Abstract
The proteins of the MYB superfamily play central roles in developmental processes and defence responses in plants. Sixty unique wheat MYB genes that contain full-length cDNA sequences were isolated. These 60 genes were grouped into three categories, namely one R1R2R3-MYB, 22 R2R3-MYBs, and 37 MYB-related members. The sequence composition of the R2 and R3 repeats was conserved among the 22 wheat R2R3-MYB proteins. Phylogenetic comparison of the members of this superfamily among wheat, rice, and Arabidopsis revealed that the putative functions of some wheat MYB proteins were clustered into the Arabidopsis functional clades. Tissue-specific expression profiles showed that most of the wheat MYB genes were expressed in all of the tissues examined, suggesting that wheat MYB genes take part in multiple cellular processes. The expression analysis during abiotic stress identified a group of MYB genes that respond to one or more stress treatments. The overexpression of a salt-inducible gene, TaMYB32, enhanced the tolerance to salt stress in transgenic Arabidopsis. This study is the first comprehensive study of the MYB gene family in Triticeae.
Keywords: Abiotic stress, expression profile, MYB transcription factor, wheat
Introduction
Abiotic stresses, such as high salt, drought, and low temperature, are caused by complex environmental conditions, which adversely influence crop growth and production. To overcome these limitations, plants must respond and cope with these stresses at both the physiological and biochemical level (Zhu, 2002; Byrt and Munns, 2008; Munns and Tester, 2008). Therefore, understanding abiotic stress responses has vital significance for crop improvement regarding stress tolerance. Transcription factors (TFs) regulate gene expression, which provides plants with a complex control mechanism for responding to abiotic and biotic stresses and modulating developmental processes (Mitsuda and Ohme-Takagi, 2009). A previous study uncovered a group of TF genes, such as DREB, MYB, and bZIP, that played important roles in plant molecular stress responses (Ahuja et al., 2010).
The MYB TFs comprise one of the largest gene families (Pabo and Sauer, 1992; Riechmann et al., 2000). The distinguishing characteristic of MYB TFs is possession of a MYB domain, which consists of 1–4 imperfect tandem repeats (MYB repeat) located near the N-terminus. The MYB gene family is divided into different types according to the number of repeat(s) in the MYB domain: 4R-MYB has four repeats, 3R-MYB (R1R2R3-MYB) has three consecutive repeats, R2R3-MYB has two repeats, and the MYB-related type usually, but not always, has a single repeat (Rosinski and Atchley, 1998; Jin and Martin, 1999; Dubos et al., 2010). Typically, the MYB repeat is 50–53 amino acids in length and contains three regularly distributed tryptophan (or phenylalanine) residues, which can together form a hydrophobic core. Each MYB repeat forms three α-helices: the two that are located at the C-terminus adopt a variation of the helix–turn–helix (HLH) conformation that recognizes and binds to the DNA major groove at the specific recognition site C/TAACG/TG (Ogata and Nishimura, 1995; Ogata, 1998).
Since the first plant MYB gene, C1, was isolated in Zea mays (Paz-Ares et al., 1987), research concerning different aspects of the MYB gene family, including gene number, sequence characterization, evolution, and potential functions, has been widely conducted in plants (Chen et al., 2006; Matus et al., 2008; Wilkins et al., 2009; Dubos et al., 2010). So far, large numbers of MYB genes have been identified in different plant species, comprising 204 members in Arabidopsis, 218 members in rice, 279 members in grapevine, 197 members in Populus, and 180 members in Brachypodium (Chen et al., 2006; Velasco et al., 2007; Wilkins et al., 2009; International Brachypodium Initiative, 2010). The MYB proteins are involved in many significant physiological and biochemical processes, including the regulation of primary and secondary metabolism, the control of cell development and the cell cycle, the participation in defence and response to various biotic and abiotic stresses, and hormone synthesis and signal transduction (Stracke et al., 2001; Feller et al., 2011; Du et al., 2009; Dubos et al., 2010).
