Abstract
The rice somaclonal mutant T3612 produces small grains with a floury endosperm, caused by the loose packing of starch granules. The positional cloning of the mutation revealed a deletion in a gene encoding a protein disulphide isomerase-like enzyme (PDIL1-1). In the wild type, PDIL1-1 was expressed throughout the plant, but most intensely in the developing grain. In T3612, its expression was abolished, resulting in a decrease in the activity of plastidial phosphorylase and pullulanase, and an increase in that of soluble starch synthase I and ADP-glucose pyrophosphorylase. The amylopectin in the T3612 endosperm showed an increase in chains with a degree of polymerization 8–13 compared with the wild type. The expression in the mutant's endosperm of certain endoplasmic reticulum stress-responsive genes was noticeably elevated. PDIL1-1 appears to play an important role in starch synthesis. Its absence is associated with endoplasmic reticulum stress in the endosperm, which is likely to underlie the formation of the floury endosperm in the T3612 mutant.
Keywords: ER stress, floury endosperm, protein disulphide isomerase-like 1-1, starch synthesis
Introduction
Starch accounts for 60–80% of cereal grain endosperm. The analysis of starch synthesis mutants (commonly referred to as ‘opaque’ mutants) has shown that the products of ISA-1 (Kubo et al., 1999), AGPS2b (Kawagoe et al., 2005), Pho1 (Satoh et al., 2008; Hwang et al., 2010), SSIIIa (Fujita et al., 2007), and BEIIb (Nishi et al., 2001) are key enzymes in the process. A number of genes responsible for the floury endosperm phenotype have, however, proven to be not directly related to starch synthesis. One of these is BiP, which encodes a chaperone active in the endoplasmic reticulum (ER) where it interacts with nascent immature proteins to assist in their folding (Yasuda et al., 2009; Wakasa et al., 2011). In maize, mutated alleles of various zein-encoding genes have also been shown to generate a floury endosperm, and a number of genes, including BiP, are known to be up-regulated in the endosperms of these mutants (Hunter et al., 2002).
When an excess of unfolded or misfolded proteins aggregates in the ER, a process referred to as ER stress ensues, which is alleviated via the unfolded protein response (UPR) (Kaufman, 1999). The UPR of yeast and mammalian cells is rather well understood, but its equivalent in plant cells is still obscure. One branch of the mammalian UPR involves the protein PERK (protein kinase-like endoplasmic reticulum kinase) which acts to block protein synthesis via the phosphorylation of the eukaryotic translation initiation factor 2α (Szegezdi et al., 2006). However, no plant orthologue of PERK has yet been found. A second branch involves the transduction of the stress signal by a number of bZIP proteins from the ER to the nucleus, where a series of stress response genes are then induced (Liu et al., 2007a, b; Iwata et al., 2008), leading to the activity of various chaperones. The functions of this class of proteins have yet to be established in plants. Finally, the third branch involves programmed cell death (PCD), which is induced if the ER stress cannot be alleviated (Cacas, 2010). Protein disulphide isomerase (PDI) is a component of the UPR, and is involved via its active site WCGHC both in the catalysis of disulphide bond formation (reduction or isomerization) and in assisting polypeptide folding (Wilkinson and Gilbert, 2004; Gruber et al., 2006). Cells lacking certain UPR components, including PDI and BiP, are prone to ER stress. In rice, for example, the absence of BiP1 induces an intense ER stress response (Wakasa et al., 2011), while yeast pdi1 loss-of-function mutants are lethal (Laboissiere et al., 1995). In metazoans, persistent or severe ER stress induces PCD (Boyce and Yuan, 2006). The link between ER stress and PCD in plants has not yet been established.
Here, it is shown that abolishing the expression of a PDI protein (PDIL1-1) leads to the formation of a floury endosperm in rice, in which the frequency of short chain amylopectin molecules is markedly higher than in the wild type. The level of expression in the endosperm of a number of genes associated with starch synthesis has been compared between the mutant (T3612) and the wild type, and it has been possible to demonstrate a greater abundance of certain chaperones, co-chaperones, bZIP transcription factors, and ERAD-related proteins. Finally, it was possible to show the induction of the PCD marker gene HSR203J in the endosperm of the mutant. These results suggest that ER stress, induced by the absence of PDIL1-1, was probably the cause for the formation of a floury endosperm in T3612.
Materials and methods
Plant materials and phenotyping
The T3612 mutant was regenerated from an in vitro culture of tissue from cv. Nipponbare. Plants were grown under natural conditions at Nanjing Agricultural University. The procedure used for scanning electron microscopy (SEM) was as described by Kang et al. (2005), and that for transmission electron microscopy (TEM) was as described by Wang et al. (2009). The starch content in the rice flour was measured with a starch assay kit (Megazyme, Wicklow, Ireland), according to the manufacturer's protocol. Amylose content was assessed following Liu et al. (2009), and that of lipids and proteins following Kang et al. (2005).
RNA extraction, RT-PCR, and qRT-PCR
Total RNA was extracted by means of an RNAprep pure Plant kit (TIANGEN Biotech, Beijing, China). A 1 μg aliquot of total RNA was reverse transcribed by priming with oligo(dT18) in a 20 μl reaction based on the PrimeScript Reverse Transcriptase kit (Takara, Otsu, Japan). The rice Actin1 sequence (GenBank accession no. NM_001057621) was employed as the RT-PCR reference, and the rice Ubiquitin sequence (NM_001056014) as the qRT-PCR reference. Primer sequences used for both RT-PCR and qRT-PCR for the ER stress-related genes are given in Supplementary Table S1 available at JXB online. The qRT-PCR primers used to assay starch synthesis and grain protein storage synthesis-related genes have been documented elsewhere (Ohdan et al., 2005; She et al., 2010), as have those targeting HSR203J, BiP-3, and Hsp70-2 (Wakasa et al., 2011). qRT-PCRs were performed on an ABI PRISM 7900 device using SYBR Premix Ex Taq (Takara).
Enzyme assays and native PAGE/activity staining
A sample of 100 mg of developing endosperm [9 or 12 days after flowering (DAF)] was homogenized on ice in 1 ml of 50 mM HEPES NaOH (pH 7.4), 2 mM MgCl2, and 12.5% (v/v) glycerol. The homogenate was centrifuged at 20 000 g for 10 min at 4 °C, and the resulting supernatant was used for all enzyme assays. ADP-glucose pyrophosphorylase (AGPase) and UGPase activity was determined following Nishi et al. (2001), and that of sucrose synthase following Doehlert et al. (1988). Debranching enzyme (DBE) and branching enzyme (BE) isozymes were separated by native PAGE and assayed according to Fujita et al. (2009). Starch synthase (SS) and phosphorylase (Pho) activities were generated in 7.5% polyacrylamide gels containing 0.8% (w/v) oyster glycogen (Sigma), according to Satoh et al. (2008). All electrophoretic separations were conducted at 4 °C at a constant voltage of 50 V.
