Abstract
Although flowering in mature fruit trees is recurrent, floral induction can be strongly inhibited by concurrent fruiting, leading to a pattern of irregular fruiting across consecutive years referred to as biennial bearing. The genetic determinants of biennial bearing in apple were investigated using the 114 flowering individuals from an F1 population of 122 genotypes, from a ‘Starkrimson’ (strong biennial bearer)בGranny Smith’ (regular bearer) cross. The number of inflorescences, and the number and the mass of harvested fruit were recorded over 6 years and used to calculate 26 variables and indices quantifying yield, precocity of production, and biennial bearing. Inflorescence traits exhibited the highest genotypic effect, and three quantitative trait loci (QTLs) on linkage group (LG) 4, LG8, and LG10 explained 50% of the phenotypic variability for biennial bearing. Apple orthologues of flowering and hormone-related genes were retrieved from the whole-genome assembly of ‘Golden Delicious’ and their position was compared with QTLs. Four main genomic regions that contain floral integrator genes, meristem identity genes, and gibberellin oxidase genes co-located with QTLs. The results indicated that flowering genes are less likely to be responsible for biennial bearing than hormone-related genes. New hypotheses for the control of biennial bearing emerged from QTL and candidate gene co-locations and suggest the involvement of different physiological processes such as the regulation of flowering genes by hormones. The correlation between tree architecture and biennial bearing is also discussed.
Keywords: Auxin, floral induction, gibberellin, irregular production, Malus×domestica, precocity
Introduction
Once a woody perennial plant has passed the juvenile period when it cannot be induced to flower and has reached its adult phase of reproductive competence, a proportion of its meristems will initiate floral organs annually. Flowering in temperate tree species can be divided into several stages that include flower induction, flower initiation, flower differentiation, and blooming. Flower initiation is the key developmental stage for fruit trees, particularly for horticultural crops such as the apple (Malus×domestica Borkh.), because it determines the success of commercial orchards (Buban and Faust, 1982) by its influence on fruit quantity and quality (Link, 2000), as well as stability of production from year to year (Schmidt et al., 1989). Flower initiation can be strongly limited by an excessive crop, leading to the phenomenon known as biennial bearing (Jonkers, 1979; Monselise and Goldschmidt, 1982). Commonly used terms related to alternate bearing include biennial bearing and irregular bearing. Biennial bearing is characterized by large yields of small sized fruit in ‘on’ years, and low yields, sometimes even no fruit, in ‘off’ years. This alternation is a widely spread phenomenon, occurring in both deciduous and evergreen trees, and in different tree families and species such as nuts (hazelnuts, pecans, pistachios, and walnuts), temperate fruits (apple, apricot, pears, and prunes), subtropical fruits (avocados, citrus, and olives), tropical fruits (litchis and mangos), and forest trees (beeches, oaks, pines, and spruces) (Monselise and Goldschmidt, 1982).
Several reliable parameters have been proposed to phenotype biennial bearing, its intensity, and the synchrony in different parts of the tree (Monselise and Goldschmidt, 1982). Hoblyn et al. (1936) originally proposed an index to estimate the intensity of deviation in yield during successive years that has been renamed by Wilcox (1944) as the Biennial Bearing Index (BBI). The BBI has been widely used to study fruit yield (i.e. mass of fruit) over orchards, individual trees, or branches (Wilcox, 1944; Singh, 1948; Pearce and Dobersek-Urbane, 1967; Jonkers, 1979). Recent examples used BBI in apple (Barrit et al., 1997), mango (Reddy et al., 2003), coffee (Cilas et al., 2011), citrus (Smith et al., 2004), pecan (Wood et al., 2004), and pistachio (Rosenstock et al., 2010).
Although generations of scientists have tried to understand this phenomenon, the cause of alternate bearing is still largely unknown (Singh, 1948; Hoad, 1978; Jonkers, 1979; Monselise and Goldschmidt, 1982; Bangerth, 2006, 2009). External factors (photoperiod, temperature, and water stress), internal factors such as the carbon-to-nitrogen ratio and hormones [auxins, cytokinins (CKs), abscisic acid, ethylene, and gibberellins (GAs)], as well as interaction with other organs (leaves, terminal shoot growth, and fruit) affect flower formation in apple (for reviews, see Hanke et al., 2007; Bangerth, 2009). The negative relationship between fruit development and flower bud differentiation is one of the most investigated causes of flower set variability in apple, as the differentiation of flower buds in apple overlaps with embryo development in the previous season's fruit (Harley, 1942; Foster et al., 2003), leading to competition between flower initiation and fruit formation.
Experiments using ‘Spencer Seedless’, which can bear both parthenocarpic and seeded fruit, suggested that seed development rather than nutritional competition may be a factor in alternate bearing (Chan and Cain, 1967; Neilsen and Dennis, 2000). The number of seed per fruit or per bourse (flowering growth unit) has an effect on biennial bearing, which can be overcome by a high vegetative growth rate of the bourse shoot itself (Chan and Cain, 1967; Grochowska and Karaszewska, 1976; Hoad, 1978; Neilsen and Dennis, 2000). Seed are known to contain relatively large amounts of hormones (Luckwill, 1974), and auxin [indole acetic acid (IAA)], GA, and CK have been implicated separately, and in combination, as being responsible for hormonal control of floral induction (FI). IAA and GA may act together or independently to inhibit FI in perennial fruit trees, whereas CK is likely to be the hormone enhancing FI (Bangerth, 2006). Although the spur (short fruiting shoot) tissues of biennial bearing cultivars receive more GA through the pedicel than annual bearing cultivars do (Hoad, 1978), and the peak activity of GA in apple seed coincides with FI (Luckwill, 1970), it has been difficult to obtain convincing evidence for the transport of GA from seed in sufficient quantities to inhibit FI. Bangerth (2006) proposed that auxin could be the mobile signal and might stimulate GA synthesis in the meristem. In this model, GA and auxin could potentially act as FI-inhibiting signals working in concert, GA as the primary messenger that stimulates the synthesis/transport of the second messenger auxin. However, characterization and quantification of both GA and auxin in the meristem still need to be performed and, moreover, an inhibitory effect of GA/auxin and stimulation by CK on the expression of genes related to FI remain to be demonstrated (Bangerth, 2006).
Since regular bearing appears to be related to FI rather than floral organ differentiation, it may be hypothesized that floral integrator and floral meristem identity genes are involved in this phenomenon. Key genes regulating floral development have been identified in model plants, such as Antirrhinum majus and Arabidopsis thaliana (Bernier and Périlleux, 2005; Tan and Swain, 2006; Corbesier et al., 2007). These include the flowering promoter gene, FLOWERING LOCUS T (FT), that encodes a protein which is a major component of florigen (Kobayashi et al., 1999), and the LEAFY (LFY) and APETALA1 (AP1) genes, which have been identified as necessary for the determination of the floral meristem identity (Yanofsky, 1995). Other genes such as FLOWERING LOCUS C (FLC), TERMINAL FLOWER 1 (TFL1), BROTHER OF FT (BFT), and SHORT VEGETATIVE PHASE (SVP) are known to be repressors of the floral pathway integrators (Boss et al., 2004; Yoo et al., 2010). Although there are fundamental differences in the flowering process between annual and perennial plants, the genetics of FI and floral organ formation are likely to be similar among these plants (Tan and Swain, 2006). A set of apple genes with sequence similarity to genes involved in floral meristem transition of Arabidopsis has been identified and subjected to expression studies (Jeong et al., 1999; Sung et al., 1999; Kotoda et al., 2000, 2002, 2003, 2006, 2010; Van der Linden et al., 2002; Wada et al., 2002; Kotoda and Wada, 2005; Esumi et al., 2005). Overexpression of the apple gene orthologues of LFY, AFL1, and AFL2 (APPLE FLORICAULA/LFY) (Wada et al., 2002), as well as MdMADS2 and MdMADS5, orthologues of the Arabidopsis FRUITFULL (FUL) and AP1, resulted in early flowering in heterologous systems (Sung et al., 1999; Kotoda et al., 2002). Conversely, overexpression of the TFL1 orthologue gene of apple, MdTFL1, in Arabidopsis delayed flowering (Kotoda and Wada, 2005). Kotoda et al. (2006) further showed that transgenic ‘Orion’ apple trees with a reduced MdTFL1 transcript level flowered 8 months after grafting, whereas non-transformed ‘Orion’ plants still had not flowered nearly 5 years after grafting.
Both progeny segregation patterns and differences in bearing behaviour among cultivars strongly suggest the involvement of alleles transmissible by both regular and non-regular types, together with a possible modification of expression by a genotype by environment interaction effect (Monselise and Goldschmidt, 1982). However, there has been no attempt to identify the genetic and molecular determinants of biennial bearing, and apple flowering genes and their allelic variants have never been evaluated for phenotypic variations in segregating populations or within the wider Malus germplasm.
The goal of the present study was to investigate the genetic determinants of biennial bearing in a segregating population using a combination of quantitative genetics analysis, quantitative trait locus (QTL) detection, and candidate gene mapping. A segregating population from a cross between contrasted genotypes for bearing behaviour, ‘Starkrimson’ and ‘Granny Smith’ (STK×GS) (Segura et al., 2006, 2008, 2009), was phenotyped over six consecutive years and quantification of biennial bearing was based on yield at the whole-tree scale. QTLs and candidate genes for the control of flowering and its regularity in apple were identified and mapped. It was demonstrated that candidate genes involved in flowering do not co-locate with these QTLs, whereas several genes related to control of amounts of the hormones auxin and GA co-located with the QTL intervals for biennial bearing. Although flowering genes may not directly determine biennial bearing, their control by plant hormones might be one of the processes leading to biennial bearing.
Materials and methods
Plant material
The F1 progeny used in this study were previously used for studying tree architecture during the juvenile phase (Segura et al., 2006, 2007, 2008, 2009). The population was derived from a cross between two cultivars with contrasted tree and fruiting habits: ‘Starkrimson’ and ‘Granny Smith’ (STK×GS). The female parent is characterized by an erect growth habit with many short shoots and a tendency to biennial bearing, whereas the male parent, GS, has a weeping growth habit with long shoots and exhibits fruit-bearing regularity (Lespinasse, 1992). This population consists of 122 genotypes and each is replicated twice. However, three genotypes had only one tree replicate since the second tree replicate died at the beginning of the experimentation. Two-year-old seedlings were grafted on the semi-dwarfing rootstock ‘Pajam 1’ and the grafted trees planted in March 2004 at the Melgueil INRA Montpellier Experimental station using a random experimental design. These trees were not pruned and neither were the fruit thinned.
Phenotyping
The 122 genotypes were observed during six consecutive years, from their second to their seventh years. A total of 241 trees were phenotyped in 2005 and 239 trees in 2010, since two trees died during the 5 years of the study. From 2005 to 2010, the number of inflorescences and the number of fruit were recorded, and the harvested mass of fruit determined at the whole-tree scale. These variables were recorded for each year of production during the 6 years of the experiment, except for the harvested mass of fruit, which was not available for year 2.
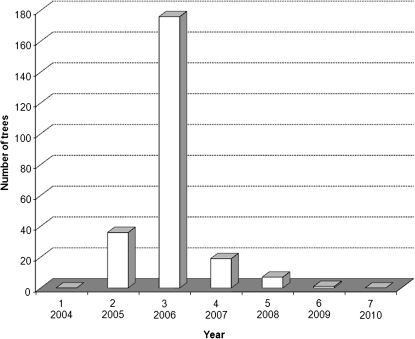
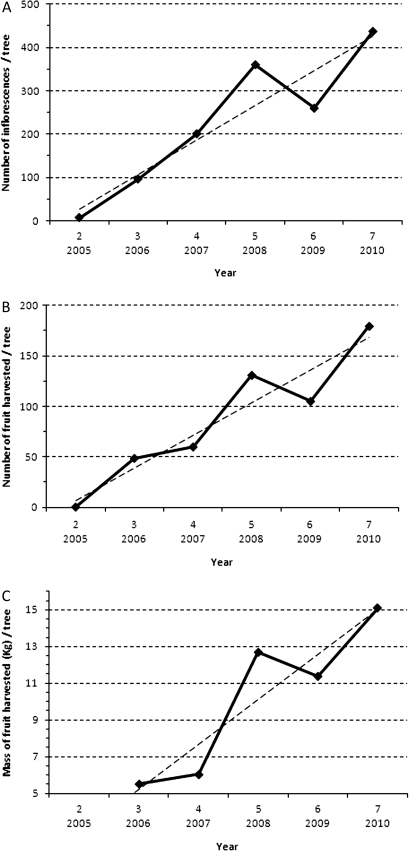
A range of descriptors was calculated from the variables measured (number of inflorescences, number of harvested fruit, and harvested mass) (Table 1). The cumulative yield (CY) is a sum of the production for each year during the whole length of the experiment (Smith et al., 2004). The Precocity Index (PI) was calculated by applying Bartlett's index for earliness of germination (Sivasubramanian, 1962) also called the Earliness Index (EI) by Cilas et al. (2011) (Table 1). This index weights yields according to the year considered, giving a higher weight to the early years of production and less to the latter ones. The alternate bearing behaviour of each genotype was quantified by the BBI, since trees exhibit a biennial pattern. The BBI was calculated using the formula developed by Hoblyn et al. (1936). BBI values vary from 0 to 1, where 0 denotes equal yields in successive years and 1 alternate yield (Hoblyn et al., 1936). CY indexes were calculated from 6 years of data, from 2005 to 2010, whereas PIs were calculated from 5 years of data (from 2005 to 2009), since no trees flowered for the first time during the seventh year of the study (Fig. 1). Because only 36 trees flowered in 2005, compared with 212 in 2006, the BBI was calculated on 5 years of data (from year 3 to year 7) excluding year 2 in order to treat genotypes equally. Genotypes that began to flower during year 4, 5, or 6 were excluded from the BBI calculation. Bearing behaviour has been graphically represented for the whole population based on the average of phenotypic values for the total number of inflorescences and fruit, and the harvested mass of fruit per tree, per year (Fig. 2).