Extensive studies of the MYB gene family in various plant species have provided a better understanding of this gene family; however, little is known about this gene family in bread wheat. Until now, only one R2R3-MYB gene, TaMYB1, whose expression occurs in response to abiotic stresses, was reported in wheat (Lee et al., 2007). As one of the most important agricultural crops, wheat is a staple for a large portion of the world's population. Unfortunately, its production is severely affected by adverse environmental stresses. Therefore, the identification and functional study of stress responses and tolerance genes will elucidate the molecular mechanisms of the plant stress response and tolerance, and will ultimately lead to improvement of stress tolerance in wheat. As a hexaploid species (Triticum aestivum L., 2n=6x=42, AABBDD), bread wheat has a huge genome of 17 000 Mb in size that comprises a high percentage of repetitive sequences (Arumuganathan and Earle, 1991; Li et al., 2004). At present, it is a great challenge to sequence the wheat genome. As a result, the full-length cDNA is an important resource for isolating the functional genes in wheat (Manickavelu et al., 2010).
Considering the multiple functions of MYB proteins, especially their important roles in response to abiotic stresses in plants, research was conducted concerning several aspects of the MYB gene family in wheat. In this study, 60 full-length cDNA sequences encoding wheat MYB proteins were isolated. A phylogenetic tree combining wheat, rice, and Arabidopsis MYB proteins was constructed to examine their evolutionary relationships and the putative functions of wheat MYB proteins based on Arabidopsis MYB proteins with known functions. Tissue-specific analysis was conducted and abiotic stress response expression profiles were generated to find potential genes that participate in the stress signal transduction pathway in wheat. The function of a salt stress-inducible gene was studied in transgenic Arabidopsis. This extended analysis is the first comprehensive study of the MYB gene family in bread wheat and provides valuable information for further exploration of the functions of this significant gene family in Triticeae.
Materials and methods
Plant materials
Several wheat materials were used for different experiments in this study. For the tissue-specific analysis, roots, stems, leaves, anthers, and pistils were harvested from Chinese Spring. Wheat cv. Chadianhong (salt resistance) was used for the high salt treatment, wheat cv. Hanxuan 10 (drought resistance) was used for the drought treatment, and wheat cv. Chinese Spring was used for low temperature and exogenous abscisic acid (ABA) treatments. Seedlings were grown in liquid culture at 22 °C under a 16 h light/8 h dark photoperiod. For the high salt, drought, and ABA treatments, 10-day-old seedlings were treated with 250 mM NaCl, 16.1% polyethylene glycol (PEG; –0.5 MPa), and 200 μM ABA, respectively. Their root tissues were collected at 0, 1, 3, 7, 12, and 24 h. For the cold treatment, 10-day-old seedlings were transferred from 22 °C to 4 °C, and the samples were harvested at 0, 1, 3, 7, 12, 24, and 72 h.
Identification of wheat MYB genes
Previous studies had generated 10 wheat full-length wheat cDNA libraries, which included 120 000 expressed sequence tag (EST) sequences (unpublished data). First, all of the EST sequences were subjected to six-frame translation. Then, the deduced translated amino acid sequences were screened using the HMMER2.2.1 software package: profile PF00249 was used to identify the wheat MYBs, and an E-value of 0.01 was adopted (Eddy, 1998; Finn et al., 2008). Full-length cDNA sequences of the screened putative MYBs were obtained by sequencing the cDNA plasmids using the ABI 3730×L 96-capillary DNA analyser (Applied Biosystems). To remove redundancy, the sequences were assembled using the SeqMan function of the DNAStar software package and adjusted manually. Only the sequences that shared >95% matches were considered redundant. The open reading frames (ORFs) were predicted using FGENESH (http://www.softberry.com) and were translated into amino acid sequences. Finally, to confirm that the obtained sequences were MYB members, all of the non-redundant amino acid sequences of the primary identified MYB members were submitted to the website http://pfam.sanger.ac.uk to predict the MYB domains. Only the sequences that shared the MYB domain were confirmed to be MYB members (Coggill et al., 2008). Wheat MYB sequences reported in this work have been submitted to GenBank with accession numbers JF951900–JF951957.
Conservative analysis of R2R3-MYB proteins within MYB domains
To analyse the features of the MYB domain of wheat R2R3-MYB proteins, the sequences of R2 and R3 MYB repeats corresponding to 22 R2R3-MYB proteins were aligned with the ClustalW method using BioEdit software and adjusted manually. The sequence logos for R2 and R3 MYB repeats were obtained by submitting the multiple alignment sequences to the website http://weblogo.berkeley.edu/logo.cgi (Crooks et al., 2004).
