions would be the strategy to increase rice productivity in drought-prone areas.
Molecular tools facilitate the identification and genomic locations of genes controlling traits related to drought tolerance using quantitative trait loci (QTL) analysis (Lanceras et al., 2004). Transcriptome analyses using microarrays have been used successfully to explore the genes associated with stress responses in rice (Kathiresan et al., 2006; Degenkolbe et al., 2009). The nearly complete sequence information of the rice genome, the structural/functional annotation of most of their genes (Kikuchi et al., 2003), the availability of ∼51K transcripts representing two rice cultivars (japonica and indica) (www.affymetrix.com) (Hazen et al., 2005), and the availability of various mutant resources in rice (Krishnan et al., 2009) offers a unique opportunity to explore and dissect the regulatory networks involved in the complex trait of drought tolerance.
Plant farnesyltransferases identified to date consist of a heterodimer of an ‘a and b’ subunit, each of which belongs to a single-gene family. In Arabidopsis, the a-subunit (AtFTA) was reported to be involved in meristem growth and development (Running et al., 2004). Previous genetic studies have implicated the involvement of the b-subunit of Arabidopsis farnesyltransferase (ERA1) in the regulation of ABA sensing and drought tolerance (Cutler et al., 1996). (Wang et al. 2005) 2009b) reported that down-regulation of either AtFTA or AtFTB resulted in both increased ABA sensitivity and improved drought stress tolerance in Arabidopsis and canola. Squalene synthase (SQS) is one of the farnesyl-diphosphate farnesyltransferase that catalyses the first reaction of the branch of the isoprenoid metabolic pathway committed specifically to sterol biosynthesis (Tansley and Shechter, 2001). Regulation of SQS is thought to direct proximal intermediates in the pathway into either sterol or non-sterol branches in response to changing cellular requirements (Wentzinger et al., 2002). SQS activity has been localized to the smooth endoplasmic reticulum in animals (Stamellos et al., 1993) and in the shoot apical meristem in plants (Devarenne et al., 2002). Early work with plant cell suspension cultures has already emphasized its involvement in plant defence reactions (Vogeli and Chappell, 1988; Haudenschild and Hartmann, 1995). The role of squalene synthases in drought tolerance has not been investigated before.
Disrupting the gene function by modulating RNA processing provides excellent tools to uncover novel molecular mechanisms for the regulation of RNA metabolism and of signal transduction networks in plants (Wang et al., 2005). It is demonstrated that RNAi-mediated disruption of rice farnesyltransferase/squalene synthase by maize squalene synthase (ZMSQS: NM_001111369.1) improves drought tolerance at both the vegetative and the reproductive stages. Upon recovery, the transgenic plants also recorded higher grain yield. Correlating genetic information with physiological and morphological traits related to drought tolerance explained the basis of drought resistance in these transgenic plants.
Materials and methods
Rice farnesyltransferase RNAi construction and transformation
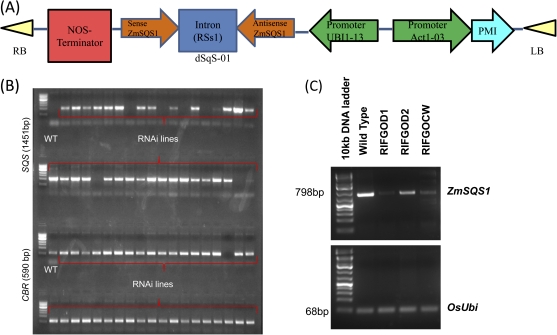
Transgenic rice plants with double-stranded squalene synthase RNAi was developed by Agrobacterium-mediated transformation. A 392 bp DNA fragment encoding partial-length of maize SQS (ZmSQS1) was then placed directly downstream of the maize Ubi1 promoter in the sense and antisense orientations spaced by an intron (rice sucrose synthase 1 (RSs1)) gene (Fig. 1A). The PMI (phospho mannose isomerase) marker gene was placed under the control of the Act1 promoter from rice. Nine independent transgenic events (RIFGOD0, RIFGOD1, RIFGOD2, RIFGOD3, RIFGOCU, RIFGOCW, RIFGOCX, RIFGOCY, and RIFGOCZ) were generated and advanced to the T1 generation.
Fig. 1.
A. Construct containing PMI marker gene under the control of Act1 promoter and SQS gene with maize UBI1-13 promoter. (LB: left border; RB: right border)B. PCR amplification of SQS in transgenic RNAi lines (lane 1 and 2) with WT in well 1. Lane 3 and 4 represent the PCR amplification of genomic DNA for internal control gene CBR (cytochrome b5 reductase) C. RT-PCR analysis of abundance of endogenous SQS transcript in WT and three independent RNAi lines. Equal amounts of RNA was extracted from WT and RNAi lines and were used to synthesize 1st strand cDNA. 3μl of this cDNA was amplified using gene-specific primers (see materials and methods for details) for SQS D. RT-PCR analysis of rice ubiquitin5 gene with 3μl template was used as the housekeeping control.
PCR analysis
The transgenic positives were identified from seven independent lines for vegetative stress experiments and from five independent lines for reproductive stage experiments. The presence of a gene of interest (promoter+gene+terminator) was tested by polymerase chain reaction (PCR) and those plants that amplified both the gene of interest and the internal control gene were considered to be transgenic positives and those that only amplified the internal control gene were considered to be transgenic negatives. Briefly, for each independent event, 30 plants were grown for 15 d and leaf samples were collected. Genomic DNA was isolated by a modified CTAB method (Saghai-Maroof et al., 1984) and quantification was done with a nanodrop model ND1000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). Primer was designed between the promoter–gene–terminator amplification using TGATAATCATCGCAAGACC as the forward primer and TGTTTCTTTTGTCGATGCTCAC as the reverse primer for the SQS gene. 50 ng genomic DNA was used to amplify the genomic DNA using TaKaRa ExTaq Hotstart Mastermix (Takara Bio Inc, Otsu, Shiga, Japan) along with the respective primers. The PCR conditions were denaturation at 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 90 s, and then extension at 72 °C for 5 min. The internal control gene (cytochrome b5 reductase; CBR) was also amplified using the forward primer TCATCTTTGGTTCATAGCTCATCCTG and the reverse sequence TCACCAACTTTCATCTCATGAAAATG and tested at the same time to identify transgenic negatives (Fig. 1B). The conditions of PCR are same for CBR except that the annealing temperature was 52 °C. The amplified PCR products were separated by electrophoresis with 1% agarose gel, visualized by ethidium bromide, and photographed by using an Alpha Imager Gel Documentation System (Alpha Innotech, San Leanardo, CA).