Table 1.
Descriptors used to study inflorescence and fruit production in the ‘Starkrimson’בGranny Smith’ segregating population over 6 years.
| Trait | Formula | Variable abbreviation | References | ||
| Number of inflorescences | Number of fruit harvested | Mass of fruit harvested | |||
| Yield | Y_inf_n | Y_fruit_n | Y_mass_n | ||
| Cumulative yield | 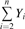 |
CY_inf | CY_fruit | CY_mass | Smith et al. (2004) |
| Precocity index | 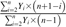 |
PI_inf | PI_fruit | PI_mass | Sivasubramanian (1962) |
| Biennial bearing index | 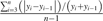 |
BBI_inf | BBI_fruit | BBI_mass | Hoblyn et al. (1936) |
The formula used to calculate each descriptor is shown in relation to the type of data measured, such as the number of inflorescences, the number of fruit harvested, and the mass of fruit harvested. Yi represents yield for year i, and n represents the number of years studied. With n=7 for Cumulative Yield and Biennial Bearing Index and n=6 for Precocity index.
Fig. 1.
Number of trees flowering for the first time according to the year after grafting. Years 1–7 on the x-axis correspond to 2004 to 2010, respectively.
Fig. 2.
Average production per tree calculated for the population (239 trees) over the 6 years of experiments. (A) Number of inflorescences, (B) number of fruit harvested, and (C) the mass of fruit harvested. Dashed lines correspond to the increasing trend estimated from a linear regression over years.
In 2010, the numbers of fruit per inflorescence (NFI) were counted for 20 inflorescences per tree. Inflorescences were sampled in the terminal position of spurs located laterally on 2- to 6-year-old wood of long axillary shoots. Then 10 fruit per tree were harvested and the number of aborted and fully developed seed per fruit (NSF) counted. To obtain the number of seed per inflorescence (NSI), phenotypic averages were calculated per tree for NFI and NSF and were multiplied together.
Clustering of bearing behaviour
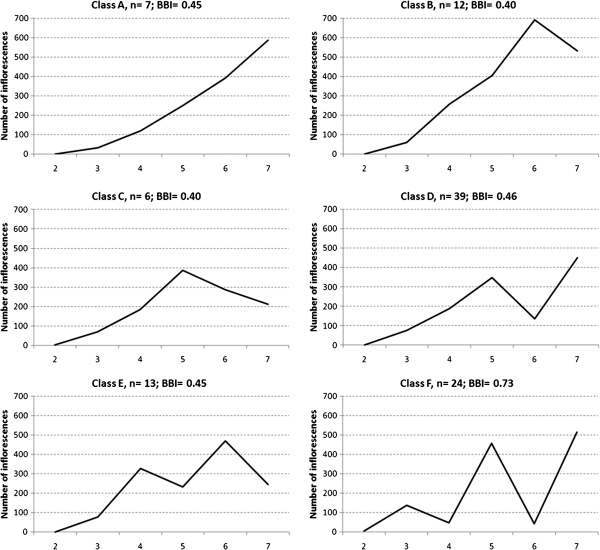
Within the population, contrasted bearing behaviours were graphically identified based on the average of phenotypic values for the total number of inflorescences per genotype and per year (Fig. 3). To establish this classification, trees that began to flower during year 4, 5, or 6 were not considered and only 114 genotypes were included (93.4% of the population) that began to flower in year 2 or 3. The classification was performed manually and relied on the study of the evolution of yield through time by the construction of sequences composed of ‘+’ and ‘–’ symbols reflecting the direction of variation of the yield for each pair of years. Yields that were higher in year n+1 than in year n were symbolized by ‘+’, whereas yields that were lower in year n+1 than in year n were symbolized by ‘–’. These sequences were used to sort genotypes into clusters with the same pattern. Finally the average yield per year of all genotypes was calculated for each cluster.
Fig. 3.
Six bearing behaviours identified among genotypes within the population based on the average phenotypic values for the number of inflorescences. Class effective and average BBI values are indicated in the legend for each graph.
Differences between tree replicates within genotypes were investigated using the same sequential method as described above. Three groups were formed: the first group included genotypes having no differences in sequence between trees, the second group was composed of genotypes for which tree replicates showed one difference in the sequence, and the third group was composed of genotypes showing at least two differences in the sequence.
Genotypes for which production during year n+1 was equal or superior to year n were considered as regular. Genotypes exhibiting production in year n+1 inferior to year n were considered as irregular, or biennial when the pattern of alternation was biennial.
Statistical analyses
Statistical analyses were performed using R software v.2.9.2. (R Development Core Team, 2009). Data sets were analysed using a two-step method: first, the statistical effects were estimated by an analysis of variance (ANOVA) and then the significant effects were used to construct a linear model that estimated the genotypic value of the trait for each genotype. Three models were considered: one for yield data, which have been observed over 6 years, a second model for the CY index, PI, and BBI, which have one value for the whole study, and a third model for NFI, NSF, and NSI, which have 1 year of data, with repetitions within the tree.
Data sets for annual yield indexes were analysed by mixed linear models that included the year (Y), the genotype (G), the interaction between genotype and year (G×Y), and the nested effect of the tree within the genotype (G[T]). For the CY index, PI, and BBI, a linear model was built considering only the genotype (G). For NFI, NSF, and NSI, the model considered the genotype (G), the nested effect of the tree within the genotype (G[T]), and the nested effect of the fruit within the tree (T[F]). Significance of the effects was estimated by a type III ANOVA (function lm) because of unbalanced data. Then, the linear models were constructed for each variable, considering the significant effects detected by the ANOVA as fixed effects (Y) and as random effects (G, G×Y, G×T, and F×T). A model selection was performed based on the Akaike Information Criterion (AIC) minimization. For each trait, when the G effect was significant in the model selected (Table 2), BLUPs (Best Linear Unbiased Predictors) were extracted using the ranef function. Normal distributions of the residual errors were analysed to control the correct estimation by the model of the genotypic value. Because of the non-significant effect of the genotype, BLUPs were not extracted for BBI_fruit and BBI_mass, and QTL detection was based on phenotypic mean values for these variables. Genetic correlations were performed based on the BLUP using the Pearson coefficient, procedure ‘cor’ (Supplementary Table S1 available at JXB online).
Table 2.
Significance of the genotype effect (G), the year (Y), the tree (T), the fruit (F), and their interactions: G×Y, G[T] (i.e. T nested in G), and T[F] (i.e. F nested in T) in type III ANOVAs performed on traits phenotyped.
| Trait | Name of variable | G | Y | G×Y | G[T] | T[F] |
| Biennial Bearing Index | BBI_inf | *** | – | – | – | – |
| BBI_fruit | NS | – | – | – | – | |
| BBI_mass | NS | – | – | – | – | |
| Yield | Y_inf | *** | *** | *** | *** | – |
| Y_fruit | *** | *** | *** | NS | – | |
| Y_mass | *** | *** | *** | * | – | |
| Cumulative yield | CY_inf | *** | – | – | – | – |
| CY_fruit | *** | – | – | – | – | |
| CY_mass | ** | – | – | – | – | |
| Precocity index | PI_inf | *** | – | – | – | – |
| PI_fruit | ** | – | – | – | – | |
| PI_mass | ** | – | – | – | – | |
| Number of fruit per inflorescence | NFI | *** | – | – | *** | *** |
| Number of seed per fruit | NSF | *** | – | – | *** | NS |
| Number of seed per inflorescence | NSI | *** | – | – | NS | – |
NS, non-significant; *P <0.05; **P <0.01; ***P <0.001).
Variable nomenclature
All BLUP variable names, except for NFI, NSF, and NSI, are composed of a short trait name followed by a suffix indicating if the variable was based on the number of inflorescences (inf), the number of harvested fruit (fruit), or the mass of harvested fruit (mass) (Table 2). BLUP variable names for annual yield indexes are followed by a suffix representing the year of the measurement. For example, the inflorescence yield measured during the second year is Y_inf_2. No numbers were attributed to BLUP variables that are independent of the year effect (e.g. Y_inf).
QTL mapping
The QTL analysis was performed using BLUP values extracted per genotype for each variable. The consensus and the parental genetic maps of STK and GS were used for QTL mapping. QTL analyses were carried out using MapQTL® 5.0. (Van Ooijen, 2004). First, a permutation test was performed to determine the logarithm of the odds (LOD) threshold at which a QTL was declared significant, using a genome-wide error rate of 0.01, 0.05, and 0.1 with 1000 permutations of the data (Van Ooijen, 2004). In the second step, an interval mapping analysis was carried out, with a step size of 1 cM, to detect potential genomic regions associated with the trait, with a LOD score higher than the threshold. The nearest marker to each QTL peak was then selected as a cofactor to perform multiple QTL mapping (MQM), with a step size of 1 cM (Van Ooijen, 2004). Each significant QTL was characterized by its LOD score, its percentage of explained phenotypic variation, and its confidence interval in cM corresponding to a LOD score drop of 1 or 2 on either side of the likelihood peak.
Allelic effects were estimated as Af=[(μac+μad)–(μbc+μbd)]/4 for female additivity, Am=[(μac+μbc)–(μad+μbd)]/4 for male additivity, and D=[(μac+μbd)–(μad+μbc)]/4 for dominance, where μac, μad, μbc, and μbd are estimated phenotypic means associated with each of the four possible genotypic classes ac, bc, ad, and bd, deriving from a <ab×cd> cross.
When a multilocus QTL was detected with at least two cofactors, models considering markers and their interactions as cofactors were constructed using a backward procedure under R software v2.8.1. Models were selected based on the AIC values. The location of QTLs was illustrated on the genetic maps based on the peak LOD–1 and LOD–2 intervals using MapChart® (Voorrips, 2002).
Two rounds of QTL detection were performed. The first round was performed on a genetic map comprising simple sequence repeats (SSRs) only, in order to detect the genomic regions of interest for the candidate gene mapping. The second round of QTL detection was performed on the genetic map including the candidate genes, and the results of this QTL detection are presented in Fig. 4.
Fig. 4.
Genomic positions of the QTLs detected on the consensus ‘Starkrimson’בGranny Smith’ (STK×GS) and parental-maps: ‘Starkrimson’ maternal map (STK) and ‘Granny Smith’ pollen parent map (GS). QTLs are represented by boxes, in which length represents the LOD–1 confidence interval and extended lines represent the LOD–2 confidence interval. Boxes representing QTLs for the number of inflorescences are white, number of harvested fruit traits are black, mass of harvested fruit traits are hatched, and number of fruit and seed per inflorescence are double hatched. For trait abbreviations, see Table 2. Mapped candidate genes are in bold underlined. For candidate gene abbreviations, see Supplementary Table S3 at JXB online.