DSA-FACE assessment of amylopectin chain length distribution
DSA-FACE (DNA sequencer-assisted flurophore-assisted carbohydrate electrophoresis) separation of mature grain starch was performed according to Nishi et al. (2001) and O'Shea and Morell (1996), with minor modifications. A 5 mg sample of rice flour was suspended in 200 μl of ultrapure water and boiled for 10 min. Then 800 μl of 50 mM sodium acetate was added, and the suspension was re-boiled for a further 10 min. After cooling, 0.1 U of Pseudomonas amyloderamosa isoamylase (ISA; Seikagaku Corporation, Tokyo) was added, and the mixture was incubated for 24 h at 37 °C. The resulting solution was then held at 100 °C for 2 min, and a 1 μl aliquot was combined with 1 μl of 0.1 M 8-aminopyrene-1,3,6-trisulphonic acid trisodium salt and 2 μl of 1 M sodium cyanoborohydride, and incubated for 24 h at 37 °C in the dark. The final solution was diluted with 21 μl of 6 M urea, 0.04 M boric acid, 0.04 M TRIS, pH 8.6. A 1 μl aliquot was loaded onto a 7.5% polyacrylamide gel and electrophoresed using an ABI PRISM 377 DNA sequencing device.
Genetic mapping and transformation
Ten F2 progeny of the cross T3612×cv. NJ11 producing floury endosperms were genotyped with 242 genome-wide microsatellite markers. Fine mapping required the derivation of de novo molecular markers based on a comparison of the local genome sequences of cv. Nipponbare and cv. 93-11, and included both microsatellite and indel markers (Supplementary Table S1 at JXB online). For the complementation of the T3612 mutation, the wild-type PDIL1-1 cDNA sequence was cloned into the binary vector pPZP2Ha3 (+) (Fuse et al., 2001) to generate the transformation cassette pPZP-35S-PDIL1-1. This plasmid was introduced into Agrobacterium tumiefaciens strain EHA105, which was then used to agroinfect rice calli (Hiei et al., 1994). Hygromycin-resistant calli were regenerated and seedlings were grown in a greenhouse.
Heterologous expression of PDIL1-1
PDIL1-1 cDNA was cloned into the pGEX-4T-2 expression vector, which was introduced into Escherichia coli strain BL21 (DE3). Heterologous expression was induced by the addition of 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG), and shaking the cultures at 28 °C for 12 h. The cells were pelleted by centrifugation (12 000 g, 5 min, 4 °C) and then resuspended in 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.5, lysed ultrasonically and recentrifuged (13 000 g, 10 min, 4 °C). The supernatant was used for the PDIL1-1 assay. Various concentrations of recombinant PDIL1-1 were added to 100 mM Na2HPO4, 2 mM EDTA, 1 mg ml−1 insulin, 2 mM dithiothreitol (DTT), kept at 30 °C, and the OD650 measured at 3 min intervals to assay for disulphide reductase activity. Its molecular chaperone activity was assayed in a 1 ml solution of 500 nM PDIL1-1, 40 mM HEPES NaOH, pH 7.5, 150 nM citrate synthetase, incubated at 42 °C. The OD360 of this solution was measured at 10 min intervals.
Detection of PDIL1-1 product in rice
To generate polyclonal antibodies recognizing PDIL1-1, recombinant PDIL1-1 protein was gel purified from lysed E. coli cells by SDS–PAGE. The gels were immersed in 250 mM KCl to allow excision of the band corresponding to the PDIL1-1 protein, and the excised gel fragment was then ground in liquid nitrogen. The powdered gel fragment was suspended in 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.5, and the suspension of 1 mg ml−1 PDIL1-1 was used as antigen for the production of rabbit polyclonal antibodies against PDIL1-1, using a standard protocol (Wang et al., 2010). Western blot assays were based on frozen plant tissue (500 mg) ground in liquid nitrogen and homogenized in 1 ml of buffer with 50 mM TRIS-HCl, pH 8.0, 0.25 M sucrose, 2 mM DTT, 2 mM EDTA, and 1 mM phenylmethylsulphonyl fluoride (PMSF). The protein concentration in these preparations was determined using a commercial kit (Bio-Rad, Herclues, CA). A 10 μg aliquot of protein was resolved by electrophoresis (10% SDS–PAGE), and transferred electrophoretically to polyvinylidene difluoride membrane (0.45 μm; Millipore, www.millipore.com/) using a semi-wet transfer method (Wang et al., 2010). The membranes were incubated in a 1:300 dilution of the polyclonal anti-PDIL1-1 antiserum in 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.5, and the hybridization signal detected by ECL via conjugation with an anti-rabbit IgG linked to horseradish peroxidase (BeyoECL Plus kit, obtained from Beyotime Institute of Biotechnology, Haimen, China).
Results
Phenotype of the T3612 mutant
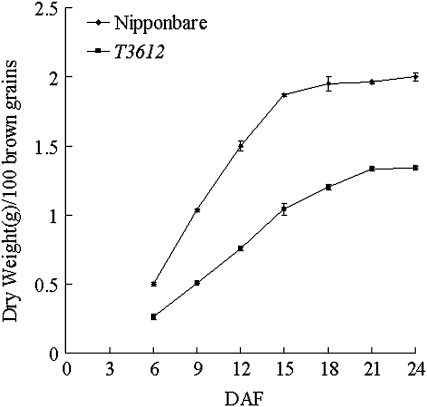
The T3612 mutant, recovered from cultured tissue of the japonica type rice cv. Nipponbare, was similar with respect to its general morphology and panicle size to the wild type (Fig. 1A, B), but produced grain with a floury endosperm (Fig. 1C) and achieved a markedly slower grain-filling rate (Fig. 2). SEM revealed that the starch granules in the mutant's endosperm were round and only loosely packed (Fig. 1E, F). Its mature grain weight was ∼66% (Table 1) and its amylose content ∼80% that of cv. Nipponbare. Starch contents per dry weight were similar for both the wild type and T3612. However, both the total protein and lipid content of the mutant's endosperm were ∼15% above those of the wild type. Selfed progeny of plants heterozygous for the mutation segregated in the monogenic ratio three normal to one floury (Fig. 1D), showing that the mutation was based on a single recessive allele.