Analysis of SQS gene expression by RT-PCR
Total RNA from 15-d-old seedlings of the wild type and three independent transgenic lines (RIFGOD1, RIFGOD2, and RIFGOCW) was extracted by using the RNEasy Plant minikit (Qiagen Inc,Valencia, CA, USA). The total RNA content was measured with a nanodrop model ND1000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). The total RNA was then digested in 50 μl of turboDNA-free DNase kit (Ambion, Austin, TX) according to the manufacturer’s instructions to remove genomic DNA. RNA was quantified by nanodrop again. cDNA synthesis from 500 ng of DNAse-treated total RNA was performed using Superscript III first-strand cDNA synthesis system (Invitrogen) in a reaction volume of 20 μl according to the manufacturer’s protocol. Gene-specific primers were designed using Primer Express 2.0 software with 5′-AAGGTTATCAAGAGGCAATTGAAG-3′ as the forward primer and 5′-ACAATAGCCACCAACAGAAGTACA-3′ as the reverse primer. The rice ubiquitin gene (OSUBQ5: Gene bank ID: AK061988) was selected as a housekeeping control. The cDNA was amplified with TaKaRa Hotstart primer mix for 28 cycles and the amplified products were separated on a 1.8% agarose gel, visualized by ethidium bromide staining, and photographed by using an Alpha Imager.
ABA sensitivity assays
Germination and seedling growth in Petri plates:
For the ABA sensitivity test of the transgenic rice at germination, around 30 seeds per event for three T2 generation independent events and Nipponbare WT plants were surface-sterilized in 50% (v/v) chlorac for 1 min and then washed thoroughly with distilled water to remove residue. Seeds were placed in each Petri dish and 20 ml of autoclaved water containing 0, 3, 5, 7, and 10 μmol ABA was applied. Germination percentage was calculated on 3, 5, and 7 d after sowing. Germination was based on the protrusion of the radicle. On the seventh day, plates were opened and measurement of root length, shoot length, and root number was done. To confirm that the reaction to ABA is not related to an inherent ABA content in seeds, imbibed seeds in water for 24 h were supplied with 3 μmol ABA and measurements were taken 7 d and 14 d after germination.
Germination and seedling growth in ABA-infused agar plates:
To test the growth of seedlings in response to ABA, seeds were germinated on plexi glass plates containing 1.2% agar supplemented with Yoshida’s nutrient solution (Yoshida et al., 1976) infused with different concentrations of ABA (0 and 5 μM). 14 d after growing with the temperature at 26 °C, plant height and root length were measured, and the number of lateral roots was counted for each seedling.
Drought stress assays
Vegetative stage drought:
The T1 seeds of seven independent events with 12 replications per event along with the wild type Nipponbare with 14 replications were sown in Ray-Leach Cone-tainers (SC-10, from Stuwe & Sons Inc, Oregon, USA). The experiment was conducted in an environmentally controlled walk-in growth chamber with a temperature of 29/21 °C day/night, a 10 h day length, 65% RH, and a light intensity of 375 μmol s−1. Pre-weighed cones were filled with oven-dried potting mix. The cones were arranged in a tray so that each tray holds 98 cones. The weight of each cone with dry potting mix was determined prior to sowing. Leaf samples (2 cm) were taken from 10 d-old plants and DNA was extracted. They were checked for the presence of the gene of interest by PCR and marked as positive or negative. At 20 d after sowing, the cones were saturated with water and the weight of individual cones along with plants was measured. Water was withheld from 21 d after sowing until the wild-type plants showed complete wilting or reached a leaf rolling score of 5. The trays were rotated every day to avoid any position effects. Measurement of plant height and leaf rolling score (scale of 0–5 as per the Standard Evaluation system of IRRI) was performed every week after withholding water. Individual cone weight was taken every day to assess the water loss. At the end of the 32 d drought period, plants were watered and recovery was recorded 2 d later.
Reproductive stage drought:
The experiment was conducted in a greenhouse between September 2007 and April 2008 at the University of Missouri, Columbia, MO (33°98′ N, 88°38′ W). Greenhouse air temperatures were maintained at 29 °C during the day and 21 °C during the night with a thermostatically controlled combination of fan–evaporative pad cooling and electric heating. Photoperiod was 12 h with overhead 400 W metal halide lamps that produced a supplementary photosynthetic photon flux density (PPFD) of approximately 1620 μmol m−2 s−1 at the tops of plants. Thirty plants per independent event were established from seeds in 18 l pots and leaf material was collected at 10 d after sowing to identify positives for the presence of the gene of interest by PMI strip and by PCR. Twenty days after sowing, the transgenic positive plants (six replications), negative plants (three replications) and the non-transgenic (WT, Nipponbare, eight replications) lines with two treatments [well-watered (WW) and water-stressed (WS)] were transplanted to large volume (10 l capacity) polyvinyl pots (21 cm diameter at the top, 14 cm diameter at the bottom, and 43.5 cm height) each filled with 2 kg of air-dried commercial potting mixture containing peat moss, perlite, bark, and vermiculite (Promix from PremierTech Horticultural Company, Canada). The pots were arranged in a completely randomized design. This growing condition was earlier found to be suitable for a slow stress development cycle in rice (Babu et al., 2004). Plants were watered daily up to 90 d after sowing (DAS) and supplemented with Yoshida’s nutrient solution (Yoshida et al., 1976) at 30 d intervals. Drought stress was imposed at the reproductive stage by withholding water to the WS treatment pots determined by the distance between the flag leaf and the penultimate leaf on the primary shoot. Primary tillers were marked on each plant and the progress of reproductive development was monitored in terms of the interauricle length when the interauricular distance between the flag leaf and the penultimate leaf is zero (d=0) ∼10 d before heading; Praba et al., 2009). Stage d=0 coincides with the pollen meiosis stage. Water was withheld until WT pots reached 30% field capacity (FC) or leaf wilting was observed, and then reapplied. Plants in both transgenic and WT lines were maintained under non-stress condition with regular irrigation for WW treatment. Development of stress in the treatment plants was routinely monitored by visual symptoms of leaf rolling and leaf drying and by measurements of daily soil moisture content and plant water status indicator, leaf relative water content (RWC) at pre-determined intervals.
Soil and plant water status, stomatal conductance, and cell membrane stability determinations
The volumetric soil water status was monitored daily by using the profile probe PR2 from Delta-T Devices, Cambridge, UK. Briefly, access tubes were inserted at the time of filling into the pots designated for the water-stress treatments. The PR2 profile probe was inserted into the access tubes and daily soil moisture content (% v/v) was recorded at two places in each pot and the average was taken. Similarly, the abaxial stomatal conductance from the top-most second fully expanded leaf from the primary tiller was recorded using an SC-1 porometer (Decagon Devices, Pullman, WA, USA) at 3 d intervals starting from day three after withholding water until the 23rd day of stress, after which no difference was noticed between WT and transgenics for stomatal conductance.
Non-stress measurements of RWC were made on the day after the last irrigation and then at 7 d intervals until the plants wilted under the stress treatment. When the complete rolling of the youngest leaves and desiccation of older leaves was observed, it was considered as wilting. On each sampling date, leaves were sampled in the morning between 10.00 h and 11.00 h and RWC was determined using the midsection of the second-youngest fully expanded leaf blade using the standard method and until 28 d after stress (Babu et al., 2004).