Phylogenetic analysis
The amino acid sequences of Arabidopsis (five R1R2R3-MYB, 126 R2R3-MYB, and 73 MYB-related members) and rice (four R1R2R3-MYB, 123 R2R3-MYB, and 91 MYB-related members) MYB proteins were downloaded from the PlantTFDB website (He et al., 2010). The complete amino acid sequences of MYB proteins were used to construct phylogenetic trees. Sequence alignments were performed with ClustalW using BioEdit software and adjusted manually. Neighbor–Joining trees for 3R-MYB and R2R3-MYB subfamilies and the MYB-related subfamily combining wheat, rice, and Arabidopsis MYB members were constructed individually using the MEGA4.1 program (Tamura et al., 2007), and internal branch support was estimated with 1000 bootstrap replicates.
Semi-quantitative RT-PCR and real-time PCR analysis
Total RNA was extracted using TRIZOL reagent and treated with DNase I. For each sample, 10 μg of total RNA was used to synthesize first-strand cDNA using SuperScriptII reverse transcriptase (Invitrogen). For semi-quantitative reverse transcription-PCR (RT-PCR), a 20 μl reaction volume that included 1 μl of cDNA template was used. The gene-specific primers for the 60 wheat MYB genes can be found in Supplementary Table S1 available at JXB online. The PCR parameters were 95 °C for 5 min; followed by 32 cycles of 95 °C for 30 s, 55 °C for 30 s, 72 °C for 40 s, and a final step at 72 °C for 5 min. Each PCR was replicated three times. The concentration of the cDNAs was adjusted using the wheat tubulin gene with the primer sequences 5′-TGAGGACTGGTGCTTACCGC-3′ and 5′-GCACCATCAAACCTCAGGGA-3′; this PCR was run for 25 cycles and the other parameters were the same as for the amplification of the MYB genes. For real-time PCR, each reaction contained 12.5 μl of 2× PCR Master Mix (Tiangen), 2.0 μl of cDNA, 5 μl of SybrGreen I, and 3 μl of gene-specific primers (2.0 μM) in a final volume of 25 μl. The PCR parameters were 95 °C for 2 min; followed by 40 cycles of 95 °C for 20 s, 57 °C for 30 s, and 72 °C for 31 s. The gene-specific primers that were used for real-time PCR were the same as those used in the semi-quantitative RT-PCR. The wheat tubulin gene was used as a reference gene, and the reactions were performed using the ABI Prism 7000 real-time PCR system (Applied Biosystems). The reactions were repeated three times. The quantitative analysis used the 2–ΔΔCT method.
Transformation of TaMYB32 in Arabidopsis
The full-length coding sequence (ATG to TAA) of TaMYB32 was amplified from plasmid cDNA by PCR using attB sites containing the following gene-specific primers: 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTCAATGGTGAGGGCTCCTT-3′ and 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTGTTAAATCT-GTGACAAACTCTGTAT-3′ (attB sites are underlined). The PCR product was cloned behind the Cauliflower mosaic virus (CaMV) 35S promoter in the plant expression vector pLEELA by the Gateway cloning method. The construct was confirmed by sequencing and was transferred into Agrobacterium GV3101::Pmp90RK. The Arabidopsis were transformed by the floral dip method (Clough and Bent, 1998). Positive transgenic plants were first screened by spraying with Basta and then tested by RT-PCR.
Salt treatment in transgenic Arabidopsis
The surface-sterilized seeds from transgenic and wild-type plants were sown on MS medium plates with 3% (w/v) sucrose and 0.8% agar (w/v). The plates were placed at 4 °C for 2 d and then the plates were moved to a vertical position under a long-day photoperiod (16 h light/8 h dark) at 22 °C. Subsequently, 4-day-old seedlings were transferred onto vertical MS agar plates, supplemented with 0, 100, or 200 mM NaCl. Four days after growth in the treatment medium, plants with absent green and albinotic cotyledons were scored. The root length of the seedlings was measured. The salt treatment experiment was performed in triplicate.
Results
Isolation of 60 genes encoding MYB proteins in wheat
To identify the wheat MYB genes, 120 000 wheat EST sequences from full-length cDNA libraries were subjected to six-frame translation and were screened using the HMMER program: profile PF00249 was used to identify the members of the MYB gene family, and only the hits with E-values <0.01 were considered putative MYB genes. The primary search revealed 103 hits, of which 43 were removed, as they either did not contain the conserved MYB domain signature or were redundant with other sequences. The remaining 60 members represented the unique wheat MYB genes (Supplementary Table S2 at JXB online). A keyword search in the NCBI database revealed 14 previously annotated wheat MYB sequences (Supplementary Table S3). Comparing the 60 sequences identified in this study with those of the 14 sequences in the NCBI database revealed two sequences that were identical to the NCBI annotated genes TaMYB1 and TaMYB7. To create consecutive nomenclature, the remaining 58 sequences were designated as TaMYB15–TaMYB72.