Fig. 1.
The phenotype of the T3612 mutant. (A) cv. Nipponbare and T3612 plants. (B) Panicles of cv. Nipponbare and T3612. (C) The floury endosperm of T3612 which (D) segregates in a monogenic fashion. Scanning electron micrographs of mature endosperms of (E) cv. Nipponbare and (F) T3612.
Fig. 2.
The grain-filling process (weight in g per 100 grains) in T3612 and cv. Nipponbare. DAF, days after flowering.
Table 1.
Key characteristics of the cv. Nipponbare and T3612 grain
| Weight (g) | Amylose (%) | Starch (%) | Protein (%) | Lipid (%) | |
| Nipponbare | 18.69±0.18 | 14.68±0.26 | 75.00±0.42 | 10.88±0.02 | 2.56±0.06 |
| T3612 | 12.39±0.25 | 12.24±0.39 | 74.55±0.64 | 12.5±0.02 | 2.95±0.07 |
| Difference | −33.70% | −19.93% | −0.6% | +14.89% | +15.23% |
Fine mapping and complementation of the T3612 mutation
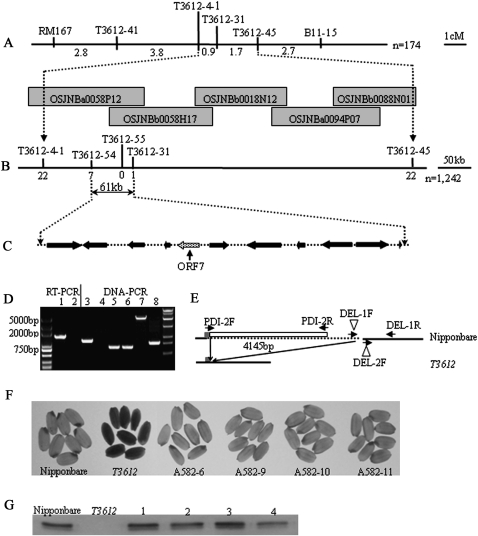
An F2 population was created from the cross of T3612 with the indica type cv. NJ11. The genotyping using 242 genome-wide markers of a sample of 10 progeny expressing the floury endosperm trait suggested the presence of linkage between the trait and the markers RM167 and B11-15, both of which are located on the short arm of chromosome 11 (Fig. 3). A subsequent fine-mapping exercise (details of all the markers used are listed in Supplementary Table S1 at JXB online) based on 1242 progeny located the mutation to a chromosomal segment bounded by T3612-31 and T3612-54, and identified T3612-55 as a co-segregating marker. This defined the position of the target gene as lying within a 61 kb region of bacterial artificial chromosome (BAC) OSJNBa0058P12. Gene prediction based on RiceGAAS software (ricegaas.dna.affrc.go.jp/) indicated the presence of 11 open reading frames (ORFs) in this segment (Supplementary Table S2). A set of RT-PCR analyses based on these 11 genes showed that the PDIL1-1 transcript was absent in T3612 (Fig. 3D). A genomic PCR (based on the primer pair DEL-1F/1R; the sequences of these, as well as of other primers referred to below, are given in Supplementary Table S1) showed that no amplicon was generated in T3612, while a PCR based on DEL-2F/-1R amplified the same size product in both T3612 and cv. Nipponbare. PDI-2F/DEL-1R amplified an ∼5 kb product from cv. Nipponbare, but an ∼1 kb amplicon was produced from T3612 (Fig 3D). The derived sequence of the ∼1 kb T3612 amplicon differed from the wild-type sequence by a 4145 bp deletion, starting 124 nucleotides downstream of the PDIL1-1 ATG start codon. The section of T3612 sequence around the deletion boundary is shown in Supplementary Fig. S1.
Fig. 3.
OsPDIL1-1 is the gene responsible for the T3612 mutant phenotype. (A) The mutated gene was mapped to the short arm of chromosome 11 between molecular markers T3612-4-1 and T3612-45. Genetic distances and names of the molecular markers are indicated (n =174). OSJNBa0058P12, OSJNBb0058H17, OSJNBb0018N12, OSJNBa0094P07, and OSJNBb0088N01 are BAC clones covering this locus. (B) The location of the mutation was narrowed to a 61 kb genomic region between T3612-54 and T3612-31. The number of recombinants is indicated below the molecular marker (n = 1242). (C) Gene prediction indicates 11 open reading frames in the critical region. The candidate gene (ORF7) is shown by an arrow. (D) PCRs based on cDNA (lanes 1,2) and genomic DNA (lanes 3–8) reveal the deletion in the T3612 PDIL1-1 allele. Lanes 1 and 2, PDI-2F/PDI-2R; lanes 3 and 4, DEL-1F/DEL-1R; lanes 5 and 6, DEL-2F/DEL-1R; lanes 7 and 8, PDI-2F/DEL-1R; lanes 1, 3, 5, and 7, cv. Nipponbare; lanes 2, 4, 6, and 8, T3612. (E) The deletion sequence (broken line) and primer annealing sites (indicated by arrows). The white box shows the deleted part of PDIL1-1. (F) Phenotypes of T1 kernels in four independent complemented lines. (G) PDIL1-1 protein was present in vitreous T1 grains. Lanes 1–4 represent four complementation lines shown in (F).
To show that the absence of the PDIL1-1 transcript was responsible for the floury endosperm trait, its ORF was fused to the Cauliflower mosaic virus (CaMV) 35S promoter and then introduced into the T3612 mutant. Grain harvested from T1 plants segregated for endosperm type (Fig. 3F). Using a screen based on the hygromycin resistance selection marker, it was possible to show that the vitreous grains carried the transgene, while the floury ones did not. Two genomic PCRs—the first based on the PDIL1-1 coding region targeted primer pair PDI-2F/-2R, and the second, on PDI-2F/DEL-1R, targeted the T3612 deletion—produced the expected amplicons in the primary transformants (Supplementary Fig. S2 at JXB online). A western blot analysis showed that the PDIL1-1 protein was present in vitreous grain harvested from T1 plants (Fig. 3G). Thus the transgenic constitutive expression of PDIL1-1 was able to complement the floury endosperm trait.