Yield and yield components
The plant traits and yield components of rice namely, panicle length (cm), number of spikelets per panicle, spikelet sterility (%), grain yield per plant (g), and plant height at maturity (cm) were measured as described by Yoshida et al. (1976).
Statistical analysis
Data were analysed using SAS (version 9.2, by SAS Institute, Inc., Cary, NC, USA). The Proc GLM procedure was used to estimate the differences between genotypes. Data were analysed by a three factor ANOVA model (event, plant type, and treatment). Means were tested for significant differences based on least significant difference (LSD). The differences between events and plant type across different treatments were estimated by using the Tukey test. To analyse the relationship between stomatal conductance and soil water content in the stress treatments, trend surface analysis, as described by Davis (1986), was used. The order of polynomial expressions for stomatal conductance, plotted as a function of volumetric soil water content was two for all the lines. A scatter plot was used for the well-watered treatments as no regression was observed between conductance and saturated soil water content above 30% v/v.
Results
Phylogenetic analysis of rice SQS
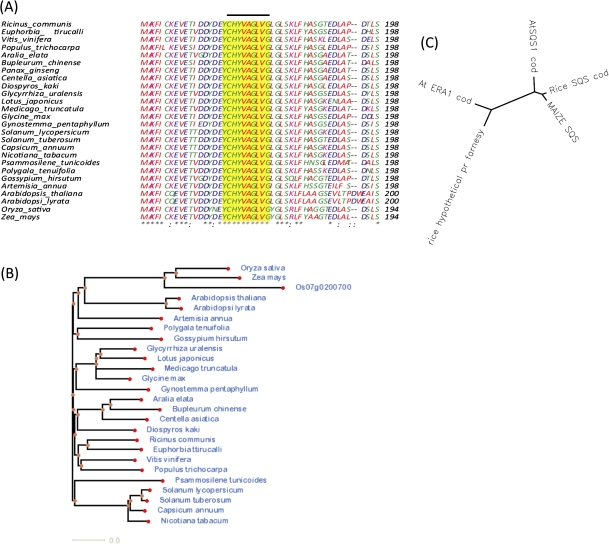
Squalene synthase (SQS) catalyses the condensation of two molecules of farnesyl diphosphate (FPP) to produce squalene (SQ), the first committed precursor for sterol, brassinosteroid, and triterpene biosynthesis. The rice squalene synthase gene SQS (Os03g0805100) is conserved in human, chimpanzee, dog, cow, mouse, rat, chicken, zebrafish, S. pombe, S. cerevisiae, K. lactis, E. gossypii, M. grisea, N. crassa, and A. thaliana. In general, most of the plant species have a single squalene synthase gene, due to a single copy of the gene in both yeast and human (Hata et al., 1997). The sequence similarity search using BLASTP revealed that rice SQS showed 87% sequence similarity with maize SQS, 74% with soybean SQS, 72% with Medicago truncatulata, and 70% with Arabidopsis SQS1. There appears to be another squalene synthase in rice (Os07g0200700) which has 78% sequence similarity with Os03g0805100. Other species like Glycyrrhiza glabra (lico rice) and Arabidopsis thaliana also have two genes for squalene synthase. It has been reported that the SQS2 of Arabidopsis has no SQS activity and SQS1 is the only functional form in Arabidopsis (Busquets et al., 2008). Multiple sequence alignments of divergent protein sequences of SQS were done by using ClustalW.2 and through the next generation biology workbench (http://www.ngbw.org). The most highly conserved regions of proteins in the sequences were searched by using BLIMPS tool (Henikoff et al., 2000), and it showed that the squalene synthase motif was highly conserved among the species (Fig. 2A). Phylogenetic analysis based around the Neighbor–Joining method of Saitou and Nei (1987) was constructed using Clustal W tree (Larkin et al., 2007). The tree results revealed that SQS genes of monocots (rice, maize) are more closely related. It also indicated that the SQS genes from monocots and dicots form distinct groups, and among dicots the clustering of families is seen (Fig. 2B). In addition, the analysis also revealed that the farnesyl transferase in Arabidopsis (ERA1) and a putative farnesyl transferase in rice are completely divergent from rice squalene synthase (Fig. 2C).
Fig. 2.
A. Multiple sequence alignment of SQS genes in crop species producing significant alignments with rice SQS using clustalwP.Due to space restriction only the sequences with SQS motif is presented. The line indicate SQS motif B. Phylogenetic analysis of SQS gene done with next generation biology workbench presented in Newick (Phylip) Tree formatC. Phylogenetic relationship between rice, arabidopsisfarnesyltransferase (ERA1) and rice, arabidopsis and maize squalene synthase.
Transgenic plant identification
The T1 transgenic seeds from seven independent positive plants and five independent plants were used in the drought characterization screening at the vegetative and reproductive stages, respectively. PCR amplification of DNA from 15 d-old plants with promoter+gene of interest+terminator combination resulted in amplification only in transgenic positive plants (Fig. 1B, lanes 1 and 2), while no amplification of the gene of interest was observed in wild-type plants (well no.1, lane 1). The transgenic plants with no amplification of SQS and with amplification of internal control gene (CBR) (Fig. 1B, lanes 3 and 4) were marked as transgenic negatives and used for phenotypic analysis.
Expression patterns of SQS in transgenic plants by RT-PCR analysis
The abundance of rice SQS in wild-type and transgenic plants was analysed by RT-PCR in three independent events (Fig. 1C). A distinct reduction in the expression of SQS in the RNAi lines compared with the control was noticed. The reduced expression of the SQS gene did not result in any alteration in plant morphology except for two plants in the event RIFGOCW.
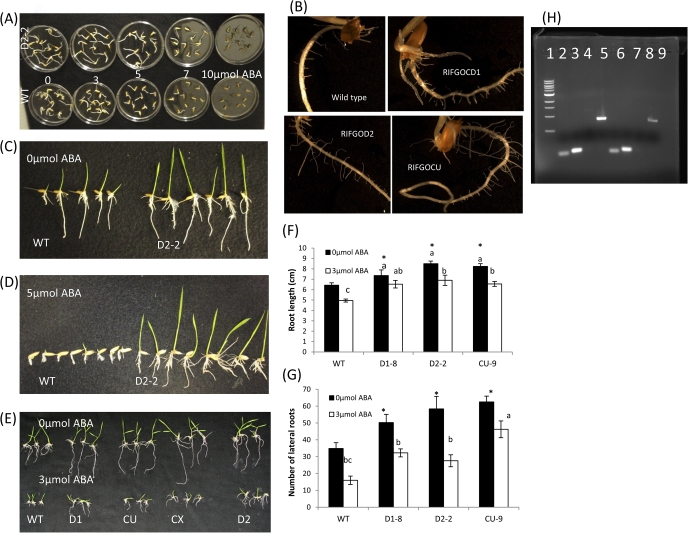
Seedling growth in agar plates and sensitivity to ABA
The T2 seeds of SQS RNAi transgenics were subjected to ABA sensitivity assay. The positive transgenic plants showed less sensitivity to ABA at 0–5 μmol concentration (Fig. 3A) and showed less reduction in shoot and root growth at 3 d and 7 d after germination than WT plants in 0 μmol ABA (Fig. 3B). When grown in Yoshida solution agar plates infused with 0 and 5 μmol ABA, there was a significant difference between the wild type and transgenics for root morphology (Fig. 3C, D; see Supplementary Fig. S1A, B at JXB online). To discount the effect of ABA on germination, in another assay, pre-soaked and germinating seeds were transferred to 3 μmol ABA and similar differences in growth was detected at 14 d after sowing (Fig. 3E, F, G). A comparable distinction in growth response was also found when plants were grown on agar plates infused with 10% polyethylene glycol (see Supplementary Fig. S1C, D at JXB online). When RNA was extracted from the roots of WT and one independent transgenic event (RIFGOD2) that showed better root growth than wild-type, a reduced expression of SQS in transgenic plant roots was observed (Fig. 3H).