Candidate gene mapping
An exhaustive in silico inventory of floral and hormone-related genes in apple was performed in order to establish a list of candidate genes that are possibly involved in biennial bearing. Protein sequences of Arabidopsis corresponding to genes involved in floral integration and meristem identity were retrieved from the NCBI database (http://www.ncbi.nlm.nih.gov/). A number of genes involved in plant response, synthesis, and transport of GA and CK were also selected. In total, sequences from 196 accessions from Arabidopsis were searched in silico within the ‘Golden Delicious’ whole-genome sequence (Velasco et al., 2010) using BLASTP (protein–protein BLAST) versus apple gene predictions (amino acid). Ten gene predictions having the best BLAST (Basic Local Alignment Search Tool) expected values were selected for each Arabidopsis gene searched. Their positions and their protein sequences were retrieved on the Malus×domestica genome browser (http://genomics.research.iasma.it/gb2/gbrowse/apple/), and then their protein sequences were blasted against the Swiss-Prot protein reference sequences (http://expasy.org/tools/blast/) in order to identify the best Arabidopsis protein and Malus cDNA related to each apple gene prediction (E-value <1E-30). Alignments and phylogenetic trees analyses were carried out using the deduced amino acid sequences in order to remove redundant gene predictions and to determine the number of gene copies present in the apple genome per Arabidopsis gene (Supplementary Figs S1, S2 at JXB online).
A physical map was generated that included the positions in Megabases (Mb) of the predicted genes and of the SSR markers present on the STK×GS genetic map published by Segura et al. 2007 (Supplementary Fig. S3 at JXB online). The positions of the QTLs detected on the STK×GS consensus map without candidate genes were then compared with the physical map. Candidate genes co-locating with QTLs were studied in detail: phylogenetic trees were built in order to clarify the relationships among the members of each family and predicted genes were named based on their similarity with Arabidopsis proteins.
Amino acid sequences were analysed using the Phylogeny.fr platform (http://www.phylogeny.fr) including the pipeline chaining programs: MUSCLE 3.7 for multiple alignment, Gblocks 0.91b for automatic alignment curation, PhyML 3.0 for tree building, and TreeDyn 198.3 for tree drawing (Dereeper et al., 2008). The tree building was based on an approximation of the standard likelihood ratio test.
An exhaustive inventory of genes related to auxin (ARF, AUX/IAA, and TIR) in the ‘Golden Delicious’ whole-genome sequence was performed by R Schaffer and K David (2011, unpublished). The position of the genes on the genome provided by this study was compared with the position of the QTLs.
The genes co-locating in silico with QTLs were considered as potential candidates, and specific markers were developed in order to position them on the genetic map to test their relationship with the QTLs. PCR primer pairs were designed for the candidate genes using Primer 3Plus software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi). The conditions were set up to amplify short fragments (100–200 bp), if possible spanning putative single nucleotide polymorphisms (SNPs). These potential SNPs were detected within the ‘Golden Delicious’ contig sequences from the apple genome primary assembly (Velasco et al., 2010) or by aligning expressed sequence tag (EST) sequences from different cultivars. The primer pairs for NZmsMdMYB12 and MdCENa were as in (Chagné et al. 2007) and (Mimida et al. 2009), respectively. PCRs were carried out with a real-time PCR instrument (LightCycler 480®, Roche), combined with high resolution melting (HRM) analysis for the detection of DNA polymorphisms (Liew et al., 2004).
The PCRs were performed in the presence of a generic double-stranded DNA dye (LCGreen), which binds to double-stranded DNA only (Wittwer et al., 2003), using a total volume of 7 μl for each well (1× LightCycler® 480 HRM Mastermix, with 2.5 mM MgCl2, 0.2 mM for each primer, and 2 ng of genomic DNA). After activation at 95 °C for 5 min, the reactions underwent 40 PCR cycles of: 95 °C for 10 s; 55 °C for 30 s; 72 °C for 15 s. The HRM analysis was performed immediately after the PCR amplification, with single steps at 95 °C for 1 min; 40 °C for 1 min; 65 °C for 1 s; and then a slow increase of the temperature to reach 95 °C over 15 min, with continuous measurement of the fluorescence intensity (25 data points per degree Celsius). The Roche software identified sequence variants as groups that exhibit similar melting profiles (Hoffmann et al., 2008).
Genetic map construction
One hundred and twenty-three individuals of the population and the parents were genotyped using 168 genetic markers, of which 107 were microsatellite markers (SSRs) and 61 were SNPs. STK and GS parental maps comprised 119 and 124 markers, respectively. Marker names were followed by the suffix ‘SG’ when polymorphic for both parents and followed by ‘S’ or ‘G’ when polymorphic for STK or GS, respectively. JoinMap 3.0 (Van Ooijen and Voorips, 2001) was used for constructing linkage maps using five segregation types: ab×cd, ef×eg, hk×hk, lm×ll, and nn×np for the consensus map. Two segregation types were used to build parental maps, hk×hk for both maps, and lm×ll and nn×np for the STK and GS map, respectively. Linkage groups (LGs) were constructed using a LOD score of 6 for grouping both the STK and GS maps. The data were analysed as population CP, and map distances were calculated using the Kosambi function.
Results
Phenotypic expression of biennial bearing in a segregating population
Of the 242 trees observed in the STK×GS segregating population, 36, 176, 19, and seven set flowers for the first time during the second to fifth year after grafting, respectively (Fig. 1). Only one tree set flowers during the sixth year and three trees still did not flower during their seventh year after grafting. The average production of the population for both numbers of inflorescences and harvested fruit and mass of harvested fruit increased continuously from the second to the seventh year after grafting. However, compared with the upward trend of production in the segregating population, a biennial pattern was observed, as the fifth and seventh years were above the average trend, whereas the fourth and sixth years were below (Fig. 2).
Within the subset of 114 genotypes that had set flowers during the second to seventh year after grafting, six bearing behaviours were graphically identified between genotypes, based on the average phenotypic values per year for the number of inflorescences for each genotype (Fig. 3). Bearing patterns A–F include genotypes that began to flower during year 2 or 3. A fraction of the population (5.7%) was regular bearing (class A), with production increasing consistently during the experiment. The remaining five classes were alternate bearing, and different ‘on’ and ‘off’ years were identified. Class B (9.8% of the population) were characterized by only one ‘off’ year during the seventh year. Class C (4.9% of the population) increased their production until the fifth year and then decreased it during the sixth and seventh year. Class D (32% of the population) began their biennial pattern during the sixth year and were characterized by one ‘off’ year and two ‘on’ years. Classes E (10.6% of the population) and F (19.7% of the population) had a clear biennial pattern but were in opposite phase. The remaining 10.6% of the population had irregular production, where ‘on’ and ‘off’ years were identified, but could not be grouped to form a homogenous class.
Differences between tree replicates within genotypes were investigated based on inflorescence yield per tree. The results showed that tree replicates of 48 genotypes had identical bearing patterns, 47 genotypes had tree replicates discriminated by 1 year of production, and 15 genotypes by 2 years of production (Supplementary Fig. S4 at JXB online).
Significance of genotypic and year effects on production traits
The genotypic effect on biennial bearing was evaluated using a set of measured variables including inflorescence, fruit, and mass annual yields, and indexes calculated from the measured variables. Annual yield consisted of 17 measured variables: six annual yields from year 2 to year 7 for both the number of inflorescences (Y_inf) and the number of fruit harvested (Y_fruit), and five annual yields from year 3 to year 7, for the mass of fruit harvested (Y_mass). Different effects were considered in ANOVAs depending on the measured variables and indexes. On annual variables, ANOVA using years and trees as repetitions showed that G, Y, G×Y, and G[T] effects were highly significant (P <0.001; Table 2). G[T] was less significant for fruit mass yield (Y_mass) and not significant for fruit yield (Y_fruit). As a result of the significant G×Y effect, BLUPs including this interaction were extracted for each measured year (n) (Y_inf_n, Y_fruit_n, and Y_mass_n), as well as BLUPs specific to the genotype effect (Y_inf, Y_fruit, and Y_mass).
Calculated variables such as the CY, PI, and BBI showed highly significant G effects in the ANOVA (Table 2). When calculated on the number and the mass of fruit harvested (BBI_fruit and BBI_mass), BBIs showed G effects slightly above the chosen P-value threshold. The PIs for number and mass of fruit harvested (PI_fruit and PI_mass) and the CY for mass of fruit harvested (CY_mass) exhibited a moderate genotypic effect compared with inflorescence variables. BLUPs were calculated for each index with significant genotype effect (BBI_inf, PI_inf, PI_fruit, PI_mass, CY_inf, CY_fruit, and CY_mass).
The NFI and NSF measured in the seventh year showed highly significant effects of G, G[T], and T[F], except for NSF, for which T[F] was not significant. For the NSI, only the genotypic effect was significant (Table 2). BLUPs specific to the genotype effect were extracted for NFI, NSF, and NSI.
Correlations between variables
Negative moderate correlations were found for the number of inflorescences of a given year to the number of fruit harvested for the previous year (–0.18 to –0.54) and to the mass of fruit harvested the previous year (–0.20 to –0.59) (Supplementary Table S1 at JXB online). The NFI and NSI were positively correlated to variables related to fruit yield (NFI and Y_fruit, 0.55; and NSI and Y_fruit, 0.43). High correlations were observed for indices that were calculated from a set of measured variables. The PI was positively correlated with the mass of fruit harvested in the first year of significant production (i.e. third year, 0.74) and negatively with the mass of fruit harvested in the sixth year (–0.72). The BBI was positively correlated with annual inflorescence yields of years identified as ‘on’ in the population, such as the third and fifth years (Fig. 2) (0.33 and 0.42, respectively), whereas it was negatively correlated with annual inflorescence yields of ‘off’ years, such as the fourth and sixth years (–0.62 and –0.50, respectively).
Candidate gene identification, phylogenetic analysis, and genetic mapping
Candidate genes were selected on the basis on their known function in Arabidopsis, and the list of selected genes included 114 genes related to flowering and 73 related to metabolism and catabolism of plant hormones. A search of the apple whole-genome sequence (Velasco et al., 2010) for candidate genes using BLASTP analysis allowed the establishment of the number of members for each family and ascertainment of the location in the genome of 120 genes putatively involved in flowering and meristem identity, 41 in metabolism and catabolism of GA and CK, and 14 in branching. Phylogenetic analyses performed for 12 gene families (six flowering, two hormones, and one branching) to determine the putative function for apple gene sets are illustrated in Supplementary Figs S1 and S2 at JXB online. Genes were named based on their similarity with Arabidopsis proteins and with cDNA from apple. The genetic map for the STK×GS population was updated from its initial version (Segura et al., 2006) by mapping 64 genetic markers located in candidate genes (Fig. 4). The improved consensus STK×GS map comprised 176 genetic markers including 107 SSRs and 69 SNPs, covered all 17 apple chromosomes, and encompassed 1057 cM. It was noted that 15 (23.4%) of the 64 candidate genes did not map at the position predicted from the genome assembly.
Candidate genes related to flowering:
In total, 114 sequences of genes related to flowering were retrieved from GenBank (http://www.ncbi.nlm.nih.gov/) and used to search homologous proteins predicted from the apple genome sequence (Velasco et al., 2010) using BLASTP analysis (i.e. Arabidopsis protein queries versus the predicted protein gene set). One hundred and twenty gene predictions related to flowering were identified, including 12 FT/TFL1, 49 MADS-box, 23 SQUAMOSA protein-like, 22 flowering genes that belong to different gene families, seven PHYTOCHROME genes, and seven CONSTANS (CO)-like (COL) genes. These 120 gene predictions showed highly significant similarity (E-value <1E-30) to 55 Arabidopsis reference protein sequences (Supplementary Table S2 at JXB online). No gene prediction was found for FLC, FLD, FLK, FRIGIDA, GLOBOSA, HASTY, and ZIPPY. Two paralogues were identified in duplicated genomic regions of apple for 39 genes, such as AP1 (LG13 and LG16), EFL3 (LG8 and LG15), and LFY (LG6 and LG14), whereas three gene copies were found for SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1; LG1, LG2, and LG7) and only one copy for PISTILLATA (PI: LG8) (Supplementary Table S2).