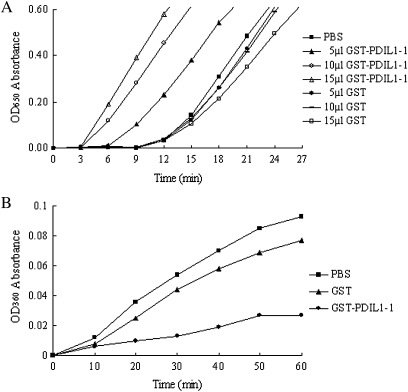
In order to determine whether PDIL1-1 protein possessed any disulphide reductase and/or molecular chaperone activity it was provided with either insulin or citrate synthetase as a substrate. In the presence of DTT, PDI reduces the intermolecular disulphide bonds between the insulin A and B chains. Precipitation of the insoluble B chain can be measured photometrically by an increase in the absorbance at 650 nm. The assay was performed with recombinant glutathione S-transferase (GST)-tagged PDIL1-1. As shown in Fig. 4A, GST–PDIL1-1 was able to accelerate the reduction of insulin, and the rate of this reduction was intensified by increasing the concentration of GST–PDIL1-1, clearly demonstrating that PDIL1-1 possesses disulphide reductase activity in vitro. The GST–PDIL1-1 was also able to protect citrate synthetase effectively from thermal denaturation at 42 °C. Precipitation of citrate synthetase decreased by 64.9% in contrast to GST control (Fig. 4B).
Fig. 4.
Assay for recombinant GST-tagged PDIL1-1 activity. (A) Assay for disulphide reductase activity of GST–PDIL1-1 using the turbidimetric assay of insulin reduction. Phosphate-buffered saline (PBS) and GST are the controls. (B) Assay for molecular chaperone activity of GST–PDIL1-1. PDI can protect citrate synthetase from thermal denaturation in 42 °C. PBS and GST are the controls.
Expression of PDIL1-1 in plants
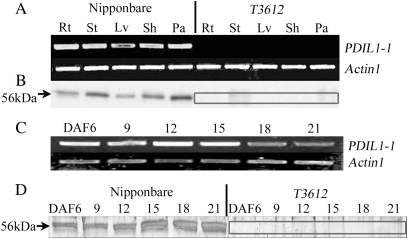
RT-PCR analysis showed that PDIL1-1 was expressed in the root, culm, leaf, sheath, and young panicle of cv. Nipponbare. Among these organs, the level of expression was lowest in the leaf and highest in the young panicle. T3612 was free of PDIL1-1 transcript (Fig. 5A). In the developing endosperm, peak expression occurred at 12 DAF (Fig. 5C). Immunoblotting showed that PDIL1-1 expression in the wild type occurred in the culm, young panicle, and developing endosperm at 12 DAF (Fig. 5B, D), but was absent from any part of the T3612 plant.
Fig. 5.
Expression of PDIL1-1. (A) Expression as assayed by RT-PCR. Actin1 was used as the reference. Rt, root; St, stem; Lv, leaf; Sh, sheath; Pa, panicle. (B) Western blot analysis based on anti-PDIL1-1 polyclonal antibodies. The absence of PDIL1-1 protein in the mutant is highlighted. (C) Expression of PDIL1-1 in the developing [6–21 days after flowering (DAF)] endosperm of cv. Nipponbare assayed by RT-PCR. (D) Western blot analysis of the presence of PDIL1-1 in the developing endosperm. The absence of PDIL1-1 protein in the mutant is highlighted.
The absence of PDIL1-1 affects major starch synthesis enzyme activity
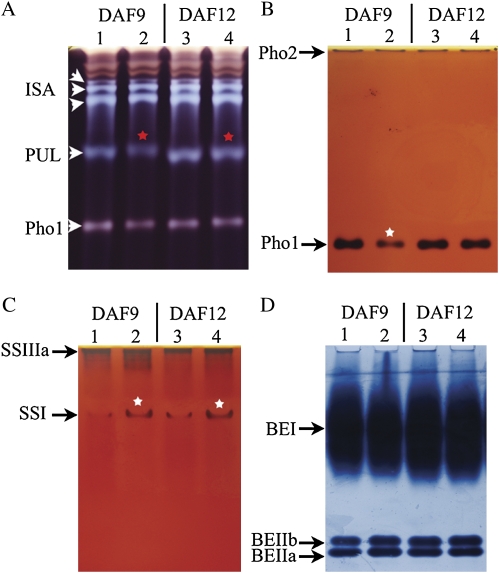
A comparison between levels of enzyme activity in the mutant and the wild type showed that there were no significant differences with respect to ISA, Pho2, SSIIIa, or BE (Fig. 6). However, Pho1 activity appeared to be lower in the mutant than in the wild type at 9 DAF. Pullulanase (PUL) activity was greatly reduced in the mutant, and the proteins appeared to have a lower electrophoretic mobility. The activity of SSI was substantially increased. However, qRT-PCR-based evaluations of gene expression suggested that PUL and SSI levels were similar in the mutant and wild type, while that of Pho1 was increased by only 1-fold (Supplementary Fig. S3 at JXB online). Thus it appears possible that the effect of PDIL1-1 is felt post-transcriptionally. UGPase activity was somewhat lower in the mutant than in the wild-type endosperm, while those of SS and AGPase were, respectively, ∼30% and 146% higher than in the wild type (Table 2). The expression of DPE-1 was only ∼50% of the level in the wild type. Thus, overall, it was clear that the deletion of PDIL1-1 affected the activities of certain starch synthesis enzymes, and that the loss of Pho1 activity may be a major determinant of the formation of the floury endosperm in the T3612 mutant.
Fig. 6.
Enzyme activity in the developing endosperm. (A) Native PAGE/debranching enzyme activity. ISA, PUL, and Pho activity are indicated by arrowheads. (B) Native PAGE/starch phosphorylase activity staining. Pho1 and Pho2 are indicated by arrowheads. (C) Native PAGE/starch synthase activity. SSIIIa and SSI activity are indicated by arrowheads. (D) Native PAGE/branching enzyme activity. BEI, BEIIa, and BEIIb activity are indicated by arrowheads. Lanes 1 and 3, cv. Nipponbare; lanes 2 and 4, T3612.
Table 2.
The effects of the T3612 mutation on the activities of enzymes involved in starch and sucrose metabolism in the developing endosperm (12 d after flowering)
| Enzyme activity (nmol min−1 mg−1 FW) |
|||
| AGPase | UGPase | Sucrose synthase | |
| Nipponbare | 5.15±0.40 | 25.07±1.35 | 2.88±0.05 |
| T3612 | 12.7±1.86 | 23.28±0.25 | 3.73±0.01 |
Values represent the means ±SD from two replicates.