Fig. 3.
Seedling development of SQSRNAi lines under ABA treatment.A. Event D2 (top) and WT grown in different concentrations of ABA (0, 3, 5,7, 10 μmol ABA) B. Lateral root growth in 3 events three days after sowing (DAS) C&D.WT and transgenic line D2-2 grown in agar plates infused with 0 and 5 μmol ABA 7DAS E, F, G. growth, root length and lateral root number of 3 events in 0 and 3μmol ABA H. Reduced SQS gene expression on root tissues of transgenic event D2 in 0 μmol ABA Lanes 2,3,6,7: internal controls Act, ElFα, for WT and D2 Lanes 5 and 9 SQS in WT and D2 4 and 8 no DNA* significantly different (p<0.05 level) from WT and bars followed by different letters differ significantly at p<0.05 level.
Plant survival and recovery over vegetative stage drought
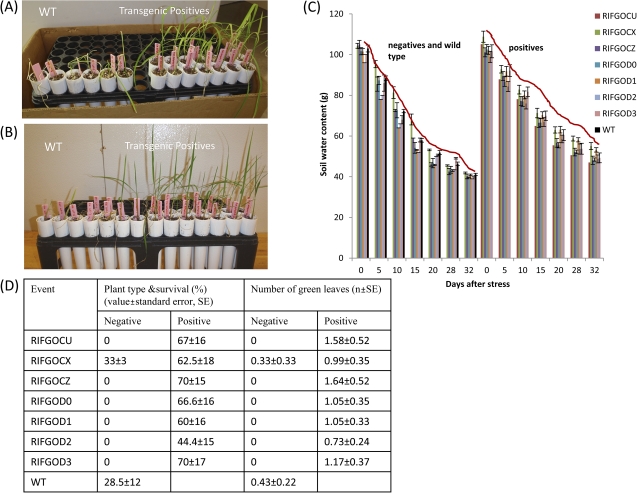
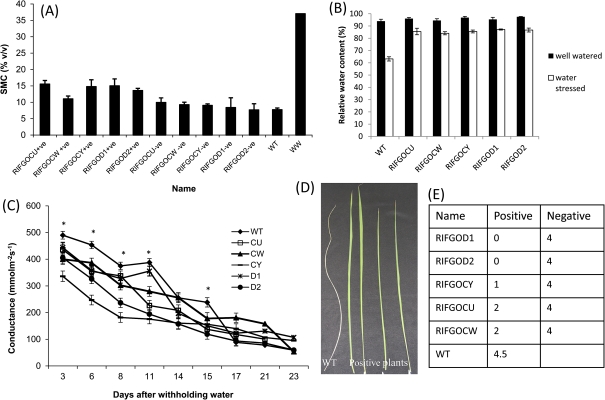
When subjected to seedling stage drought at 15 d after sowing, the transgenic plants showed delayed wilting (Fig. 4A) and continued to maintain open leaves until 20 d after drought. The plants from seven independent events exhibited a remarkably slow reduction in water loss compared with wild-type plants and maintained more water content (Fig. 4B). Thirty-two days after drought in a growth chamber, transgenic plants also recovered better than wild-type plants (Fig. 4C). At 32 d after water withdrawal, most of the WT plants died or showed severe leaf drying, whereas the transgenic plants exhibited slightly rolling symptoms. Two days after re-watering, only 28% of WT plants recovered, but transgenic positives showed >50% recovery. In addition, none of the negative plants survived except for one event, and the transgenic positive plants retained 3-fold greener leaves than the WT and negatives (Fig. 4D).
Fig. 4.
Improved drought resistance of SQS RNAi rice at vegetative stage.A. Transgenic plants showing less drying at 15 days after drought. B. Recovery after 32 days drought and 2 days after rewatering C. Reduced water loss of transgenic plants depicted by weight of soil+plant D. Survival rate and number of green leaves retained by transgenics in comparison with WT and negative controls.
Stress tolerance and yield improvement at reproductive stage drought
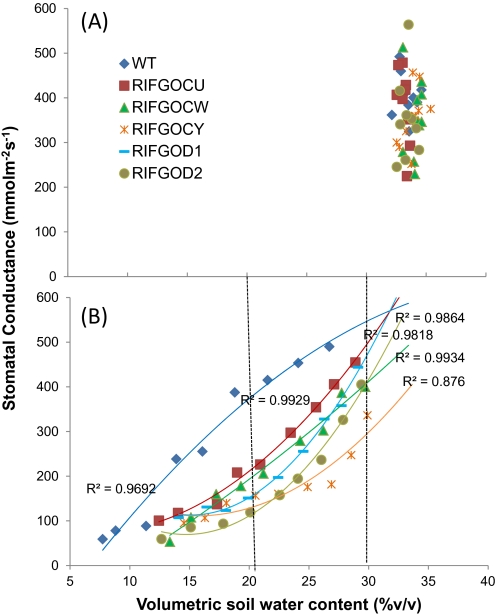
Phenotypic evaluation of five independent transgenic positive events along with negative and wild-type plants was conducted in an environmentally controlled greenhouse. Except for two positive plants of event CW, other plants showed similar growth and tiller production as WT plants. When drought was imposed at the reproductive stage (d=0), transgenic plants lost less water than WT plants which was evident from the volumetric soil water content. On the last day of the 28 d slow progressive drought, the transgenic positive plants maintained a higher soil water content (Fig. 5A). At 28 d after drought the transgenic plants also maintained greater flag leaf relative water content (∼80%) than wild-type plants (60%) as depicted in Fig. 5B. This was associated with reduced stomatal conductance of the stressed plants from the beginning of drought itself (Fig. 5C). The wild-type plants had significantly greater stomatal conductance (P <0.05) than transgenics on 3, 6, 8, 11, and 15 d after withholding water and showed a much steeper reduction at 15 d after drought. The transgenic plants had lower stomatal conductance and maintained a progressive reduction until 23 d after drought. There was no difference in conductance between WT and transgenics after 23 d (Fig. 5C). In addition, the flag leaves of wild-type plants dried and died much faster but transgenic plants showed a rolled flag leaf with a lower leaf rolling score and recovered after re-watering (Fig. 5D, E). The trend surface analysis of stomatal conductance as a function of soil water content noticeably differentiated the stomatal behaviour under water-stress conditions. There was no distinct difference in stomatal conductance between WT and transgenic plants in the fully saturated well-watered treatment (Fig. 6A). As well-watered plants were maintained at >30% volumetric soil moisture, the conductance was ranging from 245–657 mmol m−2 s−1 at different time points and there was no correlation between water content and stomatal conductance. When subjected to water stress, within a 10% reduction in soil moisture content (30–20%), the transgenic events exhibited a remarkable decrease in stomatal conductance (Fig. 6B). Interestingly, a parabolic shape response curve was observed in water-stressed wild-type plants, which lost soil moisture quickly with greater stomatal conductance initially and later showed reduced soil water content and lower stomatal conductance than WT plants.