These 60 full-length MYB sequences belonged to three subfamilies, namely one R1R2R3-MYB member, 22 R2R3-MYB members, and 37 MYB-related members; no 4R-MYB was found in this study (Supplementary Table S2 at JXB online). The R2R3 family contained fewer members of MYB genes than the MYB-related family: this result is not consistent with the annotation results for the Arabidopsis, rice, and Brachypodium genomes. The 37 MYB-related proteins contained one or two MYB repeat (s). For those six members that had two MYB repeats, their sequences did not have the normative character of the R2R3-MYB family, so they were included in the MYB-related family. The other 31 MYB-related members contain a single MYB repeat.
Sequence conservation within the MYB domain of R2R3-MYB proteins
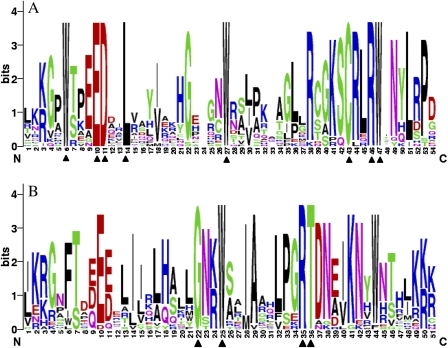
The R2R3-MYB proteins share significant sequence conservation within the MYB domain regions. To gain insight into the wheat R2R3-MYB domains, sequence logos were produced to examine the level of conservation of R2 and R3 repeats in the R2R3-MYB proteins within each residue position. The results reveal that eight and four conserved amino acid residues were identical among all the members detected in the R2 and R3 MYB repeat regions, respectively. The residues in the other positions had different levels of conservation, and approximately half of them had >50% appearance at the respective position (Fig. 1). The typical MYB protein contains three regularly spaced tryptophan residues, which play significant roles in interaction between the MYB protein and specific DNA sequences (Ogata and Nishimura, 1995). For the 22 wheat R2R3-MYB proteins, all of the R2 repeat sequences contained three tryptophan residues: however, in the R3 repeats, the first tryptophan residue was replaced by phenylalanine. The second and third tryptophan residues were conserved in all the members, except TaMYB30 whose third tryptophan residue was replaced by phenylalanine. These results are consistent with those from Arabidopsis (Stracke et al., 2001).
Fig. 1.
Sequence logos for the R2 (A) and R3 (B) repeat of wheat R2R3-MYB proteins. The overall height of each stack indicates the conservation of the sequence at that position, whereas the height of letters within each stack represents the relative frequency of the corresponding amino acid. The triangle indicates the position of the conserved amino acid that is identical among all the 22 wheat R2R3-MYB proteins. (This figure is available in colour at JXB online.)