The effect of the PDIL1-1 deletion on amylopectin structure
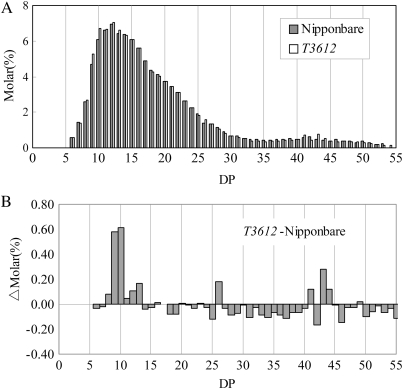
DSA-FACE (O'Shea and Morell, 1996; Nishi et al., 2001) was applied to contrast the chain length distribution in the amylopectin synthesized in the T3612 and wild-type endosperm. The analysis suggested that the mutant amylopectin consisted of a higher proportion of short chains in the range degree of polymerization (DP) 8–13 and a lower proportion of DP 6–7 chains (Fig. 7B). This shift in amylopectin chain lengths agrees with the enhanced SSI and depressed PUL activities in the mutant (Fig. 6A,C).
Fig. 7.
Amylopectin chain length distribution in the T3612 and cv. Nipponbare endosperm. (A) Estimated molar chain length distribution for DP 6–56 starch chains. (B) Difference in the amylopectin chain length distribution between T3612 and cv. Nipponbare.
Analysis of the ER stress-responsive genes
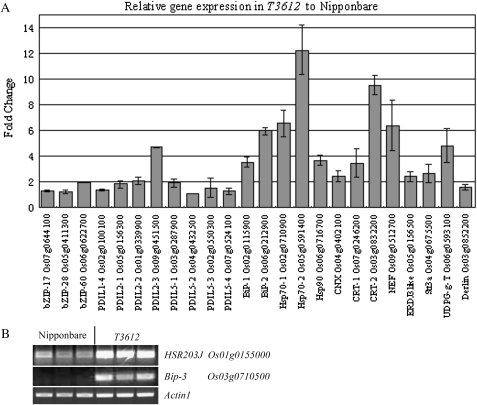
As PDIL1-1 is a chaperone protein involved in the formation of inter- and intramolecular disulphide bridges (Wilkinson and Gilbert, 2004; Satoh-Cruz et al., 2010), qRT-PCR and semi-quantitative RT-PCR were used to investigate the expression of a set of ER stress- and PCD-associated genes. Genes encoding 17 chaperone-related (eight PDILs, three BiPs, two Hsp70s, one Hsp90, one CNX, and two CRTs) and four co-chaperone-related (NEF, ERDJ3-like, Stt3a, and UDP glucose transporter) proteins, three membrane-associated bZIP transcription factors (bZIP17, bZIP28, and especially bZIP60), the ERAD-related protein (Derlin), and the PCD marker HSR203J were more highly expressed in T3612 than in wild-type endosperms (Fig. 8). The level of expression of BiP-2, both Hsp70 genes, CRT-2, and NEF was >5-fold higher in the mutant than in the wild-type endosperm. This suggested that the absence of PDIL1-1 induced a level of ER stress. Of the remaining PDIL genes, only PDIL2-3 was noticeably up-regulated (4-fold). Nevertheless, large amounts of a 57 kDa proglutelin accumulated in the T3612 endosperm (Supplementary Fig. S4 at JXB online), which suggested that PDIL2-3 and PDIL1-1 each have their own distinct role in the folding of storage proteins (Onda et al., 2011).
Fig. 8.
Expression levels of selected ER stress-associated genes (A) and RT-PCR analysis of expression of the chaperone BiP-3 and the PCD marker HSR203J in T3612 and cv. Nipponbare (B). Each value reported is the mean ±SD of three biological replicates.
Discussion
The deletion of PDIL1-1 results in the formation of a floury endosperm
The current picture of cereal starch synthesis has been established from the study of opaque mutants, largely in maize and rice (Myers et al., 2000; Toyota et al., 2006; Hennen-Bierwagen et al., 2008, 2009). However, as further opaque endosperm mutants continue to be uncovered and analysed, a number of genes have been identified which cannot readily be incorporated into the established scheme, thus requiring a re-assessment of the model. The T3612 mutant is distinct from previous floury endosperm mutants, and has implicated a role for PDIL1-1 in the starch synthesis network operating in the rice endosperm. The only documented association of PDI with starch synthesis in plants relates to the product of AtPDIL1-3 which is involved in the synthesis of transitory starch in the leaves of Arabidopsis thaliana (Lu and Christopher, 2006).
The level of activity of most of the known starch synthesizing enzymes in the T3612 endosperm was hardly different from the wild-type levels—with the exception of that of Pho1, AGPase, SSI, and PUL. The activity of Pho1 was markedly diminished in T3612. Although the specific molecular mechanism of Pho1 in the synthesis of starch is still controversial, circumstantial evidence suggests the involvement of Pho1 in starch synthesis in storage tissues. Pho1 enzyme activity and protein levels are coordinated with starch production in potato tuber (Kossmann et al., 1991; St-Pierre and Brisson, 1995), maize (Zea mays) endosperm (Ozbun et al., 1973), and wheat endosperm (Schupp and Ziegler, 2004).
Another gene which was substantially down-regulated in T3612 was DPE-1; the primary expression site of this gene is the amyloplast in developing endosperm cells (Ohdan et al., 2005; Bresolin et al., 2006). In Chlamydomonas reinhardtii, lack of D-enzyme activity (the sta11 mutant) results in significantly reduced starch deposition (Colleoni et al., 1999a). Although in higher plants the role is unclear, the partially purified active D-enzyme from the endosperm of developing wheat grain was able to elongate glycogen or amylopectin (Bresolin et al., 2006), which share the same function as D-enzyme in C. reinhardtii (Colleoni et al., 1999b). Future studies on the mutants that lack D-enzyme in rice grains will help elucidate its role in starch synthesis in vivo.
Both AGPase and SSI activities in T3612 were higher than those of the wild type, and similar observation was made in the rice SSIIIa mutant (Fujita et al., 2007). High levels of sugars enhance the expression of the maize genes sh2 and bt2 that encode the large and small subunits of AGPase (Giroux and Hannah, 1994). Therefore, the high level of sugars in T3612 (data not shown) may enhance the expression of the genes OsAGPL2, OsAGPL3, OsAGPS2b, and OsAGPS2a which led to the higher AGPase activity.