Fig. 5.
Soil and plant water-relation parameters during 28 day progressive drought of WT, negative and positive SQS RNAi lines. A. Soil moisture content B. Flag leaf RWC C. stomatal conductance (WS treatment) D. leaf drying and E. leaf rolling scores *Significantly higher conductance of WT than transgenics (p<0.01); LSD 5%-75.97
Fig. 6.
Stomatal conductance as a function of soil water content in wild-type and transgenic lines during a 23 day stress period. A) Well-watered B) Water-stressed
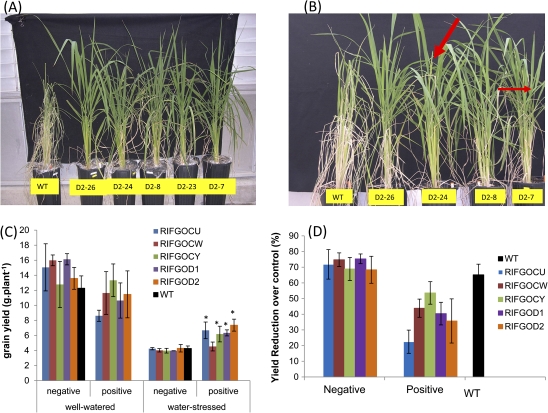
Wild-type plants reached around 10% volumetric SMC at 23 d after drought and showed complete wilting (Fig. 7A), at which point transgenic plants exhibited no wilting symptoms. When re-watered at 29 d after progressive drying, 5 d after re-watering, transgenic plants showed 90% recovery and even started flowering (Fig. 7B), whereas wild-type plants recovered only 50% biomass and showed a long delay in flowering (data not shown).
Fig. 7.
Response of RNAi lines to reproductive stage drought. A. morphology at 23 days after withholding water B. Recovery 5 days after rewatering (arrows indicate flowering) C. grain yield (filled grain weight) per plant of different treatments/events D. Percentage reduction of grain yield over control (well-watered) * indicates significantly higher yield than WT under WS at p<0.05 level.
At maturity, there was a significant difference (P <0.05 level) between the transgenic RNAi lines and WT for filled grain weight. The filled grain weight of wild-type plants was 12 g plant−1 under the well-watered treatment and was 4.2 g plant−1 under the water-stressed treatment which was only 30% of the non-stressed yield (Fig. 7C). There was no significant difference between the wild type, negatives, and positive plants for yield except for events CU and CW. Under water-stressed conditions independent transgenic events recorded a higher filled grain weight than WT plants. No significant difference in yield between wild-type and vector control plants was observed under the water-stress treatment. This is further supported by the lower percentage reduction in yield of stressed transgenic lines over the control (Fig. 7D), whereas negatives and the wild type showed a higher yield reduction from the well-watered counterparts. The other components of yield namely plant height, panicle length, and spikelet sterility are presented in Table 1. There was significant difference between transgenic positives and WT for spikelet sterility which ranged from 29–37% in positives in the drought treatment, whereas the spikelet sterility of vector control plants and WT were 46–59% and 52%, respectively. No significant difference between the wild type, negatives, and positives was noticed for plant height. There was difference between treatments for panicle length.
Table 1.
Yield components of WT, negative control and positive transgenics (mean ± SE) under normal growth and drought conditions.
| Event/Treatment | Plant Height |
Panicle length |
Spikelet sterility (%) |
|||
| Well watered (WW) | Water stressed (WS) | Well watered | Water stressed | Well watered | Water stressed | |
| WT | 117 ± 3.4 | 108 ± 3.7 | 20.3 ± 1.1b | 17.2 ±0.3c | 14.3 ± 2.4 | 52.1 ± 2.5d,e |
| Negatives | ||||||
| RIFGOCU | 110.3 ±3.8 | 113.3 ± 4.3 | 21.5 ± 1.7a | 20.5 ± 0.3a,b | 17.6 ±3.5 | 46.1 ± 1.5c |
| RIFGOCW | 111.6 ± 3.2 | 118 ± 2.8 | 20.5 ± 0b | 19.4 ± 2.1b,c | 16.5 ± 4.3 | 45.4 ± 1.1c |
| RIFGOCY | 106.6 ± 4.2 | 107.3 ±3.3 | 28.6 ±0a | 22 ± 0b | 10.7 ±0.5a | 59.3 ± 0f |
| RIFGOD1 | 106 ± 2.1 | 113.6 ±12.8 | 24.5 ± 0a | 18.7 ±0.7b,c | 12.4 ±0.6 | 50.2 ± 2.6d,e |
| RIFGOD2 | 117.3 ±0.6 | 117 ±3.7 | 23.7 ± 0a | 19.7 ± 1.1b,c | 12.9 ±0.9 | 53.5 ± 3.1d,e |
| Positives | ||||||
| RIFGOCU | 114.4 ±4.9 | 108.7 ± 3.1 | 22 ± 1.5a | 18.8 ±1.5b | 15.7 ±2.8 | 31.9 ± 3.4a |
| RIFGOCW | 114.8 ± 2.3 | 113.8 ±4.3 | 24.9 ± 1.1a | 19.5 ±1.06c | 16.2 ± 3.4 | 37.9 ± 3.3b |
| RIFGOCY | 101 ± 16.5 | 111.8 ±2.8 | 26.3 ± 0.5a | 18.5 ± 0.5b,c | 10.4 ± 2.1a | 35.3 ± 3.2b |
| RIFGOD1 | 100.5 ±12.1 | 110 ± 8.3 | 21.4 ±0.9b | 21.82 ± 0.9b | 12.8 ±1.2 | 32.9 ± 3.5a |
| RIFGOD2 | 107.8 ±8 | 116.2 ± 6.8 | 24.8 ±1b | 20.9 ±1.0b,c | 14.4 ± 4.7 | 29.1 ± 2.7a |
| LSD0.05 | 5.50 | 1.602 | 3.368 | |||
| Event | ns | ns | <0.01 | |||
| Type | ns | ns | <0.0001 | |||
| (positive/negative) | ||||||
| Treatment | ns | <0.0001 | <0.0001 | |||
| (trt WW/WS) | ||||||
| Event×trt | ns | ns | <0.001 | |||
| Type×trt | ns | ns | <0.001 | |||
| Event×type×trt | ns | ns | ns | |||
Discussion
The maize squalene synthase is the smallest among the enzymes of various species, from yeast to man (Hata et al., 1997). Protein sequence analysis revealed that rice SQS has two amino acids (401 aa) less than maize (403 aa) with a large number of conserved blocks. The coding sequence size of rice SQS is 1882 bp and that of maize SQS is 1206 bp. The RNAi was created by the insertion of a 392 bp nucleotide of maize SQS. The novelty of the result in this study is that disruption of gene function in one monocot (rice) by another monocot gene (maize) did not result in any adverse effects on plant architecture, growth, and development. Hence, this concept would be a convenient tool for studying gene function across the species.