Phylogenetic comparison of the MYB TFs from wheat, rice, and Arabidopsis
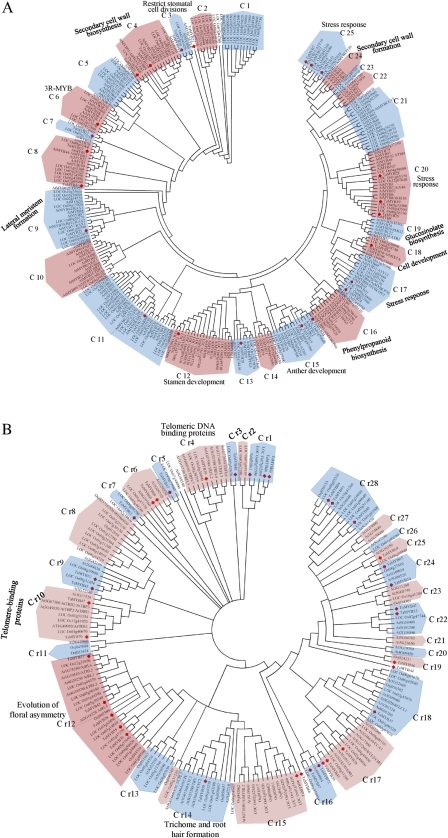
The functions of several Arabidopsis MYB proteins has been well characterized experimentally, and phylogenetic analysis of Arabidopsis MYB proteins has identified several functional clades (Matus et al., 2008; Dubos et al., 2010). Thus, a phylogenetic tree combining wheat, rice, and Arabidopsis MYB proteins would not only help to understand the phylogenetic relationships among the wheat, rice, and Arabidopsis MYB proteins, but would also allow speculation on the putative functions of the wheat MYB proteins based on the functional clades that are identified in Arabidopsis. Previous studies indicated that MYB-related proteins had undergone differentiation and evolution to a larger extent than R2R3-MYB and 3R-MYB in plants (Chen et al., 2006). Therefore, the phylogenetic trees for R2R3-MYB and 3R-MYB subfamilies and the MYB-related subfamily were constructed individually (Fig. 2A, B). The resulting trees generated 25 clades for R2R3-MYB and 3R-MYB subfamilies (named C1–C25), and 28 clades for the MYB-related subfamily (named Cr1–Cr28), respectively. A total of 57 of the 60 wheat MYB proteins were clustered into 33 clades. The other three wheat MYB sequences did not fit into any clade. Remarkably, 27 clades included different numbers of MYB proteins from all of the three species, while another six clades only contained wheat and rice MYB members. In some cases, it was easy to distinguish the putative orthologous genes of wheat MYBs in rice and Arabidopsis, since they were grouped in pairs within a clade, such as clade 7 and clade r25. In other situations, taking clade 1 and clade r27 as examples, homologues were grouped by species within a clade.
Fig. 2.
(A) Phylogenetic relationships of wheat, rice, and Arabidopsis R2R3-MYB and 3R-MYB proteins. The complete amino acid sequences of the 23 wheat, 131 Arabidopsis, and 127 rice R2R3-MYB and 3R-MYB proteins were aligned by ClustalW, and the Neighbor–Joining tree was constructed using MEGA 4.1 with 1000 bootstrap replicates. The functions of some clades were annotated. (B) Phylogenetic relationships of wheat, rice, and Arabidopsis MYB-related proteins. The complete amino acid sequences of the 37 wheat, 73 Arabidopsis, and 91 rice R2R3-MYB and 3R-MYB proteins were aligned by ClustalW, and the Neighbor–Joining tree was constructed using MEGA 4.1 with 1000 bootstrap replicates. The functions of some clades were annotated. (This figure is available in colour at JXB online.)
Some wheat MYB proteins were clustered into Arabidopsis functional clades, which provided an excellent reference to explore the functions of the wheat MYB genes. For example, TaMYB30 grouped together with Arabidopsis AtMYB88 and AtMYB124 (Lai et al., 2005; Xie et al., 2010) into clade 3, referring to the regulation of stoma aperture development. Wheat TaMYB25 was clustered into clade 9, sharing a high level of sequence similarity with three Arabidopsis meristem formation regulation proteins AtMYB37/RAX1, AtMYBYB38/BIT1/RAX2, and AtMYB84/RAX3 (Keller et al., 2006; Nilsson et al., 2007; Hong et al., 2008), implying that the possible functions of TaMYB25 are related to the development process of the meristem. TaMYB15, TaMYB34, and TaMYB26 were assembled together with AtMYB33/AtMYB65, AtMYB21, AtMYB24, and AtMYB108 (Millar and Gubler, 2005; Mandaokar et al., 2006; Reyes and Chua, 2007; Mandaokar and Browse, 2009) in clade 12 and clade 15, which represent the functional clades of the regulation of anther or stamen development. TaMYB16 was grouped into clade 18 with two Arabidopsis proteins AtMYB16 and AtMYB106 (Baumann et al., 2007; Jakoby et al., 2008), representing the functional clade with proteins responsible for cell development or morphogenesis. When plants are exposed to unfavourable conditions, a number of genes play roles in the response and tolerance to stresses. Clade 17, clade 20, and clade 25 contained many of the Arabidopsis proteins that are involved in these processes. For example, AtMYB30 is an activator of the hypersensitive cell death programme in response to pathogen attack (Vailleau et al., 2002; Raffaele et al., 2008; Li et al., 2009). AtMYB96 mediated the ABA signal network that confers abiotic and/or biotic stress tolerance (Seo et al., 2009; Seo and Park, 2010). The other five proteins, AtMYB3, AtMYB4, AtMYB8, AtMYB13, and AtMYB15, are regulatory factors related to different biotic or abiotic stress responses (Kirik et al., 1998; Jin et al., 2000; Hemm et al., 2001; Zhu et al., 2005; Agarwal et al., 2006; Zhao et al., 2007; Bang et al., 2008). Eight wheat R2R3-MYB proteins were grouped into these clades, which thus provided significant guidancee to identify the wheat genes that play roles in the response or tolerance to stress conditions.