Not only was PUL activity lowered in the mutant, but its electrophoretic mobility was also changed. A similar change to PUL has been noted in an ISA1 mutant (Kubo et al., 2005). The PUL-null mutation had no effects on the seed morphology and crystallinity of starch granules, except that the short chains (DP ≤13) of amylopectin were increased. The T3612 mutant overproduced amylopectin chains in the range DP 8–13 and underproduced the shorter DP 6–7 chains. The change in the mature endosperm amylopectin structure was consistent with the observed increase in SSI activity and decrease in PUL activity (Figs 6, 7).
Therefore, further work will be needed to elucidate whether the reduced activity of Pho1 and the down-regulation of DPE-1 had a major effect on starch synthesis in the T3612 endosperm.
The absence of PDIL1-1 had a deleterious effect on storage protein synthesis
The fine mapping and complementation analysis provided ample evidence that the floury endosperm phenotype of the T3612 mutant was due to the absence of PDIL1-1. The rice esp2 mutant also apparently lacks a functional allele of PDIL1-1 (Takemoto et al., 2002), while its mature endosperm accumulates large quantities of a 57 kDa proglutelin, which is mislocalized to an unusual, small ER-derived protein body I (PBI). This observation has been taken to suggest that PDIL1-1 has a role in grain storage protein trafficking in the developing endosperm. No mention has been made as to whether the esp2 mutant produces floury endosperm. The T3612 endosperm accumulated large quantities of a 57 kDa proglutelin (Supplementary Fig. S4 at JXB online) and produced an unusually small PBI (Supplementary Fig. S5). In addition, the expression of a number of grain storage protein genes was markedly suppressed in the mutant's endosperm at 9 DAF, a time when grain storage proteins began to appear in abundance in cv. Nipponbare (Supplementary Fig. S6). PDIL1-1 was thus clearly involved in the synthesis of both grain storage protein and starch.
In the ER lumen, PDIL proteins are thought to assist in the folding of immature secretory proteins, in co-operation with other chaperones such as the BiPs, CNX, and CRT. PDI and BiP are the two main classes of chaperone in the ER lumen involved in the folding of grain storage proteins (Satoh-Cruz et al., 2010; Wakasa et al., 2011). Glutelin is abundant enough in the endosperm to allow defects, such as in T3612 and esp2, to be readily detectable by SDS–PAGE. Of course, many other proteins—perhaps including those involved in starch synthesis—may be similarly misfolded, but this can be hard to identify. In A. thaliana, a proteomics approach has shown that >8% of the chloroplast proteins carry signal peptides directing translocation to the ER (Kleffmann et al., 2004). One suggestion, therefore, is that some at least of such proteins are incorrectly folded in the T3612 mutant as a result of the absence of PDIL1-1, with consequences for starch metabolism in the plastids.
ER stress interacts with grain starch synthesis
The severe suppression of BiP1 is known to result in the formation of a floury endosperm; along with a reduction in starch accumulation, these endosperms suffer from acute ER stress. Microarray and RT-PCR-based analyses have shown that many of the genes involved in ER stress are strongly up-regulated (Wakasa et al., 2011), and many of these same genes were also up-regulated in the T3612 endosperm. It is assumed, therefore, that the deletion of PDIL1-1 resulted in severe ER stress. In the maize opaque mutants fl2, Mc, and De-B30, ER stress is induced by the accumulation of several abnormal zeins (Lending and Larkins, 1992; Kim et al., 2004). The expression of chaperone genes such as BiP and PDI is also increased in these mutants (Hunter et al., 2002). The overexpression of BiP1 or human amyloid β also generates ER stress and produces floury endosperms, which achieve a reduced level of starch accumulation (Oono et al., 2010; Wakasa et al., 2011). In all of these examples, opaque mutant endosperm cells are prone to ER stress, which suggests that it may be the ER stress itself which contributes to the formation of the opaque phenotype. The overall conclusion is that the mutant endosperm suffers from persistent and/or acute ER stress, and this both serves as a trigger for PCD and compromises the normal starch synthesis process. The forward focus is on the identification of the amyloplast proteins which interact with PDIL1-1 and the demonstration that PCD is occurring in the T3612 endosperm.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Part of the sequence of the deletion boundary in T3612.
Figure S2. Genomic PCR from four independent T1 complemented lines (A582-6, -9, -10, and -11). Lane 1, cv. Nipponbare; lane 2, T3612.
Figure S3. Expression levels of various genes involved in starch synthesis. Each value reported is the mean ±SD of three biological replicates.
Figure S4. SDS–PAGE analysis of total mature grain protein.
Figure S5. TEM images of the developing endosperm of (A) cv. Nipponbare and (B) T3612.
Figure S6. Expression levels of various genes involved in grain storage protein synthesis. Each value reported is the mean ±SD of three biological replicates.
Table S1. Primer sequences used.
Table S2. The 11 predicted genes present in the candidate region of chromosome 11 containing the floury endosperm mutation of T3612.
Acknowledgments
We are grateful to Professor Masahiro Yano (National Institute of Agrobiological Sciences, Ibaraki, Japan) for providing us with the binary vector pPZP2Ha3 (+). This work was supported by the 973 Program of China (2007CB108802), a project from the Ministry of Agriculture of China for Transgenic Research (2009ZX08009-114B and 2008ZX08009-003), the National Natural Science Foundation of China (30971757), the Earmarked Fund for Modern Agro-industry Technology Research System, and a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions.