Sensitivity to ABA
In Arabidopsis, knockout mutations in ERA1 (Enhanced Response to ABA 1, At5g40280), a farnesyl transferase gene caused an enhanced response to ABA in seeds (Wang et al., 2005) suggesting that at least one farnesylated protein functions as a negative regulator of ABA signalling. By contrast, in the present study, it was found that RNAi-mediated silencing of farnesyl transferase in rice did not show any ABA sensitivity. The germination and growth of Petri-plate-grown and agar-plate-grown transgenics showed better germination and root growth with different concentrations of ABA. It was reported that over-expression of the GGB (geranylgeranyl transferase beta, At2g39550) gene partially suppresses the era1 phenotype (Johnson et al., 2005). Knockout mutants in the GGB gene did not cause developmental phenotypes but, like era1 mutants, ggb mutants exhibited an enhanced response to ABA in stomata, which was also found in our study. In seeds, however, ggb plants did not show an enhanced response to ABA, similar to our results. Compared with wild-type plants, the transgenic plants even showed enhanced lateral root number and root length in 0 μM plates. Brady et al. (2003) found that era1 mutants exhibited increased lateral root number, possibly because ERA1 attenuates the expression of ABI3, a positive regulator of ABA and auxin signalling. In addition, concomitant with our study, ggb mutants also exhibited an enhanced response to auxin-induced lateral root formation without exhibiting the auxin-induced inhibition of primary root growth (Johnson et al., 2005). These results establish that geranylgeranylated proteins have a role in the negative regulation of tissue-specific ABA and auxin responses (Crowell and Huizinga, 2009). The contribution of roots to drought tolerance was not explained in this study. However, increased rooting depth and number might be important in the field where larger volumes of soil could be accessed by larger root systems. Studies should be conducted to confirm the impact of the enhanced root growth in relation to drought tolerance which could not be explored in this paper.
Water relations traits
The unrestricted gas exchange through fully open stomates would accelerate water loss during drought and therefore increase photorespiration (Araus et al., 2008). One basic mechanism for reducing the impact of drought is early stomatal closure at the beginning of a period of water deficit. Under water-deficit conditions, ‘conservative’ plants grow more slowly and have a lower stomatal conductance. These plants perform better as they achieve a larger biomass accumulation per gram of water transpired (Araus et al., 2008). In the current study, the transgenic RNAi lines exhibited a low stomatal conductance from the beginning of stress itself, which explained their water conservation through restricted gas exchange which prevented excessive water loss in early drought (Fig. 6B). A negligible variation in stomatal conductance was noted between the wild type and transgenic positives under well-watered conditions (Fig. 6A). The plant biomass was not estimated in the study to help prove this claim, but we did not see any differences in plant height (Table 1) between non-transgenic and transgenic plants. The estimation of total biomass produced by the plants per unit of water applied and the determination of photosynthetic rate of these lines in the future is suggested in order to understand the actual role of SQS in stomatal regulation and drought tolerance without affecting photosynthesis. The utilization of deep pots in this study facilitated a slow progressive drought of 28 d, which helped the plants to adjust transpiration progressively than to a sudden soil-drying. These results are concomitant with Hu et al. (2006) where transgenic rice over-expressing the NAC transcription factor (sNAC1) showed reduced water loss through restricted stomatal opening and enhanced drought tolerance. Recently, Huang et al. (2009) showed that loss of an unknown rice zinc finger protein (DST) enhanced drought tolerance in rice by increased stomatal closure similar to our findings. The Arabidopsis farnesyl transferase deletion mutants (era1) also exhibited a decreased transpiration rate of leaves and slow dessication under drought (Pei et al., 1998). In canola, similarly, the down-regulation of a farnesyl transferase subunit resulted in a lower stomatal conductance, reduced transpiration, and increased yield under drought (Wang et al., 2005). In general, rice has been reported to be one of the hydrophilic crops that function with more open stomata above optimal. Karaba et al. (2007) found that expression of the Arabidopsis HARDY (HRD) gene in rice plants improved water use efficiency by enhancing photosynthetic assimilation and reduced transpiration. These drought-tolerant, low-water-consuming rice plants exhibited increased shoot biomass under well-watered conditions and an adaptive increase in root biomass under drought stress. Hence, further investigations on the determination of water use efficiency in the SQS RNAi lines could lead to the identification of a similar mechanism.
One of the drought-tolerance mechanisms in rice is achieved by delayed leaf rolling (Price et al., 2002). During the process of stress development, the transgenic plants showed much delayed leaf-rolling compared with the wild-type and negative control plants. The WT plants showed leaf rolling symptoms around 16 d after withholding water; whereas, leaf rolling was delayed by approximately 10 d in the RNAi lines. This was clearly explained by the maintenance of higher leaf relative water content (RWC). Under non-stress conditions, there were no obvious differences in the leaf RWC between WT and the transgenic plants, however, approximately 20% higher RWC than WT was noticed in transgenic plants under water stress. Rapid transpiration in the WT might have led to a quick loss of turgor and triggered early leaf rolling and drying in these plants. Similar findings were reported in rice (Babu et al., 2004), tobacco (Rivero et al., 2007), and maize (Zhang et al., 2010). The function of early leaf rolling as a drought avoidance trait was not reflected in the recovery or yield of WT plants in this study.
Yield components
Boyer (2010) reported that plants respond to a changing physiology imposed by the environment, which results in floral abortion, reduced fruit quality, or smaller grains. In rice, yield losses due to water shortage probably exceeds losses from all other causes combined and the extent of the yield loss depends on both the severity and duration of the water stress (Farooq et al., 2009). Therefore, the identification of genes that impart yield improvement at the reproductive stage is imperative. In the present study, water deficit was imposed at the critical stage of reproductive development (division of pollen mother cells) which continued until heading. The severe water stress resulted in one-third of grain yield in the WT, whereas significantly less reduction was noticed in the transgenic plants. This was associated with an increased number of filled grains and reduced spikelet sterility as reported in other studies (Xiao et al., 2009; Jeong et al., 2010). Although, statistically, no difference was observed between the wild type and transgenic positives under well-watered conditions due to the large error bars, there was definitely a yield penalty observed in the positives. The extent of difference was obvious at least for one event (RIFGOCU) which showed 29% yield penalty under well-watered conditions. This could be overcome by using a stress-inducible promoter.