A phylogenetic analysis of the FT/TFL1 family genes indicated the presence of five distinct clades within the apple genome, with two paralogous predicted gene copies for each family member: FT, TFL1, homologues of CENTRORADIALIS, MOTHER OF FT, and BROTHER OF FT. Apple paralogous genes shared more homology with each other than with Arabidopsis genes (Supplementary Fig. S1A). Twenty-two Arabidopsis MADS-box genes exhibiting sequence similarities with 49 apple predicted proteins were retrieved from GenBank. A phylogenetic analysis indicated that 2–4 apple putative MADS-box gene had clear orthology with one Arabidopsis MADS-box (Supplementary Fig. S1B). Only one apple gene prediction was found for five Arabidopsis MADS-box genes (AGL3, AGL12, AGL19, AGL21, and PI), whereas there were two predictions for nine Arabidopsis accessions (AGL8, AGL11, AGL24, AP1, SEP1, SEP2, SEP3, SVP, and TT16), three copies for SOC1 and AGL62, and four for AGL15, AGL80, AP2, and AP3 (Supplementary Table S2). A phylogenetic analysis of genes from distinct gene families related to flowering revealed separated clades for each Arabidopsis gene. Two paralogous genes were present in duplicated genomic regions for LFY, EFL3, FCA, GI, and VRN2 because of the Maloideae whole-genome duplication (WGD) (Velasco et al., 2010) (Supplementary Fig. S1C). A total of 23 apple predicted proteins were similar to 12 Arabidopsis SQUAMOSA protein-like (SPL) genes (Supplementary Table S2) and a phylogenetic analysis indicated the presence of nine clades (Supplementary Fig. S1D). The PHYTOCHROME family clustered in four clades with seven apple predicted genes (Supplementary Fig. S1E). The COL family comprised seven apple predicted genes that matched with three Arabidopsis genes (Supplementary Fig. S1F).
Genetic markers developed from 30 flowering genes were mapped using the STK×GS mapping population (Supplementary Table S3 at JXB online). Paralogous genes positioned in homoeologous genomic regions based on the WGD hypothesis of Velasco et al. (2010) included: MdSOC1-like, MdSOC1a, and MdSOC1b located on LG1, LG2, and LG7, MdAFL1 and MdAFL2 on LG6 and LG14, MdVRN2.1 and MdVRN2.2 on LG4 and LG6, MdVRN1a and MdVRN1b on LG5 and LG10, MdCLV1a and MdCLV1b on LG8 and LG15, and MdEFL3a and MdEFL3b on LG8 and LG15, respectively (Fig. 4). Only one copy of MdPI was mapped, on LG8.
Candidate genes related to hormones:
Seventy-three sequences of genes related to metabolism and catabolism of GAs and CKs were used to search homologous proteins predicted from the apple genome: 33 and 40 genes related to GAs and CKs, respectively.
Malus×domestica possessed several copies of gibberellin oxidases, including GA2ox, GA3ox, and GA20ox. Fourteen MdGA2ox, 10 MdGA3ox, and seven MdGA20ox were identified in the predicted apple gene set (Supplementary Table S4 at JXB online), whereas seven, four, and five copies, respectively of these genes have been reported in Arabidopsis. A phylogenetic analysis indicated four separate clades, one each for MdGA3ox and MdGA20ox, and two for MdGA2ox (Supplementary Fig. S2A). Malus×domestica paralogous gene copies shared more sequence similarity with each other than with Arabidopsis genes. Ten apple putative cytokinin oxidases showed high orthology with five Arabidopsis cytokinin oxidases, and the phylogenetic analysis indicated the presence of one clade per Arabidopsis gene (Supplementary Fig. S2B).
Eleven auxin-related genes identified in the apple genome sequence by R Schaffer and K David (unpublished) co-located in silico with the QTLs, including: MdAFB6, MdARF3, MdARF104, MdARF10, MdARF110, MdIAA4, MdIAA25, MdIAA33, MdIAA103, MdIAA106, and MdIAA127A.
Genetic markers for 22 hormone-related candidate genes positioned on the STK×GS genetic map included four MdGA2ox, two MdGA3ox, three MdGA20ox, five DELLA, two cytokinin oxidases, and six auxin-related (Supplementary Table S3 at JXB online). Six DELLA proteins were identified as proposed by Foster et al. (2006), and five of them were positioned on the STK×GS genetic map, with each subgroup of paralogous gene copies located on homoeologous genomic regions: MdRGL1a and MdRGL1b on LG16 and LG13, MdRGL2a and MdRGL2b on LG9 and LG17, and MdRGL3a and MdRGL3b on LG15 and LG2, respectively (Fig. 4).
Candidate genes related to carotenoid cleavage dioxygenase (CCD):
The CCD gene family involved in plant branching comprised 14 apple predicted genes that matched with six Arabidopsis genes (Supplementary Fig. S2C at JXB online). The phylogenetic analysis indicated the presence of five clades, including one common clade for NCED3 and NCED5. Four gene copies were mapped in silico for MdCCD4, three for MdCCD1, two for MdCCD8, and only one copy for MdCCD7a on LG2. A second copy of MdCCD7 was located on LG7 of the STK×GS map, a duplicated genomic region of LG2, whereas it was not detected in silico. Three CCD genes were mapped on the STK×GS genetic map: MdCCD8a, MdCCD8b, and MdCCD7b (Fig. 4).
QTL detection
In total, 43 QTLs spanning 12 LGs were detected on the STK×GS consensus genetic map. Twelve, 15, and eight QTLs were detected for variables related to the number of inflorescences, harvested fruit, and the mass of harvested fruit, respectively (Table 3). Seven QTLs were detected for biennial bearing, including three for the number of inflorescences (BBI_inf) and two for both the number and the mass of harvested fruit (BBI_fruit and BBI_mass). Seventeen and 10 QTLs were mapped on the STK and GS parental maps, respectively (Table 3, Fig. 4).
Table 3.
QTLs detected on the consensus STK×GS map by MQM mapping for the number of inflorescences, the number of fruit harvested, and the mass of fruit harvested phenotyped over 6 years-in the STK×GS apple progeny. For trait abbreviations, see Table 2.
| QTLs | LG | LOD | R2 | Cofactor | Allelic effect | Af | Am | D | Parental map detection |
| BBI_inf | 4 | 5.31*** | 0.157 | Hi04c10x_SG | Af | 0.039 | –0.006 | –0.008 | STK |
| 8 | 4.33** | 0.120 | Hi04b12_S | Af | –0.027 | –0.013 | –0.018 | STK | |
| 10 | 4.63** | 0.135 | MdGA2ox8a_G | Am, D | 0.013 | 0.032 | 0.023 | ||
| Y_inf_2 | 15 | 6.11*** | 0.229 | NZ02b01_S | Af, Am, D | 9.46 | –9.30 | 6.31 | STK/GS |
| Y_inf_3 | 1 | 5.18*** | 0.197 | MdGA20ox1a_S | Am | 1.958 | 16.13 | –2.149 | STK |
| Y_inf_6 | 1 | 4.51** | 0.136 | MdGA20ox1a_S | Am | –7.57 | –38.2 | 6.46 | STK |
| 8 | 5.55*** | 0.185 | Hi04b12_S | Af, Am, D | 35.0 | 26.7 | 11.96 | STK | |
| Y_inf | 15 | 6.03*** | 0.226 | NZ02b01_S | Af, Am, D | –19.3 | 19.0 | –13.2 | STK/GS |
| CY_inf | 15 | 6.35*** | 0.240 | NZ02b01_S | Af, Am, D | –117 | 103 | –71 | STK/GS |
| PI_inf | 3 | 4.51** | 0.128 | NZmsMdMYB12_S | Am, D | 0.005 | 0.013 | –0.010 | |
| 7 | 4.77** | 0.145 | MdSOC1b_S | Af, Am, D | 0.014 | –0.011 | 0.002 | ||
| 10 | 4.12** | 0.104 | MS06g03_G | Am | 0.001 | 0.017 | 0.002 | ||
| BBI_fruit | 10 | 5.16*** | 0.184 | MdAFB6_S | Am | 0.027 | 0.052 | 0.023 | GS |
| 13 | 4.87*** | 0.148 | CH03h03z_SG | Af, Am, D | 0.021 | –0.004 | –0.045 | GS | |
| Y_fruit_2 | 1 | 4.19** | 0.140 | MdGA20ox1a_S | Af, Am, D | 1.471 | –3.662 | 1.914 | STK/GS |
| 8 | 4.03** | 0.124 | MdPI_SG | Af, Am, D | 2.417 | 3.373 | –1.674 | STK | |
| Y_fruit_3 | 5 | 4.43** | 0.144 | CH04e03_SG | Af, D | –4.477 | 0.112 | 2.280 | |
| 11 | 5.46*** | 0.162 | GD_SNP01140_SG | Af, Am, D | –3.330 | 4.330 | –2.869 | ||
| Y_fruit_5 | 1 | 3.71* | 0.134 | MdSOC1-like_G | Af, Am, D | –3.674 | 5.513 | –7.144 | |
| Y_fruit_7 | 8 | 4.84** | 0.176 | MdPI_SG | Af, Am, D | –12.52 | –13.51 | 6.064 | STK |
| Y_fruit | 1 | 4.35** | 0.144 | MdGA20ox1a_S | Af, Am, D | –4.682 | 11.80 | –6.186 | STK/GS |
| 8 | 4.06* | 0.123 | MdPI_SG | Af, Am, D | –8.017 | –10.40 | 5.481 | STK | |
| CY_fruit | 1 | 5.91*** | 0.191 | MdGA3ox_like_b_S | Af, Am, D | –49.00 | 99.7 | –51.43 | STK/GS |
| 8 | 5.07** | 0.157 | MdPI_SG | Af, Am, D | –62.98 | –83.53 | 50.44 | STK | |
| PI_fruit | 3 | 5.46*** | 0.154 | CH03g07_SG | D | 0.001 | 0.003 | –0.012 | |
| 5 | 5.37*** | 0.170 | CH03a04_S | Af, D | –0.010 | 0.003 | 0.007 | ||
| 11 | 4.80** | 0.163 | NZ04h11y_G | Am | 0.002 | 0.011 | –0.005 | ||
| BBI_mass | 10 | 4.18** | 0.144 | MdAFB6_S | Af, Am, D | 0.030 | 0.045 | 0.016 | |
| 13 | 4.89*** | 0.160 | CH03h03z_SG | Af, D | 0.021 | –0.003 | –0.049 | ||
| Y_mass_3 | 1 | 4.92** | 0.179 | B2-T7_S | Am | –0.088 | 0.363 | –0.063 | |
| 5 | 4.96** | 0.151 | CH02a08z_S | Af | –0.314 | 0.076 | 0.103 | ||
| Y_mass_7 | 2 | 3.78* | 0.161 | NH033b_SG | Am | –0.013 | 0.818 | 0.181 | |
| PI_mass | 3 | 4.96** | 0.154 | CH03g07_SG | D | 0.001 | 0.004 | –0.013 | |
| 5 | 4.74** | 0.156 | CH03a04_S | Af | –0.011 | 0.005 | 0.007 | ||
| 11 | 4.28* | 0.143 | NZ04h11y_G | Am | 0.000 | 0.013 | –0.004 | ||
| NFI | 8 | 4.64** | 0.215 | MdPI_SG | Af, Am, D | –0.057 | –0.155 | 0.038 | STK |
| NSF | 3 | 7.33*** | 0.210 | CH03e03_SG | Af, Am, D | –0.406 | 0.640 | –0.395 | |
| 3 | 5.14*** | 0.148 | MdCENa_S | Af, Am, D | 0.603 | –0.245 | –0.228 | ||
| 17 | 5.42*** | 0.179 | MS06g03_G | Af, Am | 0.575 | –0.446 | –0.115 | ||
| NSI | 3 | 5.54*** | 0.154 | NZmsMdMYB12_S | Af, Am, D | 0.071 | 1.120 | –0.664 | |
| 3 | 4.03** | 0.102 | MdCENa_S | Af, D | 0.788 | –0.360 | 0.782 | ||
| 10 | 4.48** | 0.155 | MS06g03_G | Am, D | 0.354 | –1.311 | –1.807 | ||
| 17 | 6.38*** | 0.191 | MdLD_G | Af, Am | 1.047 | –1.016 | –0.237 |
QTLs for traits related to the number of inflorescences:
The 12 QTLs detected for characters related to the number of inflorescences were spread across seven different LGs (Table 3). The explained genetic variability (R2) for each of the 12 QTLs ranged from 10.4% (precocity, PI_inf) to 24% (cumulative yield, CY_inf). Nine and four inflorescence QTLs were detected on the STK and GS parental genetic maps, respectively, with eight and three of them confirming positions identified using the consensus map. No significant QTL was detected for inflorescence yield for the years 4, 5, and 7 (Y_inf_4, Y_inf_5, and Y_inf_7).
Using the consensus map, three QTLs were detected for biennial bearing (BBI_inf): at the top of LG4 and LG8 and at the bottom of LG10. The global linear model indicated an interaction between the BBI_inf QTLs on LG8 and LG10 and explained 50% of the genetic variability (Table 4). The two BBI_inf QTLs mapped on LG4 and LG8 exhibited female effect and were confirmed on the STK parental map using the same cofactors as the consensus map, Hi04c10x_SG and Hi04b12_S, respectively. The third BBI_inf QTL detected on LG10 mainly resulted in male effect and co-located with a QTL for precocity (PI_inf) (Fig. 4).