Although the MYB-related subfamily seemingly expanded and evolved more rapidly than the R2R3-MYB subfamily, the phylogenetic analysis of the Arabidopsis MYB-related proteins also identified some functional clades (Jin and Martin, 1999; Chen et al., 2006). In this study, several wheat MYB proteins were clustered into those clades. Clade r4, which included wheat TaMYB39 and Arabidopsis TRFL proteins, and clade r10, which included wheat TaMYB47 and TaMYB70 and three interactional Arabidopsis proteins of the TRB subfamily, represented two telomeric DNA binding clades (Schrumpfova et al., 2008). Clade r14, containing wheat TaMYB50 and Arabidopsis proteins ETC2, TCL1, TRY, CPC, and ETC1, represented the clades for the regulation of trichome and root hair formation (Kirik et al., 2004). These results undoubtedly provide important clues for studying the function of the wheat MYB proteins.
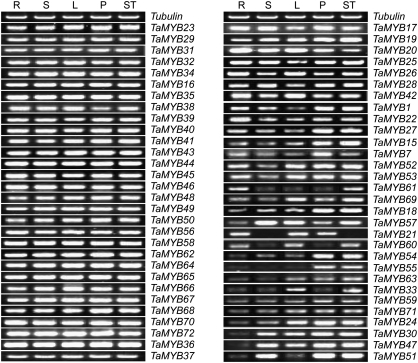
Expression profiles of wheat MYB genes in different tissues
Many important genes are selectively expressed in specific tissues involving some physiological and developmental processes. To investigate the spatial transcript profiles of the wheat MYB genes, semi-quantitative RT-PCR was used to detect the expression patterns of all wheat MYB genes in the roots, stems, leaves, pistils, and stamens. The expression profiles of the 60 wheat MYB genes showed different patterns of tissue-specific expression (Fig. 3). Half of the wheat MYB genes investigated were expressed in all of the tested tissues with similar high expression levels, indicating that the wheat MYB genes worked in various aspects of physiological and developmental processes. In contrast, the rest of the genes showed spatial variations of expression, with high levels of expression in one or some tissues, and low expression in others. For example, TaMYB30 and TaMYB47 showed high levels of expression in stems, leaves, pistils, and stamens, but clearly have low expression in the roots. TaMYB15 is significantly expressed in pistils and stamens and was an orthologous gene of AtMYB33 and AtMYB65, which function in facilitating proper anther development in Arabidopsis, indicating that TaMYB15 might have an important role in the process of flower development. Only one gene, TaMYB21, showed tissue-specific expression and was only detected in roots, leaves, and pistils.
Fig. 3.
Tissue-specific gene expression of 60 wheat MYB genes. Total RNA was isolated from roots (R), stems (S), leaves (L), pistils (P), and stamens (ST). The wheat tubulin gene was used to adjust cDNA concentrations. Gene-specific primers were used for semi-quantitative RT-PCR analysis of wheat MYB genes.