Conclusion
The impact of global climate change in the future will result in the frequent occurrence of drought/heat waves or the combination of both. Drought and heatwaves resulted in 25.3% damage (180.2 billion dollar loss in 18 years) in the USA. In developing countries, a decline of 9–21% overall potential agricultural productivity was recorded as a result of drought and heat (FAO Media Center Newsletter, 30 September 2009). One of the most important mechanisms contributing to drought tolerance at least in rice is inferred to be dehydration avoidance (Serraj et al., 2009; Guan et al., 2010). Results from the present study indicate that down-regulation of squalene synthase reduces stomatal conductance and conserved more moisture that led to increased grain yield. Because the RNAi lines did not show any reduction in biomass under control conditions, exploiting this gene for improved dehydration avoidance and/or better ability to extract soil water show promise for genetic engineering of rice for drought tolerance.
Supplementary data
Supplementary data can be found at JXB online.
Supplementary Fig. S1. Growth of WT and transgenic events 14 days after sowing in agar plates infused with A&C. 0 μmol ABA; B. 5 μmol ABA and D. 10% PEG (Polyethylene Glycol). Insert in C and D represent the results of PMI positive transgenics.
Supplementary Fig. S2. Root Growth of WT and transgenic positive events (D1 and CU) 15 days after sowing in soil (profile mix) under well-watered conditions. (n=5).
Acknowledgments
We thank Professor JS Boyer for his time to read the manuscript and his valuable suggestions to improve the manuscript. The assistance of Allison Thiele in growing the plants and scientific inputs from Dr Anjanasree Neelakandan and Dr Rajesh Kumar is greatly acknowledged. We appreciate Theresa Musket for proofreading the manuscript.
References
- Araus JL, Slafer GA, Royo C, Serret D. Breeding for yield potential and stress adaptation in cereals. Critical Reviews in Plant Science. 2008;27:377–412. [Google Scholar]
- Babu RC, Zhang JX, Blum A, Ho THD, Wu R, Nguyen HT. HVA1, a LEA gene from barley confers dehydration tolerance in transgenic rice (Oryza sativa L.) via cell membrane protection. Plant Science. 2004;166:855–862. [Google Scholar]
- Boyer JS. Drought decision-making. Journal of Experimental Botany. 2010;61:3493–3497. doi: 10.1093/jxb/erq231. [DOI] [PubMed] [Google Scholar]
- Brady SM, Sarkar SF, Bonetta D, McCourt P. The ABSCISIC ACID INSENSITIVE 3 (ABI3) gene is modulated by farnesylation and is involved in auxin signaling and lateral root development in Arabidopsis. The Plant Journal. 2003;34:67–75. doi: 10.1046/j.1365-313x.2003.01707.x. [DOI] [PubMed] [Google Scholar]
- Busquets A, Keim V, Closa M, del Arco A, Boronat A, Arró M, Ferrer A. Arabidopsis thaliana contains a single gene encoding squalene synthase. Plant Molecular Biology. 2008;67:25–36. doi: 10.1007/s11103-008-9299-3. [DOI] [PubMed] [Google Scholar]
- Crowell DN, Huizinga DH. Protein isoprenylation: the fat of the matter. Trends in Plant Science. 2009;14:163–170. doi: 10.1016/j.tplants.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Cruz RT, O’Toole JC. Dry land rice response to an irrigation gradient at flowering stage. Agronomy Journal. 1984;76:178–183. [Google Scholar]
- Cutler S, Ghassemian M, Bonetta D, Cooney S, McCourt P. A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science. 1996;273:1239–1241. doi: 10.1126/science.273.5279.1239. [DOI] [PubMed] [Google Scholar]
- Davis JC. Statistics and data analysis in geology. New York: Wiley; 1986. [Google Scholar]
- Degenkolbe T, Do PT, Zuther E, Repsilber D, Walther D, Hincha DK, Köhl KI. Expression profiling of rice cultivars differing in their tolerance to long-term drought stress. Plant Molecular Biology. 2009;69:133–153. doi: 10.1007/s11103-008-9412-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devarenne TP, Ghosh A, Chappell J. Regulation of squalene synthase, a key enzyme of sterol biosynthesis, in tobacco. Plant Physiology. 2002;129:1095–1106. doi: 10.1104/pp.001438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evenson RE, Herdt RW, Hossain M. Rice research in Asia: progress and priorities. Wallingford, UK: CAB International; 1996. [Google Scholar]
- Farooq M, Wahid A, Ito O, Lee DJ, Siddique KHM. Advances in drought resistance of rice. Critical Reviews in Plant Sciences. 2009;28:199–217. [Google Scholar]
- Guan YS, Serraj R, Liu SH, Xu JL, Ali J, Wang WS, Venus E, Zhu LH, Li ZK. Simultaneously improving yield under drought stress and non-stress conditions: a case study of rice (Oryza sativa L.) Journal of Experimental Botany. 2010;61:4145–4156. doi: 10.1093/jxb/erq212. [DOI] [PubMed] [Google Scholar]
- Hata S, Sanmiya K, Kouchi H, Matsuoka M, Yamamoto N, Izui K. cDNA cloning of squalene synthase genes from mono- and dicotyledonous plants, and expression of the gene in rice. Plant and Cell Physiology. 1997;38:1409–1413. doi: 10.1093/oxfordjournals.pcp.a029137. [DOI] [PubMed] [Google Scholar]
- Haudenschild C, Hartmann M- A. Inhibition of sterol biosynthesis during elicitor-induced accumulation of furanocoumarins in parsley cell suspension cultures. Phytochemistry. 1995;40:1117–1124. [Google Scholar]
- Hazen SP, Pathan MS, Sanchez A, Baxter I, Dunn M, Estes B, Chang HS, Zhu T, Kreps JA, Nguyen HT. Expression profiling of rice segregating for drought tolerance QTLs using a rice genome array. Functional and Integrative Genomics. 2005;5:104–116. doi: 10.1007/s10142-004-0126-x. [DOI] [PubMed] [Google Scholar]