Table 4.
Global model estimations for traits with several QTLs detected by MQM with P, the effect probability, and global R2, the proportion of variation explained by the model.
| Trait | LG | Effects | Cofactor | P-value | Global R2 |
| BBI_inf | 4 | Hi04c10×_SG | Hi04c10x_SG | 0.0017 | 0.49 |
| 8 | Hi04b12_S | CH02g09_SG | 0.0068 | ||
| 10 | MdGA2ox8a_G | COL_SG | 3.81E-05 | ||
| 8*10 | CH02g09_SG*COL_SG | 0.0134 | |||
| Y_inf_6 | 1 | MdGA20ox1a_S | CH05g08_SG | 6.54E-05 | 0.24 |
| 8 | Hi04b12_S | CH02g09_SG | 0.0539 | ||
| PI_inf | 3 | NZmsMdMYB12_S | CH03e03_SG | 9.72E-05 | 0.31 |
| 7 | MdSOC1b_S | Hi03a10_SG | 0.0024 | ||
| 10 | MS06g03_G | COL_SG | 0.0094 | ||
| BBI_fruit | 10 | MdAFB6_S | COL_SG | 0.0002 | 0.37 |
| 13 | CH03h03z_SG | CH03h03z_SG | 0.0003 | ||
| 10*13 | CH03h03z_SG*COL_SG | 0.0267 | |||
| Y_fruit | 1 | MdGA20ox1a_S | CH05g08_SG | 4.11E-05 | 0.29 |
| 8 | MdPI_SG | CH02g09_SG | 0.0022 | ||
| Y_fruit_2 | 1 | MdGA20ox1a_S | CH05g08_SG | 4.83E-05 | 0.29 |
| 8 | MdPI_SG | CH02g09_SG | 0.0026 | ||
| Y_fruit_3 | 5 | CH04e03_SG | CH04e03_SG | 5.12E-05 | 0.49 |
| 11 | GD_SNP01140_SG | GD_SNP01140_SG | 1.95E-06 | ||
| 5*11 | CH04e03_SG*GD_SNP01140_SG | 3.37E-05 | |||
| CY_fruit | 1 | MdGA3ox_like_b_S | CH05g08_SG | 2.90E-05 | 0.40 |
| 8 | MdPI_SG | CH02g09_SG | 0.0010 | ||
| 1*8 | CH05g08_SG:CH02g09_SG | 0.0626 | |||
| PI_fruit | 3 | CH03g07_SG | CH03g07_SG | 0.0001 | 0.71 |
| 5 | CH03a04_S | CH04e03_SG | 0.0033 | ||
| 11 | NZ04h11y_G | CH04g07_SG | 0.1705 | ||
| 3*5*11 | CH03g07_SG:CH04e03_SG:CH04g07_SG | ||||
| BBI_mass | 10 | MdAFB6_S | COL_SG | 0.0042 | 0.23 |
| 13 | CH03h03z_SG | CH03h03z_SG | 0.0006 | ||
| Y_mass_3 | 1 | B2-T7_S | CH05g08_SG | 0.0338 | 0.15 |
| 5 | CH02a08z_S | CH05f06_SG | 0.0056 | ||
| PI_mass | 3 | CH03g07_SG | CH03g07_SG | 0.0002 | 0.70 |
| 5 | CH04e03_SG | CH04e03_SG | 0.0098 | ||
| 11 | CH04g07_SG | CH04g07_SG | 0.1999 | ||
| 3*5*11 | CH03g07_SG:CH04e03_SG:CH04g07_SG | ||||
| NSF | 3 | CH03e03_SG | CH03e03_SG | 0.0002 | 0.65 |
| 3 | MdCENa_S | Hi04c10y_SG | 0.0000 | ||
| 17 | MdLD_G | CH05d08y_SG | 0.0002 | ||
| 3*3*17 | CH03e03_SG:Hi04c10y_SG:CH05d08y_SG | ||||
| NSI | 3 | NZmsMdMYB12_S | CH03e03_SG | 0.0037 | 0.85 |
| 3 | MdCENa_S | Hi04c10y_SG | 0.0010 | ||
| 10 | MS06g03_G | COL_SG | 0.2828 | ||
| 17 | MdLD_G | CH05d08y_SG | 0.0031 | ||
| 3*3*10 | CH03e03_SG:Hi04c10y_SG:COL_SG | ||||
| 3*3*17 | CH03e03_SG:Hi04c10y_SG:CH05d08y_SG |
Models were selected according to AIC values. Some of the markers used in MapQTL as cofactors were replaced by their nearest marker with four genetic classes (ab, bc, ad, and bd, or ef, eg, fg, and ee) for the model construction. For trait abbreviations, see Table 2.
Strong effect QTLs for inflorescence yield and cumulative yield (Y_inf_2, Y_inf, and CY_inf) clustered at the top of LG15 of the STK×GS map and explained 22.9, 22.6, and 24%, respectively, of the genetic variability. These QTLs were confirmed on both parental maps in the same genomic regions (Fig. 4). Although QTLs were detected on LG15 for Y_inf_6 on both parental maps, when the consensus map was used, QTLs for this trait were identified on other genomic regions (LG1 and LG8). NZ02B01_S was used as the cofactor for the QTLs detected on LG15 on both consensus and female maps, and the MdCCD8b_SG marker was the cofactor for the QTLs detected on the male parental map.
The QTLs detected on LG1 for annual inflorescence yield for years 3 and 6 (Y_inf_3 and Y_inf_6) mapped in the same genomic region on the consensus map and used the same cofactor (MdGA20ox1a_S) (Table 3). They explained 19.7% and 13.6% of the variability, respectively, and both were confirmed on the STK genetic map using CH05g08_SG as the cofactor. A second QTL identified on LG8 for Y_inf_6 was confirmed on the STK map; both maps used Hi04b12_S as the cofactor and co-located with a QTL for BBI_inf. The two QTLs on LG1 and LG8 were not involved in any epistasic effect and explained 24% of the genetic variability (Table 4).
Three QTLs were detected on the consensus genetic map for precocity (PI_inf) on LG3, LG7, and LG10 (Table 3). None of these QTLs was confirmed on parental maps. The selected global linear model for this character showed no interactions among the three QTLs and they together explained 31% of the variability (Table 4). The LG3 and LG10 QTLs resulted in a male additivity effect, whereas the QTL mapped on LG7 mainly resulted in female effect.
QTLs for traits related to the number and the mass of harvested fruit:
Fifteen and eight QTLs were mapped on the consensus map for variables related to the number and to the mass of harvested fruit, respectively (Table 3). For the number of harvested fruit, QTLs were spread across seven different LGs, with LG1 and LG8 exhibiting the highest number of QTLs. Their explained genetic variability ranged from 12.3% (fruit yield independent of year effect, Y_fruit) to 19.1% (cumulated yield, CY_fruit). Seven and four QTLs were confirmed using the parental genetic maps STK and GS, respectively. No significant QTLs were detected for fruit yield of years 4 and 6 (Y_fruit_4 and Y_fruit_6). For the mass of harvested fruit, no QTLs were mapped using the parental maps. QTLs were spread over six different LGs and were related to four variables: annual mass yield for years 3 and 7 (Y_mass_3, Y_mass_7), precocity (PI_mass) and biennial bearing (BBI_mass).
Two genomic regions were identified for the BBI (BBI_fruit and BBI_mass) on LG10 and LG13 (Fig. 4). QTLs for BBI_fruit and BBI_mass were similarly located and the same cofactors were used for both traits. The interactions between the cofactors were significant for BBI_fruit in the global linear model and explained 37% of the genotypic variability (Table 4). The QTLs detected on LG10 displayed mainly male and female effects, whereas on LG13 the QTLs were mainly due to dominance and female effects (Table 3). The LG10 and LG13 BBI_fruit QTLs were confirmed using the GS genetic map.
Five QTLs related to fruit yield were detected on LG1 using the consensus map: four for the yield (Y_fruit_2, Y_fruit_5, Y_fruit, and Y_mass_3) and one for the CY (CY_fruit) (Fig. 4). The QTLs detected on the consensus map for Y_fruit_2, Y_fruit, and CY_fruit were confirmed on both parental maps using MdGA3ox-like-b_S and MdSOC1-like_G markers on STK and GS maps, respectively. Both QTLs displayed female, male, and dominant effects. The Y_fruit_5 QTL resulted in female, male, and dominant effects and was not confirmed on the parental maps, probably because of its low LOD score (Table 3).
A second QTL cluster was identified on LG8 for variables related to annual and cumulated yields of harvested fruit: Y_fruit_2, Y_fruit_7, Y_fruit, and CY_fruit. Their explained genotypic variability ranged from 12.2% (Y_fruit) to 17.6% (Y_fruit_7) (Table 3). These four QTLs were confirmed on the STK genetic map and MdPI_SG was used as the cofactor on both maps.
For the Y_fruit_2, Y_fruit, and CY_fruit QTLs mapped on both LG1 and LG8, the global linear model included interaction between LG1 and LG8 only for CY (CY_fruit), explaining 40% of the genetic variability (Table 4).
The annual yield of year 3 (Y_mass_3) QTLs on LG1 mainly resulted in male additivity and explained 17.9% of the variability (Table 3). A second Y_mass_3 was detected on LG5 and displayed a female effect. For this variable, the interaction between the cofactors was not significant in the global linear model and explained 15% of the genetic variability (Table 4).
Two QTLs were detected on the consensus map for fruit yield of year 3 (Y_fruit_3) on LG5 and LG11, with epistatic effect explaining together 49% of the genetic variability. No QTLs were mapped on the parental maps for this variable. However, the QTLs for both fruit and mass yields of year 3 (Y_fruit_3 and Y_mass_3) on LG5 co-located with QTLs for precocity (PI_fruit and PI_mass) (Fig. 4).
Three QTLs were detected for the PI of fruit and mass (PI_fruit and PI_mass) on LG3, LG5, and LG11 of the consensus map. The global linear model showed that the LG11 QTL was only involved in epistasic effect and showed significant epistatic effect, with the three QTLs explaining together 71% and 70% of the genetic variability for PI_fruit and PI_mass, respectively (Table 4). The QTLs displayed female and male additivity, and also important dominant effects, and were not detected on the parental maps.
Number of fruit per inflorescences and number of seed per fruit:
The QTL detected for the NFI on LG8 resulted mainly in male additivity effect and explained 21.5% of the genetic variability (Table 3). MdPI_SG was used as the cofactor and the QTL co-located with QTLs mapped for fruit yield.
Three QTLs were mapped for the NSF: two on LG3 and one on LG17 (Fig. 4). The QTLs exhibited female, male, and dominance effects, but none of these QTLs was confirmed on parental maps. The global linear model included an interaction among the three QTLs and explained 65% of the genetic variability (Table 4). The QTL mapped on the top of LG3 co-located with the QTL for the PI of inflorescences (PI_inf).
The QTLs detected for the NSI were very similar to those described for NSF; however, a fourth QTL was detected at the bottom of LG10. The global linear model included an interaction among the four QTLs and explained 85% of the genetic variability (Table 4).
Co-location between candidate genes and QTLs
The QTL cluster detected for inflorescence and fruit yields at the bottom of LG1 overlaid four candidate genes genetically mapped in a small genomic region of 13 cM: MdSOC1-like, MdGA20ox1a, MdBFTa, and MdGA3ox-like-b. According to the ‘Golden Delicious’ genome sequence, a COL gene, MdCOL1, is also located in the same genomic region; however, this could not be genetically mapped in the present population. On the consensus map, MdSOC1-like and MdGA20ox1a were located within the QTL interval of annual yields (Y_inf_3, Y_inf_6, Y_fruit_2, Y_fruit_5, and Y_fruit), while MdBFTa and MdGA3ox-like-b co-located with the cumulative fruit yield (CY_fruit) QTL. When the parental maps were used, the candidate gene co-location with the QTL differed slightly. On the STK genetic map, only MdBFTa and MdGA3ox-like-b co-located with the QTL cluster, and on the GS parental map, MdSOC1-like was at the limit of the LOD significance for the QTL cluster.
The candidate gene MdMADS4a mapped on LG2 was located at the external border of the LOD significance for the yield mass QTL of year 7 (Y_mass_7).