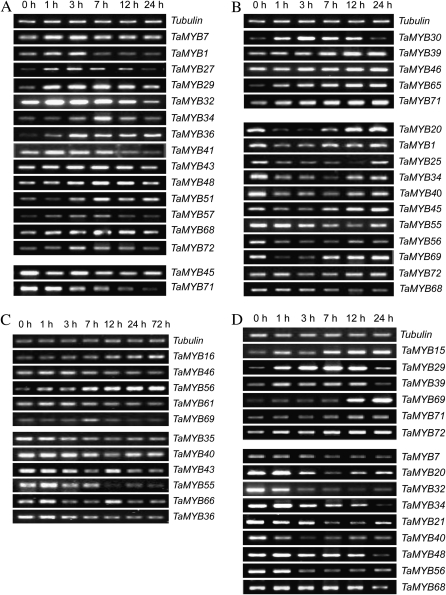
Screening the abiotic stress-responsive genes by the expression patterns of the 60 wheat MYB genes under different stress treatment conditions
Increasing evidence suggests that MYB transcription factors play important roles in the response to abiotic stresses (Dubos et al., 2010). The stress responses of the 60 wheat MYB genes were therefore screened using the results from the semi-quantitative RT-PCR of their expression profiles under different abiotic stress conditions. As a whole, 32 genes were detected to respond to at least one treatment, which included 16 genes treated with high salt, 16 genes treated with PEG, 11 genes treated with low temperature, and 15 genes treated with Exogenetic ABA (Fig. 4; Supplementary Table S4 at JXB online). Among them, 20 genes were able to respond to more than one treatment, and another 12 genes only responded to a single treatment. For example, TaMYB29 was induced by both high salt and exogenous ABA, while TaMYB30 was only induced by PEG. Interestingly, some genes behaved oppositely to their expression profile when subjected to different treatments. For instance, TaMYB1, TaMYB34, TaMYB68, and TaMYB72 were induced by high salt but were repressed by PEG; and TaMYB56 and TaMYB69 were repressed by PEG but induced by low temperature. When exposed to different treatments, the expression profiles of the wheat MYB genes seemed to be inconsistent. Under the high salt condition, most of the responsive genes (14 of 16) were induced. In contrast, under PEG treatment, more genes (11 of 16) were repressed than induced. However, nearly equal numbers of genes were induced and repressed after treatment with low temperature or exogenous ABA.
Fig. 4.
Expression patterns of wheat abiotic-responsive MYB genes under different treatment conditions. For high salt (A), PEG (B), and exogenous ABA (D) treatments, total RNA was isolated from roots of wheat seedlings at the indicated times, and for low temperature treatment (C), total RNA was isolated from leaves of wheat seedlings at the indicated times. In all, 5 μg of total RNA was reverse-transcribed into first-strand cDNA for RT-PCR. The wheat tubulin gene was used to adjust cDNA concentrations. Gene-specific primers were used for semi-quantitative RT-PCR analysis of wheat MYB genes.
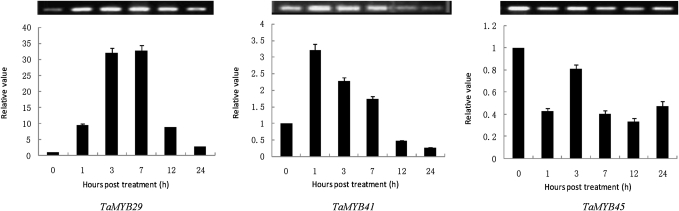
To validate the expression profiles obtained by semi-quantitative RT-PCR, three salt-inducible genes, TaMYB29, TaMYB41, and TaMYB45, were randomly selected and their expression patterns were analysed by real-time PCR. The results of the real-time PCR and the semi-quantitative RT-PCR are consistent (Fig. 5).
Fig. 5.
Expression patterns of three randomly selected high salt-responsive MYB genes. The expression patterns of three high-salt responsive genes, TaMYB29, TaMYB41, and TaMYB45, were detected by real-time PCR. The results of real-time PCR and semi-quantitative RT-PCR are consistent.
The overexpression of TaMYB32 confers increased tolerance to salt stress in transgenic Arabidopsis
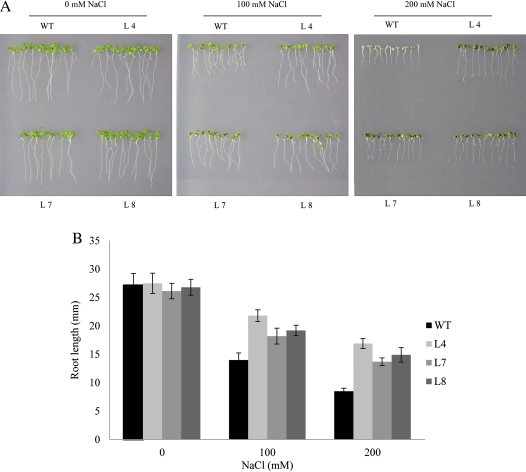
To understand the roles of wheat MYB genes in plant abiotic stress responses, a salt stress-inducible gene TaMYB32 was overexpressed in Arabidopsis under the control of the CaMV 35S promoter. For the salt tolerance analysis of TaMYB32-overexpressing Arabidopsis, three transgenic lines of T3 progeny and wild-type plants were grown in normal and high salt conditions. Under normal conditions, no obvious difference in the phenotype between wild-type and transgenic seedlings was observed. After treatment with 100 mM NaCl, both wild-type and transgenic seedlings grew normally, but the roots of the transgenic seedlings were longer than those of the wild-type. When the NaCl concentration was increased to 200 mM, the growth of the wild-type seedlings was completely inhibited, and the cotyledons showed evident albinism, while the transgenic seedlings were still green and continued growing (Fig. 6). These results indicate that the overexpression of TaMYB32 enhanced tolerance to salt in Arabidopsis. Additionally, the responses of the transgenic Arabidopsis to other abiotic stresses including drought, cold, and ABA treatment were also investigated; however, no phenotypic difference was observed between transgenic plants and the wild-type (results not shown).