- Henikoff JG, Greene EA, Pietrokovski S, Henikoff S. Increased coverage of protein families with the Blocks Database servers. Nucleic Acids Research. 2000;28:228–230. doi: 10.1093/nar/28.1.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Dai M, Yao J, Xiao B, Li X, Zhang Q, Xiong L. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proceedings of the National Academy of Sciences, USA. 2006;103:12987–12992. doi: 10.1073/pnas.0604882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang XY, Chao DY, Gao JP, Zhu MZ, Shi M, Lin HX. A previously unknown zinc finger protein, DST, regulates drought and salt tolerance in rice via stomatal aperture control. Genes and Development. 2009;23:1805–1817. doi: 10.1101/gad.1812409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JS, Kim YS, Baek KH, Jung H, Ha SH, Do Choi Y, Kim M, Reuzeau C, Kim JK. Root-specific expression of OsNAC10 improves drought tolerance and grain yield in rice under field drought conditions. Plant Physiology. 2010;153:185–197. doi: 10.1104/pp.110.154773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CD, Chary SN, Chernoff EA, Zeng Q, Running MP, Crowell DN. Protein geranylgeranyltransferase I is involved in specific aspects of abscisic acid and auxin signaling in Arabidopsis. Plant Physiology. 2005;139:722–733. doi: 10.1104/pp.105.065045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaba A, Dixit S, Greco R, Aharoni A, Trijatmiko KR, Marsch-Martinez N, Krishnan A, Nataraja KN, Udayakumar M, Pereira A. Improvement of water use efficiency in rice by expression of HARDY, an Arabidopsis drought and salt tolerance gene. Proceedings of the National Academy of Sciences, USA. 2007;104:15270–15275. doi: 10.1073/pnas.0707294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kathiresan A, Lafitte HR, Chen JX, Mansueto L, Bruskiewich R, Bennett J. Gene expression microarrays and their application in drought stress research. Field Crops Research. 2006;97:101–110. [Google Scholar]
- Kikuchi S, Satoh K, Nagata T, et al. Collection, mapping, and annotation of over 28 000 cDNA clones from japonica rice. Science. 2003;301:376–379. doi: 10.1126/science.1081288. [DOI] [PubMed] [Google Scholar]
- Krishnan A, Guiderdoni E, An G, et al. Mutant resources in rice for functional genomics of the grasses. Plant Physiology. 2009;149:165–170. doi: 10.1104/pp.108.128918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanceras J, Pantuwan G, Jongdee B, Toojinda T. Quantitative trait loci associated with drought tolerance at reproductive stage in rice. Plant Physiology. 2004;135:384–399. doi: 10.1104/pp.103.035527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. Clustal W and Clustal X version 2. Bioinformatics. 2007;223:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lin H, Ouyang S, Egan A, Nobuta K, Haas BJ, Zhu W, Gu X, Silva JC, Meyers BC, Buell CR. Characterization of paralogous protein families in rice. BMC Plant Biology. 2008;8:18. doi: 10.1186/1471-2229-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Toole JC. The first international conference on rice for the future. Bangkok, Thailand: The Rockefeller Foundation; 2004. Rice and water: the final frontier. [Google Scholar]
- Pei ZM, Ghassemian M, Kwak CM, McCourt P, Schroeder JI. Role of farnesyltransferase in ABA regulation of guard cell anion channels and plan water loss. Science. 1998;282:287–290. doi: 10.1126/science.282.5387.287. [DOI] [PubMed] [Google Scholar]
- Praba ML, Cairns JE, Babu RC, Lafitte HR. Identification of physiological traits underlying cultivar differences in drought tolerance in rice and wheat. Journal of Agronomy and Crop Science. 2009;195:30–46. [Google Scholar]
- Price AH, Cairns JE, Horton P, Jones HG, Griffiths H. Linking drought-resistance mechanisms to drought avoidance in upland rice using a QTL approach: progress and new opportunities to integrate stomatal and mesophyll responses. Journal of Experimental Botany. 2002;53:989–1004. doi: 10.1093/jexbot/53.371.989. [DOI] [PubMed] [Google Scholar]
- Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E. Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proceedings of the National Academy of Sciences, USA. 2007;104:19631–19636. doi: 10.1073/pnas.0709453104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Running MP, Lavy M, Sternberg H, Galichet A, Gruissem W, Hake S, Ori N, Yalovsky S. Enlarged meristems and delayed growth in plp mutants result from lack of CaaX prenyltransferases. Proceedings of the National Academy of Sciences, USA. 2004;101:7815–7820. doi: 10.1073/pnas.0402385101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proceedings of the National Academy of Sciences, USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The Neighbor–Joining method: a new method for reconstructing phylogenetic trees. Molecular Evolution Biology. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Serraj R, Kumar A, McNally KL, Slamet-Loedin I, Bruskiewich R, Mauleon R, Cairns J, Hijmans RJ. Improvement of drought resistance in rice. Advances in Agronomy. 2009;103:41–98. [Google Scholar]
- Stamellos KD, Shackelford JE, Shechter I, Jiang G, Conrad D, Keller GA, Krisans SK. Subcellular localization of squalene synthase in rat hepatic cells. Journal of Biological Chemistry. 1993;268:12818–12824. [PubMed] [Google Scholar]
- Tansley TR, Shechter I. Squalene synthase: structure and regulation. Progress in Nucleic Acid Research and Molecular Biology. 2001;65:157–195. doi: 10.1016/s0079-6603(00)65005-5. [DOI] [PubMed] [Google Scholar]
- Vogeli U, Chappell J. Induction of sesquiterpene cyclase and suppression of squalene synthetase activities in plant cell cultures treated with fungal elicitor. Plant Physiology. 1988;88:1291–1296. doi: 10.1104/pp.88.4.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Siopongco J, Wade LJ, Yamauchi A. Fractal analysis on root systems of rice plants in response to drought stress. Environmental and Experimental Botany. 2009a;65:338–344. [Google Scholar]
- Wang Y, Beaith M, Chalifoux M, Ying J, Uchacz T, Sarvas C, Griffiths R, Kuzma M, Wan J, Huang Y. Shoot specific down-regulation of protein farnesyltransferase (α-subunit) for yield protection against drought in canola. Molecular Plant. 2009b;1:191–200. doi: 10.1093/mp/ssn088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Yin J, Kuzma M, et al. Molecular tailoring of farnesylation for plant drought tolerance and yield protection. The Plant Journal. 2005;43:413–424. doi: 10.1111/j.1365-313X.2005.02463.x. [DOI] [PubMed] [Google Scholar]
- Wentzinger LF, Bach TJ, Hartmann M. Inhibition of squalene synthase and squalene epoxidase in tobacco cells triggers an up-regulation of 3-hydroxy-3-methylglutaryl Coenzyme A reductase. Plant Physiology. 2002;130:334–346. doi: 10.1104/pp.004655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao BZ, Chen X, Xiang CB, Tang N, Zhang QF, Xiong LZ. Evaluation of seven function-known candidate genes for their effects on improving drought resistance of transgenic rice under field conditions. Molecular Plant. 2009;2:73–83. doi: 10.1093/mp/ssn068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Forno D, Cock JH. Laboratory manual for physiological studies of rice. 1976 Los Baños, Philippines: IRRI. [Google Scholar]
- Zhang S, Li N, Gao F, Yang A, Zhang J. Over-expression of TsCBF1 gene confers improved drought tolerance in transgenic maize. Molecular Breeding. 2010;26:455–465. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.