On LG3, the peak LOD score for the QTLs for precocity (PI_inf), NSF, and NSI was located right above the transcriptional factor MdMYB12. Although QTLs for precocity (PI_fruit and PI_mass) were located in the same genomic region, MdMYB12 did not map within the QTL confidence interval. The LOD peaks of the NSF and NSI QTLs located in the middle of LG3 were positioned directly above MdCENa. For the second PI_inf QTL on LG7, the MdSOC1b candidate gene was located within the LOD score interval.
MdEFL3a mapped within the limit of the QTL intervals for Y_inf_6 and BBI_inf QTLs on LG8. In silico mapping revealed that five other candidate genes, MdARF10, MdARF110, MdIAA4, MdIAA25, and MdGA3ox1a, were present within these QTL intervals.
The candidate gene MdPI was located right in the middle of the LOD significance interval for the QTL cluster mapped on LG8 relating to fruit production and to the number of fruit per inflorescence. On LG10, the QTL cluster related to BBI and to the precocity of flowering spanned MdGA2ox8a and MdAFB6. For these four QTLs, the LOD score was higher at the MdAFB6 locus than it was at MdGA2ox8a. The NSI QTL mapped above this QTL cluster, and several candidate genes were located within the QTL interval: MdARF3, MdGA2ox2b, MdPHYEb, and MdGA2ox8a. On LG15, the LOD score of the inflorescence QTL cluster fell 2 cM before the position of MdCCD8b on both male and consensus maps. The LG17 QTLs for NSF and NSI co-located with the MdLD candidate gene.
Flowering genes such as MdFT, MdMFT, MdTFL1, MdCEN, MdLHP1, MdAFL, MdAP1, and MdMADS4, and hormone-related genes such as MdRGL did not co-locate with any QTL mapped.
Discussion
The challenge of quantifying alternate bearing
Quantifying biennial bearing, a physiological phenomenon that occurs over a range of years, is a complex task. The approach here utilized data collected over 7 years from an apple segregating population, including the juvenile phase and the entrance into mature phase. The calculation of indices to quantify biennial bearing was essential both to describe the genetic variability and to identify the genomic regions linked to this trait.
Most of the trees first flowered during their third year and their production increased during the experiment. BBI was calculated using 5 years of yield (i.e. from the third to the seventh year), although Huff (2001) recommended using BBI over a minimum of 6 years during the trees’ mature phase. Indeed, the BBI values can change, depending on the number of annual yields included in the calculation, and between the juvenile and mature phases (Smith et al., 2004). However, these recommendations do not allow the early evaluation of biennial bearing tendency among genotypes, which would be useful for breeders. The results suggest that an early evaluation is possible, since genotypes having a clear alternate behaviour; that is, characterized by two ‘on’ and two ‘off’ years, had higher BBI values than genotypes having a clear regular behaviour (Fig. 3). However, intermediate behaviours might be difficult to characterize based on BBI. One major difficulty derived from the fact that this index includes positive yield differences due to the ontogenic increasing trend of tree production. Estimation of biennial bearing during this period would certainly be improved by removing the increasing trend and accounting for yearly fluctuations only.
In addition to indexes, yearly variables were also studied. Most of them were significantly affected by the year factor effect and its interaction with the genotype factor (Table 2). However, the experimental design did not allow a distinction to be made between the ontogenic and climatic year effects within the year effect, as previously proposed by Segura et al. (2008). The interaction G×Y is illustrated by the graphic representations of bearing behaviours over years (Fig. 3), which show that ‘on’ and ‘off’ years can occur during the same climatic year, depending on the genotype. In addition, genotype by environment interactions can also be expected, since previous studies demonstrated different bearing behaviours for the same cultivar, depending on the cultivation site (for a review, see Monselise and Goldschmidt, 1982).
BLUPs were used as a tool to predict the genetic merit of trees based on their field performance for the traits studied. QTL detection was performed based on BLUP values, in order to improve the statistical power to detect significant QTLs (see Segura et al., 2009). Among the traits studied, the number of inflorescences per tree was the most accurate for quantification of production and its regularity, because this trait is less subject to environmental effect. The significance of the genetic effects was higher for the variables related to the number of inflorescences than for the variables related to the number and the mass of harvested fruit (Table 2). This might be due to environmental effects that would induce variability during fruit set, self-thinning, and fruit development.
QTL detection, clustering, and trait correlation
The higher number of QTLs for STK suggests a greater effect of this parent on these traits and is consistent with the strong tendency towards the biennial bearing characteristic of STK compared with the regular bearing GS (Lespinasse, 1992). This also suggests that biennial bearing in the studied population may be due to alleles that have a negative effect rather than to positive regular bearing alleles.
QTL clusters were identified on eight genomic regions, and several of these clusters were due to indices calculated from a set of measured variables, resulting in QTL co-location between the index and the variables. For instance, production precocity was calculated from measured yearly fruit and mass yields. Strong correlations (0.66 and 0.74 for fruit and mass, respectively) were found between the yield in the third year (i.e. the first year of significant production) and the calculated production precocity, which in turn resulted in a QTL cluster mapping on LG5 (Fig. 4; Supplementary Table S1 at JXB online). This emphasized the third year of production as being highly determinant for precocity.
However, several statistical correlations and QTL co-locations occurred for independent variables that result from common physiological processes. This can be exemplified by QTLs for inflorescence yield of a given year that co-located with QTLs for fruit yield of the next year on LG1 and were consistent with negative correlations between the variables, for example Y_fruit_5 and Y_Inf_6, –0.54. These results corroborate the main hypothesis for biennial bearing in apple that the presence of fruit influences the formation of inflorescences the following year, and point to the base of LG1 as being highly determinant for biennial bearing.
Another example is the fruit yield QTLs that clustered with a QTL for the NFI on LG8. These co-locations are supported by significant correlations between variables (ranging from 0.55 to 0.99). However, lower correlations were found between the number of inflorescences and fruit yield (from 0.18 to 0.51). Therefore, the fruiting yield appears to be influenced more by the NFI than by the number of inflorescences per tree.
Most interestingly, a number of QTL clusters resulted from non-correlated variables. For instance, the NSI mapped adjacent to QTLs for biennial bearing for inflorescence, fruit number, and mass on LG10, despite a low correlation between these variables (correlations ranging from 0.10 to 0.16). Similarly, the co-location between a QTL for flowering precocity with QTLs for biennial bearing on LG10 opens up interesting breeding perspectives. Indeed, these variables showed moderate correlations (0.32), meaning that some genotypes are precocious and regular, whereas others are precocious and biennial. Genotypes that are both precocious and irregular represent the largest proportion within the population, suggesting that in general trees producing flowers in early stages might enter in a biennial bearing cycle. Despite this, breeders wish to select genotypes that combine precocity and regularity, and these were present in the population studied.
Emerging hypotheses for the control of biennial bearing
This study used QTL detection in a segregating population combined with a candidate gene mapping strategy to identify potential genetic determinants of biennial bearing. It was demonstrated that biennial bearing involves interactions between independent genomic regions spanning genes of various functions. While stable genetic transformation to overexpress or knock out genes often provides solid proof of function for plant genes, it is believed that using genetic transformation would be extremely challenging for dissecting biennial bearing because of the control system's complexity. It is impossible to generate phenotypes equivalent to biennial bearing in annual model plants using genetic transformation. Knocking out TFL1 in apple resulted in an extreme reduction of the juvenile phase with collateral effects on inflorescence architecture and chilling requirements (Kotoda et al., 2003, 2006). However, in the present study, QTLs for precocity did not co-locate with TFL1, suggesting that mechanisms upstream of TFL1, and possibly controlling TFL1, are determinants in the decision to set flowers.
The availability of the apple genome sequence enabled a comprehensive search for a set of candidate genes involved in flowering, hormones, and branching to be performed and their position could be compared with those of the QTLs detected for biennial bearing in the STK×GS genetic map. This enabled the fact to be highlighted that co-location between QTL clusters and candidate genes, which is not definitive evidence, provides pertinent new information on putative genetic control of biennial bearing in apple. While the approach used here was based on a systematic search of candidate genes using in silico analysis, it was shown that 24% of the genetic markers mapped to a different location from that in the genome assembly, which points at discrepancies in the apple genome assembly. This indicates that some relevant candidate genes might have been left out from the QTLs because of wrong genome location.
Flowering integrator genes and biennial bearing:
It has been shown that genes described as key flowering genes in Malus, such as MdFT and MdTFL1 (Mimida et al., 2009; Kotoda et al., 2010), were not present within QTL intervals for annual yields, precocity, and biennial bearing. Although it is suggested that these genes are likely not to be directly responsible for biennial bearing in apple tree, their control and regulation could be determinant. In contrast, other flowering genes such as MdBFTa, MdSOC1-like, and MdCOL1 were located within QTL intervals for inflorescence and fruit production mapping on LG1. In Arabidopsis, BFT possesses a TFL1-like activity and functions redundantly with TFL1 in inhibition of inflorescence meristem development (Yoo et al., 2010). SOC1 co-located with QTLs on LG1 and LG7 for inflorescence and fruit production and precocity, respectively. In annual plants, SOC1 enhances FI in response to GAs (GA4) (Eriksson et al., 2006). In apple, GA4 has been shown to promote flowering during ‘off’ years when applied the year before (Looney et al., 1985). Similarly, CO positively regulates the expression of two floral integrators, LFY and SOC1, via FT in Arabidopsis (Samach et al., 2000; Parcy, 2005). However, based on QTL mapping, it cannot be determined which gene among MdSOC1, MdBFTa, and MdCOL1 is causative for the LG1 QTL. Further study, including mRNA expression during FI with different applications of hormones (e.g. different forms of GA), is needed.
Homeotic genes and fruit yield:
Homeotic genes that are involved downstream of FI would not be a priori candidate genes for FI. However, MdPI co-located with a QTL cluster for fruit production and for the number of fruit per inflorescence on LG8. Previous studies have suggested that MdPI could be responsible for seed development, after the observation that a seedless apple mutant has a mutated PI gene (Yao et al., 2001), whereas Tanaka et al. (2007) proposed that the MdPI gene was related to the development of petals and stamens and had function equal to Arabidopsis PI (Coen and Meyerowitz, 1991; Weigel and Meyerowitz, 1994). Consistently with Tanaka et al. (2007), the QTL cluster on LG8 does not control the NSF but the NFI.
Plant hormones and biennial bearing:
Several genes involved in the GA biosynthesis pathway were located in QTL cluster intervals for production and its alternation: MdGA20ox1a and MdGA3ox-like-b on LG1 and MdGA2ox8a on LG10. These genes are known to determine the final amount of bioactive GA through their influence on key steps of GA synthesis (reviewed by Hedden and Phillips, 2000). Furthermore, auxin-related genes (MdAFB6) were mapped in the interval of QTLs for biennial bearing on LG10, and in silico mapping suggested that some AUX/IAA and ARF genes might be also located on LG8 and LG13. In pea, auxin has an important role in regulating GA biosynthesis by inducing the accumulation of PsGA3ox1 mRNA and reducing the PsGA2ox1 transcript, increasing the amount of bioactive GA1 (Ross et al., 2000). Bioactive GA might also directly target key flowering genes in the shoot apical meristem, including SOC1 and LFY, which have been shown to be regulated by GA4 in Arabidopsis (Eriksson et al., 2006). LFY has been proposed to have a role in apple tree architecture and be responsible for columnar phenotype (Flachowsky et al., 2010). In the monocot Lolium temulentum, it has been shown that GA5 and GA6 are the active GAs in the induction of flowering (King et al., 2003). However, in apple, there is no evidence about which GAs are active in FI, although GAs are known to have an opposite effect on FI in perennial and annual plants (Jackson and Sweet, 1972). Indeed, applications of GA to apple trees showed that GA7 is the most inhibitory GA on FI (Tromp, 1982), and horticultural practices commonly involve the application of GA during ‘off’ years to prevent an excessive FI and so attenuate the biennial bearing cycle (Schmidt et al., 2009). Bioactive GAs might thus be expected to have an inhibitory effect on key flowering genes/steps in apple.