References
- Boyce M, Yuan J. Cellular response to endoplasmic reticulum stress: a matter of life or death. Cell Death and Differentiation. 2006;13:363–373. doi: 10.1038/sj.cdd.4401817. [DOI] [PubMed] [Google Scholar]
- Bresolin NS, Li Z, Kosar-Hasgemi B, Tetlow IJ, Chatterjee M, Rahman S, Morell MK, Howitt CA. Characterisation of disproportionating enzyme from wheat endosperm. Planta. 2006;224:20–31. doi: 10.1007/s00425-005-0187-7. [DOI] [PubMed] [Google Scholar]
- Cacas JL. Devil inside: does plant programmed cell death involve the endomembrane system? Plant, Cell and Environment. 2010;33:1453–1473. doi: 10.1111/j.1365-3040.2010.02117.x. [DOI] [PubMed] [Google Scholar]
- Colleoni C, Dauvillee D, Mouille G, et al. Genetic and biochemical evidence for the involvement of α-1,4 glucanotransferases in amylopectin synthesis. Plant Physiology. 1999a;120:993. doi: 10.1104/pp.120.4.993. -1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colleoni C, Dauvillee D, Mouille G, Morell MK, Samuel M, Slomiany MC, Lienard L, Wattebled F, d'Hulst C, Ball S. Biochemical characterisation of the Chlamydomonas reinhardtii α-1,4 glucanotransferase supports a direct function in amylopectin biosynthesis. Plant Physiology. 1999b;120:1005–1014. doi: 10.1104/pp.120.4.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doehlert DC, Kuo TM, Felker FC. Enzymes of sucrose and hexose metabolism in developing kernels of two inbreds of maize. Plant Physiology. 1988;86:1013–1019. doi: 10.1104/pp.86.4.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Toyosawa Y, Utsumi Y, et al. Characterization of pullulanase (PUL)-deficient mutants of rice (Oryza sativa L.) and the function of PUL on starch biosynthesis in the developing rice endosperm. Journal of Experimental Botany. 2009;60:1009–1023. doi: 10.1093/jxb/ern349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N, Yoshida M, Kondo T, et al. Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm. Plant Physiology. 2007;144:2009–2023. doi: 10.1104/pp.107.102533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse T, Sasaki T, Yano M. Ti-plasmid vectors useful for functional analysis of rice genes. Plant Biotechnology. 2001;18:219–222. [Google Scholar]
- Giroux MJ, Hannah LC. ADP-glucose pyrophosphorylase in shrunken-2 and brittle-2 mutants of maize. Molecular and General Genetics. 1994;243:400–408. doi: 10.1007/BF00280470. [DOI] [PubMed] [Google Scholar]
- Gruber CW, Cemazar M, Heras B, Martin JL, Craik DJ. Protein disulfide isomerase: the structure of oxidative folding. Trends in Biochemical Sciences. 2006;31:455–464. doi: 10.1016/j.tibs.2006.06.001. [DOI] [PubMed] [Google Scholar]
- Hennen-Bierwagen TA, Lin Q, Grimaud F, Planchot V, Keeling PL, James MG, Myers AM. Proteins from multiple metabolic pathways associate with starch biosynthetic enzymes in high molecular weight complexes: a model for regulation of carbon allocation in maize amyloplasts. Plant Physiology. 2009;149:1541–1559. doi: 10.1104/pp.109.135293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennen-Bierwagen TA, Liu F, Marsh RS, Kim S, Gan Q, Tetlow IJ, Emes MJ, James MG, Myers AM. Starch biosynthetic enzymes from developing maize endosperm associate in multisubunit complexes. Plant Physiology. 2008;146:1892–1908. doi: 10.1104/pp.108.116285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Hunter BG, Beatty MK, Singletary GW, Hamaker BR, Dilkes BP, Larkins BA, Jung R. Maize opaque endosperm mutations create extensive changes in patterns of gene expression. The Plant Cell. 2002;14:2591–2612. doi: 10.1105/tpc.003905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang SK, Nishi A, Satoh H, Okita TW. Rice endosperm-specific plastidial α-glucan phosphorylase is important for synthesis of short-chain malto-oligosaccharides. Archives of Biochemistry and Biophysics. 2010;495:82–92. doi: 10.1016/j.abb.2009.12.023. [DOI] [PubMed] [Google Scholar]
- Iwata Y, Fedoroff NV, Koizumi N. Arabidopsis bZIP60 is a proteolysis-activated transcription factor involved in the endoplasmic reticulum stress response. The Plant Cell. 2008;20:3107–3121. doi: 10.1105/tpc.108.061002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang H, Park S, Matsuoka M, An G. White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C4-type pyruvate orthophosphate dikinase gene (OsPPDKB) The Plant Journal. 2005;42:901–911. doi: 10.1111/j.1365-313X.2005.02423.x. [DOI] [PubMed] [Google Scholar]
- Kaufman RJ. Stress signaling from the lumen of the endoplasmic reticulum: coordination of gene transcriptional and translational controls. Genes and Development. 1999;13:1211–1233. doi: 10.1101/gad.13.10.1211. [DOI] [PubMed] [Google Scholar]
- Kawagoe Y, Kubo A, Satoh H, Takaiwa F, Nakamura Y. Roles of isoamylase and ADP-glucose pyrophosphorylase in starch granule synthesis in rice endosperm. The Plant Journal. 2005;42:164–174. doi: 10.1111/j.1365-313X.2005.02367.x. [DOI] [PubMed] [Google Scholar]
- Kim CS, Hunter BG, Kraft J, Boston RS, Yans S, Jung R, Larkins BA. A defective signal peptide in a 19-kD α-zein protein causes the unfolded protein response and an opaque endosperm phenotype in the maize De*-B30 mutant. Plant Physiology. 2004;134:380–387. doi: 10.1104/pp.103.031310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleffmann T, Russenberger D, von Zychlinski A, Christopher W, Sjolander K, Gruissem W, Baginsky S. The Arabidopsis thaliana chloroplast proteome reveals pathway abundance and novel protein functions. Current Biology. 2004;14:354–362. doi: 10.1016/j.cub.2004.02.039. [DOI] [PubMed] [Google Scholar]
- Kossmann J, Visser RGF, Muller-Rober B, Willmitzer L, Sonnewald U. Cloning and expression analysis of a potato cDNA that encodes branching enzyme: evidence for co-expression of starch biosynthetic genes. Molecular and General Genetics. 1991;230:39–44. doi: 10.1007/BF00290648. [DOI] [PubMed] [Google Scholar]
- Kubo A, Fujita N, Harada K, Matsuda T, Satoh H, Nakamura Y. The starch-debranching enzymes isoamylase and pullulanase are both involved in amylopectin biosynthesis in rice endosperm. Plant Physiology. 1999;121:399–410. doi: 10.1104/pp.121.2.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo A, Rahman S, Utsumi Y, et al. Complementation of sugary-1 phenotype in rice endosperm with the wheat isoamylase1 gene supports a direct role for isoamylase1 in amylopectin biosynthesis. Plant Physiology. 2005;137:43–56. doi: 10.1104/pp.104.051359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laboissiere MC, Sturley SL, Raines RT. The essential function of protein-disulfide isomerase is to unscramble non-native disulfide bonds. Journal of Biological Chemistry. 1995;270:28006–28009. doi: 10.1074/jbc.270.47.28006. [DOI] [PubMed] [Google Scholar]