Is there a common genetic determinism for tree architecture and biennial bearing?:
Branching intensity and spur extinction have been demonstrated to be correlated with biennial bearing in a set of apple cultivars (Lauri et al., 1995, 1997). More precisely, spur-type cultivars have often been described as having an irregular fruiting behaviour (Looney and Lane, 1984). Since the STK×GS population was previously used for dissecting the genetic control of scion architecture during the first 3 years of growth (Segura et al., 2009), the comparison of QTL positions between the two studies enabled a number of QTLs mapping to common locations to be highlighted. QTLs for branching intensity were found to co-locate with QTL clusters for biennial bearing on LG4 and LG13, as well as with QTLs for flower and fruit production on LG1. This may result from statistical correlation between traits, since an increase in branching intensity increases the number of flowering sites. This is an important component of flowering yield that can be managed by horticultural practices such as ‘spur artificial extinction’ (short fruiting shoot thinning) (Lauri, 2002), which mitigates biennial bearing. Moreover, the CCD8 gene, which is involved in branching in petunia (Snowden et al., 2005), was located at the border of the QTL cluster for inflorescence yield on LG15. Although no QTL for vegetative branching traits was located on this LG in the previous study during the first years of tree development, it is suggested that variations in the MdCCD8b gene might possibly have an effect on axillary bud activity and on the number of flowering sites.
A QTL for mean internode length of proleptic axillary shoots was co-located with a biennial bearing QTL on LG4. This corroborates the previous assumption of a positive correlation between bourse shoot length (shoots growing from inflorescence bases) and return bloom (consecutive occurrence of flowering in years n and n+1 on two successive shoots). That resulted from the observation that bourse shoot length >10 mm appeared to override the negative effect of seed on FI (Neilsen and Dennis, 2000). Finally, the biennial bearing QTLs on LG10 were located close to the genomic region that includes the columnar locus responsible for compact growth habit (Hemmat et al., 1997) and a pleiotropic effect for architectural traits in apple (Conner et al., 1998; Kenis and Keulemans, 2007). As previously discussed, this region also included candidate genes for GA biosynthesis and degradation. Numerous studies have demonstrated the implication of GAs in the cell elongation process. In particular, GA4 has been shown to be the active GA in the regulation of cell elongation and shoot growth in Arabidopsis (Xu et al., 1997), as well as in the regulation of stem elongation in L. temulentum (King et al., 2001).
Taken together, these factors, including QTL co-locations, and mapping of candidate genes associated with vegetative growth, branching, and FI to QTLs associated with biennial bearing, strongly support the hypothesis of common molecular controls for tree architecture and biennial bearing in apple.
Conclusion
This study of biennial bearing in segregating apple progeny has provided new knowledge concerning the genetic architecture of this complex character. Biennial bearing is clearly a multigenic trait that is influenced by plant age and year effect as well as genetic effects. The comparison of locations of QTLs with candidate genes has given a clear indication that biennial bearing is unlikely to be directly controlled by floral integrator or meristem identity genes. However, their control by hormones might be the determinant factor in the decision to flower, consequently leading to biennial bearing. Even if not a definite indication of the exact physiological process, several genes related to metabolism, degradation, and transport of GA and auxin co-located with QTLs for biennial bearing, and these genes could regulate the amounts of substances inhibiting floral induction in the shoot apical meristem.
It is proposed that the candidate genes may act on physiological processes believed to be involved in biennial bearing and might be the genetic determinants of biennial bearing. However, further analyses are needed to narrow down the list of candidate genes and to confirm their implication in biennial bearing.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Phylogenetic analysis of flowering genes from Arabidopsis thaliana and Malus×domestica.
Figure S2. Phylogenetic analysis of hormone- and branching-related genes from Arabidopsis thaliana and Malus×domestica.
Figure S3. Physical position (Mb) of genetic markers (black) and candidate genes on the 17 chromosomes (Chr) of the apple genome (‘Golden Delicious’).
Figure S4. Variability of the number of inflorescences per tree between replicates of the same genotype.
Table S1. Genetic correlations between variables observed for the number of inflorescences, the number of fruit harvested, and the mass of fruit harvested.
Table S2. List of 120 candidate genes related to flowering identified in silico in the ‘Golden Delicious’ apple genome.
Table S3. Accession numbers of gene predictions in the apple genome and sequences of primers developed for candidate gene mapping in the population STK×GS.
Table S4. List of 55 candidate genes related to gibberellin and cytokinin oxidases and to carotenoid cleavage dioxygenase identified in silico in the ‘Golden Delicious’ apple genome.
Supplementary Material
Acknowledgments
We thank V. Segura, G. Garcia, S. Martinez, and S. Ferals for their contribution to field measurements and constitution of the first data set. We also thank S. Bulley, V. Bus, P. É. Lauri, S. Tustin, and R. Volz for fruitful discussions, as well as T. Foster and R. Schaffer for critical reading of the manuscript. We are grateful to K. David and R. Schaffer for providing the position of the AUX/IAA and ARF genes. This work was supported by the Plant Breeding Department of the National Institute of Agronomic Research of France (INRA), by Plant & Food Research (Pipfruit Internal Investment Project), and by the New Zealand Ministry of Science and Innovation [Horticultural Genomics programme (CO6X0810)]. BG, DC, and EC would like to thank the New-Zealand–France bilateral Dumont d'Urville programme for travel support.
Glossary
Abbreviations
- AIC
Akaike Information Criterion; BBI, Biennial Bearing Index
- BLAST
Basic Local Alignment Search Tool
- BLASTP
protein–protein BLAST
- BLUP
Best Linear Unbiased Predictor
- CK
cytokinin
- CY
cumulative yield
- EST
expressed sequence tag
- FI
floral induction
- GA
gibberellic acid
- GS
‘Granny Smith’
- HRM
high resolution melting
- MRM
multiple QTL mapping; NFI, number of fruit per inflorescence
- NSF
number of seed per fruit
- NSI
number of seed per inflorescence
- PI
Precocity Index
- QTL
quantitative trait loci
- SNP
single nucleotide polymorphism
- SSR
simple sequence repeat
- STK
‘Starkrimson’
- WGD
whole-genome duplication
References
- Bangerth F. Flower induction in perennial fruit trees: still an enigma? Acta Horticulturae. 2006;727:177–195. [Google Scholar]
- Bangerth F. Floral induction in mature, perennial angiosperm fruit trees: similarities and discrepancies with annual/biennial plants and the involvement of plant hormones. Scientia Horticulturae. 2009;122:153–163. [Google Scholar]
- Barritt BH, Konishi B, Dilley M. Tree size, yield and biennial bearing relationships with 40 apple rootstocks and three scion cultivars. Acta Horticulturae. 1997;451:105–112. [Google Scholar]
- Bernier G, Perilleux C. A physiological overview of the genetics of flowering time control. Plant Biotechnology Journal. 2005;3:3–16. doi: 10.1111/j.1467-7652.2004.00114.x. [DOI] [PubMed] [Google Scholar]
- Boss PK, Bastow RM, Mylne JS, Dean C. Multiple pathways in the decision to flower: enabling, promoting, and resetting. The Plant Cell. 2004;16:18–31. doi: 10.1105/tpc.015958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buban T, Faust M. Flower bud induction in apple trees: internal control and differentiation. Horticultural Reviews. 1982;4:174–263. [Google Scholar]
- Chagné D, Carlisle C, Blond C, et al. Mapping a candidate gene (MdMYB10) for red flesh and foliage colour in apple. BMC Genomics. 2007;8:212. doi: 10.1186/1471-2164-8-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan B, Cain J. The effect of seed formation on subsequent flowering in apple. Journal of the American Society for Horticultural Science. 1967;91:63–67. [Google Scholar]
- Cilas C, Montagnon C, Bar-Hen A. Yield stability in clones of Coffea canephora in the short and medium term: longitudinal data analyses and measures of stability over time. Tree Genetics and Genomes. 2011;7:421–429. [Google Scholar]
- Coen ES, Meyerowitz EM. The war of the whorls: genetic interactions controlling flower development. Nature. 1991;353:31–37. doi: 10.1038/353031a0. [DOI] [PubMed] [Google Scholar]
- Conner PJ, Brown SK, Weeden NF. Molecular-marker analysis of quantitative traits for growth and development in juvenile apple trees. Theoretical and Applied Genetics. 1998;96:1027–1035. [Google Scholar]
- Corbesier L, Vincent C, Jang S, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science. 2007;316:1030–1033. doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- Dereeper A, Guignon V, Blanc G, et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Research. 2008;36:465–469. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S, Böhlenius H, Moritz T, Nilsson O. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. The Plant Cell. 2006;18:2172–2181. doi: 10.1105/tpc.106.042317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esumi T, Tao R, Yonemori K. Isolation of LEAFY and TERMINAL FLOWER 1 homologues from six fruit tree species in the subfamily Maloideae of the Rosaceae. Sexual Plant Reproduction. 2005;17:277–287. [Google Scholar]
- Flachowsky H, Hättasch C, Höfer M, Peil A, Hanke MV. Overexpression of LEAFY in apple leads to a columnar phenotype with shorter internodes. Planta. 2010;231:251–263. doi: 10.1007/s00425-009-1041-0. [DOI] [PubMed] [Google Scholar]
- Foster T, Johnston R, Seleznyova A. A morphological and quantitative characterization of early floral development in apple (Malus×domestica Borkh.) Annals of Botany. 2003;92:199–206. doi: 10.1093/aob/mcg120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster T, Kirk C, Jones W T, Allan A C, Espley R, Karunairetnam S, Rakonjac J. Characterisation of the DELLA subfamily in apple (Malus×domestica Borkh.) Tree Genetics and Genomes. 2006;3:187–197. [Google Scholar]
- Grochowska M, Karaszewska A. The production of growth promoting hormones and their active diffusion from immature, developing seeds of four apple cultivars. Fruit Science Reproduction. 1976;3:5–16. [Google Scholar]
- Hanke M, Flachowsky H, Peil A, Hättasch C. No flower no fruit. Genetic potentials to trigger flowering in fruit trees. Genes, Genomes and Genomics. 2007;1:1–20. [Google Scholar]
- Harley C. Investigations on the cause and control of biennial bearing of apple trees. USDA Technical Bulletin. 1942;792:1–58. [Google Scholar]
- Hedden P, Phillips AL. Gibberellin metabolism: new insights revealed by the genes. Trends in Plant Science. 2000;5:523–530. doi: 10.1016/s1360-1385(00)01790-8. [DOI] [PubMed] [Google Scholar]
- Hemmat M, Weeden NF, Conner PJ, Brown SK. A DNA marker for columnar growth habit in apple contains a simple sequence repeat. Journal of the American Society for Horticultural Science. 1997;122:347–349. [Google Scholar]
- Hoad GV. The role of seed-derived hormones in the control of flowering in apple. Acta Horticulturae. 1978;80:93–103. [Google Scholar]
- Hoblyn T, Grubb N, Painter A, Wates B. Studies in biennial bearing. I. Journal of Pomology Horticultural Science. 1936;14:39–76. [Google Scholar]
- Hoffmann M, Geulen O, Weilke C. The LightCycler 480 real-time PCR system: a versatile platform for genetic variation research. Nature Methods. 2008 5. [Google Scholar]
- Huff A. A significance test for biennial bearing using data resampling. Journal of Horticultural Science and Biotechnology. 2001;76:534–535. [Google Scholar]
- Jackson DI, Sweet GB. Flower initiation in temperate woody plants. A review based largely on the literature of conifers and deciduous fruit trees. Horticultural Abstracts. 1972;42:9–24. [Google Scholar]
- Jeong D, Sung S, An G. Molecular cloning and characterization of constans-like cDNA clones of the Fuji apple. Journal of Plant Biology. 1999;42:23–31. [Google Scholar]