- Lending CR, Larkins BA. Effect of the floury-2 locus on protein body formation during maize endosperm development. Protoplasma. 1992;171:123–133. [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH. Salt stress responses in Arabidopsis utilize a signal transduction pathway related to endoplasmic reticulum stress signaling. The Plant Journal. 2007a;51:897–909. doi: 10.1111/j.1365-313X.2007.03195.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JX, Srivastava R, Che P, Howell SH. An endoplasmic reticulum stress response in Arabidopsis is mediated by proteolytic processing and nuclear relocation of a membrane-associated transcription factor, bZIP28. The Plant Cell. 2007b;19:4111–4119. doi: 10.1105/tpc.106.050021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Ma X, Liu S, et al. Identification and characterization of a novel Waxy allele from a Yunnan rice landrace. Plant Molecular Biology. 2009;71:609–626. doi: 10.1007/s11103-009-9544-4. [DOI] [PubMed] [Google Scholar]
- Lu DP, Christopher DA. Immunolocalization of a protein disulfide isomerase to Arabidopsis thaliana chloroplasts and its association with starch biogenesis. International Journal of Plant Sciences. 2006;167:1–9. [Google Scholar]
- Myers AM, Morell MK, James MG, Ball SG. Recent progress toward understanding biosynthesis of the amylopectin crystal. Plant Physiology. 2000;12,:989–997. doi: 10.1104/pp.122.4.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishi A, Nakamura Y, Tanaka N, Satoh H. Biochemical and genetic analysis of the effects of amylose-extender mutation in rice endosperm. Plant Physiology. 2001;127:459–472. [PMC free article] [PubMed] [Google Scholar]
- Ohdan T, Francisco PB, Sawada T, Hirose T, Terao T, Satoh H, Nakamura Y. Expression profiling of genes involved in starch synthesis in sink and source organs of rice. Journal of Experimental Botany. 2005;56:3229–3244. doi: 10.1093/jxb/eri292. [DOI] [PubMed] [Google Scholar]
- Onda Y, Nagamine A, Sakurai M, Kumamaru T, Ogawa M, Kawagoe Y. Distinct roles of protein disulfide isomerase and P5 sulfhydryl oxidoreductases in multiple pathways for oxidation of structurally diverse storage proteins in rice. The Plant Cell. 2011;23:210–223. doi: 10.1105/tpc.110.079509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oono Y, Wakasa Y, Hirose S, Yang L, Sakuta C, Takaiwa F. Analysis of ER stress in developing rice endosperm accumulating β-amyloid peptide. Plant Biotechnology. 2010;8:691–718. doi: 10.1111/j.1467-7652.2010.00502.x. [DOI] [PubMed] [Google Scholar]
- O'Shea MG, Morell MK. High resolution slab gel electrophoresis of 8-amino-1,3,6-pyrenetrisulfonic acid (APTS) tagged oligosaccharides using a DNA sequencer. Electrophoresis. 1996;17:681–686. doi: 10.1002/elps.1150170410. [DOI] [PubMed] [Google Scholar]
- Ozbun JL, Hawker JS, Greenberg E, Lammel C, Preiss J. Starch synthetase, phosphorylase, ADPglucose pyrophosphorylase, and UDPglucose pyrophosphorylase in developing maize kernels. Plant Physiology. 1973;51:1–5. doi: 10.1104/pp.51.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh H, Shibahara K, Tokunaga T, et al. Mutation of the plastidial α-glucan phosphorylase gene in rice affects the synthesis and structure of starch in the endosperm. The Plant Cell. 2008;20:1833–1849. doi: 10.1105/tpc.107.054007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh-Cruz M, Crofts AJ, Takemoto-Kuno Y, Sugino A, Washida H, Crofts N, Okita TW, Ogawa M, Satoh H, Kumamaru T. Protein disulfide isomerase like 1-1 participates in the maturation of proglutelin within the endoplasmic reticulum in rice endosperm. Plant and Cell Physiology. 2010;51:1581–1593. doi: 10.1093/pcp/pcq098. [DOI] [PubMed] [Google Scholar]
- Schupp N, Ziegler P. The relation of starch phosphorylases to starch metabolism in wheat. Plant and Cell Physiology. 2004;45:1471–1484. doi: 10.1093/pcp/pch170. [DOI] [PubMed] [Google Scholar]
- She KC, Kusano H, Koizumi K, et al. A novel factor FLOURY ENDOSPERM 2 is involved in regulation of rice grain size and starch quality. The Plant Cell. 2010;22:3280–3294. doi: 10.1105/tpc.109.070821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Pierre B, Brisson N. Induction of the plastidic starch-phosphorylase gene in potato storage sink tissue. Planta. 1995;195:339–344. [Google Scholar]
- Szegezdi E, Logue SE, Gorman AM, Samali A. Mediators of endoplasmic reticulum stress-induced apoptosis. EMBO Reports. 2006;7:880–885. doi: 10.1038/sj.embor.7400779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto Y, Coughlan SJ, Okita TW, Satoh H, Ogawa M, Kumamaru T. The rice mutant esp2 greatly accumulates the glutelin precursor and deletes the protein disulfide isomerase. Plant Physiology. 2002;128:1212–1222. doi: 10.1104/pp.010624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyota K, Tamura M, Ohdan T, Nakamura Y. Expression profiling of starch metabolism-related plastidic translocator genes in rice. Planta. 2006;223:248–257. doi: 10.1007/s00425-005-0128-5. [DOI] [PubMed] [Google Scholar]
- Wakasa Y, Yasuda H, Oono Y, Kawakatsu T, Hirose S, Takahashi H, Hayashi S, Yang L, Takaiwa F. Expression of ER quality control-related genes in response to changes in BiP1 levels in developing rice endosperm. The Plant Journal. 2011;65:675–689. doi: 10.1111/j.1365-313X.2010.04453.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Ren Y, Liu X, et al. OsRab5a regulates endomembrane organization and storage protein trafficking in rice endosperm cells. The Plant Journal. 2010;64:812–824. doi: 10.1111/j.1365-313X.2010.04370.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Zhu S, Liu S, et al. The vacuolar processing enzyme OsVPE1 is required for efficient glutelin processing in rice. The Plant Journal. 2009;58:606–617. doi: 10.1111/j.1365-313X.2009.03801.x. [DOI] [PubMed] [Google Scholar]
- Wilkinson B, Gilbert HF. Protein disulfide isomerase. Biochimica et Biophysica Acta. 2004;1699:35–44. doi: 10.1016/j.bbapap.2004.02.017. [DOI] [PubMed] [Google Scholar]
- Yasuda H, Hirose S, Kawakatsu T, Wakasa Y, Takaiwa F. Overexpression of BiP has inhibitory effects on the accumulation of seed storage proteins in endosperm cells of rice. Plant and Cell Physiology. 2009;50:1532–1543. doi: 10.1093/pcp/pcp098. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.