- Jonkers H. Biennial bearing in apple and pear: a literature survey. Scientia Horticulturae. 1979;11:303–307. [Google Scholar]
- Kenis K, Keulemans J. Study of tree architecture of apple (Malus × domestica Borkh.) by QTL analysis of growth traits. Molecular Breeding. 2007;19:193–208. [Google Scholar]
- King KE, Moritz T, Harberd NP. Gibberellins are not required for normal stem growth in Arabidopsis thaliana in the absence of GAI and RGA. Genetics. 2001;159:767–776. doi: 10.1093/genetics/159.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King RW, Evans LT, Mander LN, Moritz T, Pharis RP, Twitchin B. Synthesis of gibberellin GA6 and its role in flowering of. Lolium temulentum. Phytochemistry. 2003;62:77–82. doi: 10.1016/s0031-9422(02)00447-8. [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Kaya H, Goto K, Iwabuchi M, Araki T. A pair of related genes with antagonistic roles in mediating flowering signals. Science. 1999;286:1960–1962. doi: 10.1126/science.286.5446.1960. [DOI] [PubMed] [Google Scholar]
- Kotoda N, Hayashi H, Suzuki M, et al. Molecular characterization of FLOWERING LOCUS T-Like genes of apple (Malus×domestica Borkh.) Plant and Cell Physiology. 2010;51:561–575. doi: 10.1093/pcp/pcq021. [DOI] [PubMed] [Google Scholar]
- Kotoda N, Iwanami H, Takahashi S, Abe K. Antisense expression of MdTFL1: a TFL1-like gene, reduces the juvenile phase in apple. Journal of the American Society for Horticultural Science. 2006;131:74–81. [Google Scholar]
- Kotoda N, Wada M. MdTFL1: a TFL1-like gene of apple, retards the transition from the vegetative to reproductive phase in transgenic Arabidopsis. Plant Science. 2005;168:95–104. [Google Scholar]
- Kotoda N, Wada M, Komori S, Kidou S, Abe K, Masuda T, Soejima J. Expression pattern of homologues of floral meristem identity genes LFY and AP1 during flower development in apple. Journal of the American Society for Horticultural Science. 2000;125:398–403. [Google Scholar]
- Kotoda N, Wada M, Kusaba S, Kano-Murakami Y, Masuda T, Soejima J. Overexpression of MdMADS5: an APETALA1-like gene of apple, causes early flowering in transgenic Arabidopsis. Plant Science. 2002;162:679–687. [Google Scholar]
- Kotoda N, Wada M, Masuda T, Soejima J. The break-through in the reduction of juvenile phase in apple using transgenic approaches. Acta Horticulturae. 2003;625:337–343. [Google Scholar]
- Lauri PÉ From tree architecture to tree training—an overview of recent concepts developed in apple in France. Journal of the Korean Society for Horticultural Science. 2002;43:782–788. [Google Scholar]
- Lauri PÉ Térouanne E, Lespinasse J. Relationship between the early development of apple fruiting branches and the regularity of bearing. An approach to the strategies of various cultivars. Journal of Horticultural Science. 1997;72:519–530. [Google Scholar]
- Lauri PÉ Térouanne E, Lespinasse J, Regnard J, Kelner JJ. Genotypic differences in the axillary bud growth and fruiting pattern of apple fruiting branches over several years: an approach to regulation of fruit bearing. Scientia Horticulturae. 1995;64:265–281. [Google Scholar]
- Lespinasse Y. Le pommier. In: Gallais A, Bannerot H, editors. Amélioration des espèces végétales cultivées, objectifs et critères de sélection. Paris: INRA Editions; 1992. pp. 579–594. [Google Scholar]
- Liew M, Pryor R, Palais R, Meadows C, Erali M, Lyon E, Wittwer C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clinical Chemistry. 2004;50:1156–1164. doi: 10.1373/clinchem.2004.032136. [DOI] [PubMed] [Google Scholar]
- Link H. Significance of flower and fruit thinning on fruit quality. Plant Growth Regulation. 2000;31:17–26. [Google Scholar]
- Looney NE, Lane WD. Spur-type growth mutants of McIntosh apple: a review of their genetics, physiology and field performance. Acta Horticulturae. 1984;146:31–46. [Google Scholar]
- Looney NE, Pharis RP, Noma M. Promotion of flowering in apple trees with gibberellin A4 and C-3 epi-gibberellin A4. Planta. 1985;165:292–294. doi: 10.1007/BF00395054. [DOI] [PubMed] [Google Scholar]
- Luckwill LC. The control of growth and fruitfulness of apple trees. In: Luckwill LC, Cutting CV, editors. Physiology of tree crops. London: Academic Press; 1970. pp. 237–254. [Google Scholar]
- Luckwill LC. A new look at the process of fruit bud formation in apple. Proceedings of the XIXth International Horticultural Congress. 1974;3:237–246. [Google Scholar]
- Mimida N, Kidou S, Kotoda N. Constitutive expression of two apple (Malus× domestica Borkh.) homolog genes of LIKE HETEROCHROMATIN PROTEIN1 affects flowering time and whole-plant growth in transgenic. Arabidopsis. Molecular Genetics and Genomics. 2007;278:295–305. doi: 10.1007/s00438-007-0250-0. [DOI] [PubMed] [Google Scholar]
- Monselise S, Goldschmidt E. Alternate bearing in fruit trees. Horticultural Review. 1982;4:128–173. [Google Scholar]
- Neilsen JC, Dennis FG. Effects of seed number, fruit removal, bourse shoot length and crop density on flowering in ‘Spencer Seedless’ apple. Acta Horticulturae. 2000;527:137–146. [Google Scholar]
- Parcy F. Flowering: a time for integration. International Journal of Developmental Biology. 2005;49:585–593. doi: 10.1387/ijdb.041930fp. [DOI] [PubMed] [Google Scholar]
- Pearce SC, Dobersek-Urbane S. The measurement of irregularity in growth and cropping. Journal of Horticultural Science. 1967;42:295–305. [Google Scholar]
- R Development Core Team . Vienna, Austria: R Foundation for Statistical Computing. 2009. R: A language and environment for statistical computing. [Google Scholar]
- Reddy YTN, Kurian RM, Ramachander PR, Singh G, Kohli RR. Long-term effects of rootstocks on growth and fruit yielding patterns of ‘Alphonso’ mango (Mangifera indica L.) Scientia Horticulturae. 2003;97:95–108. [Google Scholar]
- Rosenstock TS, Rosa UA, Plant RE, Brown PH. A reevaluation of alternate bearing in pistachio. Scientia Horticulturae. 2010;124:149–152. [Google Scholar]
- Ross JJ, O'Neill DP, Smith JJ, Kerckhoffs LHJ, Elliott RC. Evidence that auxin promotes gibberellin A1 biosynthesis in pea. The Plant Journal. 2000;21:547–552. doi: 10.1046/j.1365-313x.2000.00702.x. [DOI] [PubMed] [Google Scholar]
- Samach A, Onouchi H, Gold SE, Ditta GS, Schwarz-Sommer Z, Yanofsky MF, Coupland G. Distinct roles of CONSTANS target genes in reproductive development of. Arabidopsis. Science. 2000;288:1613–1616. doi: 10.1126/science.288.5471.1613. [DOI] [PubMed] [Google Scholar]
- Schmidt S, Handschack M, Katzfuß M, Sandke G. Präzisierte anwendungsempfehlungen für wachstumsregulatoren zur ertragsstabilisierung bei apfel. Gartenbau. 1989;36:47–48. [Google Scholar]
- Schmidt T, McFerson J, Elfving DC, Whiting M. Practical gibberellic acid programs for mitigation of biennial bearing in apple. Acta Horticulturae. 2009;884:663–670. [Google Scholar]
- Segura V, Cilas C, Costes E. Dissecting apple tree architecture into genetic, ontogenetic and environmental effects: mixed linear modeling of repeated spatial and temporal measures. New Phytologist. 2008;178:302–314. doi: 10.1111/j.1469-8137.2007.02374.x. [DOI] [PubMed] [Google Scholar]
- Segura V, Cilas C, Laurens F, Costes E. Phenotyping progenies for complex architectural traits: a strategy for 1-year-old apple trees (Malus× domestica Borkh.) Tree Genetics and Genomes. 2006;2:140–151. [Google Scholar]
- Segura V, Denancé C, Durel CE, Costes E. Wide range QTL analysis for complex architectural traits in a 1-year-old apple progeny. Genome. 2007;50:159–171. doi: 10.1139/g07-002. [DOI] [PubMed] [Google Scholar]
- Segura V, Durel CE, Costes E. Dissecting apple tree architecture into genetic, ontogenetic and environmental effects: QTL mapping. Tree Genetics and Genomes. 2009;5:165–179. doi: 10.1111/j.1469-8137.2007.02374.x. [DOI] [PubMed] [Google Scholar]
- Singh L. Studies on biennial bearing. III. Growth studies on ‘on’ and ‘off’ year trees. Journal of Horticultural Science. 1948;24:123–148. [Google Scholar]
- Sivasubramanian V. Selection of suitable indices for determination of earliness of cotton varieties. Madras Agricultural Journal. 1962;49:117–120. [Google Scholar]
- Smith MW, Shaw RG, Chapman JC, Owen-Turner J, Slade Lee L, Bruce McRae K, Jorgensen KR, Mungomery WV. Long-term performance of ‘Ellendale’ mandarin on seven commercial rootstocks in sub-tropical Australia. Scientia Horticulturae. 2004;102:75–89. [Google Scholar]
- Snowden KC, Simkin AJ, Janssen BJ, Templeton KR, Loucas HM, Simons JL, Karunairetnam S, Gleave AP, Clark DG, Klee HJ. The Decreased apical dominance1/ Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE8 gene affects branch production and plays a role in leaf senescence, root growth, and flower development. The Plant Cell. 2005;17:746–759. doi: 10.1105/tpc.104.027714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung S, Yu G, An G. Characterization of MdMADS2, a member of the SQUAMOSA subfamily of genes, in apple. Plant Physiology. 1999;120:969–978. doi: 10.1104/pp.120.4.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan F, Swain SM. Genetics of flower initiation and development in annual and perennial plants. Physiologia Plantarum. 2006;128:8–17. [Google Scholar]
- Tanaka N, Wada M, Komori S, Bessho H, Suzuki A. Functional analysis of MdPI, the PISTILLATA gene homologue of apple, in Arabidopsis. Journal of the Japanese Society for Horticultural Science. 2007;76:125–132. [Google Scholar]
- Tromp J. Flower-bud formation in apple as affected by various gibberellins. Journal of Horticultural Science. 1982;57:277–282. [Google Scholar]
- Van der Linden C, Vosman B, Smulders M. Cloning and characterization of four apple MADS-box genes isolated from vegetative tissue. Journal of Experimental Botany. 2002;53:1025–1036. doi: 10.1093/jexbot/53.371.1025. [DOI] [PubMed] [Google Scholar]
- Van Ooiejn J. MapQTL® 5, software for the mapping of quantitative trait loci in experimental populations. Wageningen: Kyazma BV; 2004. [Google Scholar]
- Van Ooiejn J, Voorrips RE. JoinMap® 3.0, software for the calculation of genetic linkage maps. Wageningen: Plant Research International; 2001. [Google Scholar]
- Velasco R, Zharkikh A, Affourtit J, et al. The genome of the domesticated apple (Malus× domestica Borkh.) Nature Genetics. 2010;42:833–839. doi: 10.1038/ng.654. [DOI] [PubMed] [Google Scholar]
- Voorrips RE. MapChart: software for the graphical presentation of linkage maps and QTLs. Journal of Heredity. 2002;93:77–78. doi: 10.1093/jhered/93.1.77. [DOI] [PubMed] [Google Scholar]
- Wada M, Cao Q, Kotoda N, Soejima J, Masuda T. Apple has two orthologues of FLORICAULA/ LEAFY involved in flowering. Plant Molecular Biology. 2002;49:567–577. doi: 10.1023/a:1015544207121. [DOI] [PubMed] [Google Scholar]
- Weigel D, Meyerowitz EM. The ABCs of floral homeotic genes. Cell. 1994;78:203–209. doi: 10.1016/0092-8674(94)90291-7. [DOI] [PubMed] [Google Scholar]
- Wilcox J. Some factors affecting apple yields in the Okanagan Valley: tree size, tree vigor, biennial bearing, and distance of planting. Scientia Agricola. 1944;25:189. [Google Scholar]
- Wittwer CT, Reed GH, Gundry CN, Vandersteen JG, Pryor RJ. High-resolution genotyping by amplicon melting analysis using LCGreen. Clinical Chemistry. 2003;49:853–860. doi: 10.1373/49.6.853. [DOI] [PubMed] [Google Scholar]
- Wood BW, Conner PJ, Worley RE. Insight into alternate bearing of pecan. Acta Horticulturae. 2004;636:617–629. [Google Scholar]
- Xu YL, Gage DA, Zeevaart J. Gibberellins and stem growth in Arabidopsis thaliana. Effects of photoperiod on expression of the GA4 and GA5 loci. Plant Physiology. 1997;114:1471–1476. doi: 10.1104/pp.114.4.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky MF. Floral meristems to floral organs: genes controlling early events in Arabidopsis flower development. Annual Review of Plant Physiology and Plant Molecular Biology. 1995;46:167–188. [Google Scholar]
- Yao J, Dong Y, Morris BAM. Parthenocarpic apple fruit production conferred by transposon insertion mutations in a MADS-box transcription factor. Proceedings of the National Academy of Sciences, USA. 2001;98:1306–1311. doi: 10.1073/pnas.031502498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SJ, Chung KS, Jung SH, Yoo SY, Lee JS, Ahn JH. BROTHER OF FT AND TFL1 (BFT) has TFL1-like activity and functions redundantly with TFL1 in inflorescence meristem development in Arabidopsis. The Plant Journal. 2010;63:241–253. doi: 10.1111/j.1365-313X.2010.04234.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.