Fig. 6.
Effects of TaMYB32 overexpression on salt tolerance in transgenic Arabidopsis. (A) Salt tolerance of transgenic Arabidopsis seedlings. Seeds of the wild-type and three transgenic lines were germinated on MS agar medium, and then transferred to fresh MS medium containing different concentrations of NaCl. (B) Root length of seedling in A. Error bars indicate the SD. (This figure is available in colour at JXB online.)
Discussion
Characterization of the wheat MYB gene family
The MYB superfamily has been defined as the most abundant TF family in plants, with at least 204 and 218 members in Arabidopsis and rice, respectively (Chen et al., 2006). However, no related information has been reported in Triticum. In this study, 60 wheat MYB genes containing full-length gene sequences were isolated. The common wheat (T. aestivum L.) contains a large genome of 17 000 Mb and is ∼128-fold larger than that of Arabidopsis and 40-fold larger than that of rice (Arumuganathan and Earle, 1991). However, whether wheat had more MYB genes than Arabidopsis and rice is uncertain, and the exact number of MYB genes in wheat will be determined when the whole wheat genome sequencing is completed. Considering that Arabidopsis and rice have notably more R2R3-MYB proteins than MYB-related proteins, it is interesting that the 60 wheat MYB proteins contained distinctly fewer members of the R2R3-MYB subfamily than of the MYB-related family. This observation might represent a unique characteristic of the wheat MYB gene family that should be determined after all of the wheat MYB genes are isolated from the wheat genome.
The evolution of the MYB proteins in wheat, rice, and Arabidopsis
Phylogenetic analysis of the MYB proteins had been conducted extensively in Arabidopsis, rice, grape, and Populus, and the evolutionary relationship of this gene family within and among the different species has been systematically studied. To obtain an overall picture of the 60 wheat MYB proteins and their relationships with those of rice and Arabidopsis, phylogenetic trees combining the wheat, rice, and Arabidopsis MYB proteins were constructed, which divided the 60 MYB members into 33 clades and three lines (single wheat MYB proteins). Most of the clades (27 of 33) contained different numbers of rice and Arabidopsis MYB proteins, indicating the large extent of conservation among the wheat, rice, and Arabidopsis MYB gene families. Six clades, r1, r3, r5, r7, r16, and r19, did not include any Arabidopsis MYB proteins but only members from wheat and rice, suggesting that the proteins in these clades were either acquired or expanded in wheat and rice lineages, or lost in Arabidopsis, following the divergence of wheat, rice, and Arabidopsis from the last common ancestor. Three wheat MYB proteins, TaMYB22, TaMYB44, and TaMYB66, did not cluster in any clade, implying that these MYB proteins may have specialized roles that were acquired or expanded in wheat during the process of wheat genome evolution. The 3R-MYB subfamily was considered to share the conservative function of controlling cell development between plants and animals (Jin and Martin, 1999). So far, five Arabidopsis and four rice 3R-MYB proteins have been identified, and, among them, AtMYB3R-1 and AtMYB3R-4 positively regulate cytokinesis (Haga et al., 2007). In this study, only one wheat MYB protein, 3R-MYB, was grouped together with all the five Arabidopsis and four rice 3R-MYB members. It seems that the 3R-MYB subfamily has sequence conservation among wheat, rice, and Arabidopsis, which may be related to their conserved functions in different species.
Several wheat MYB proteins were clustered into Arabidopsis functional clades, which provides valuable information for studying the functions of wheat MYB genes. Interestingly, none of the wheat and rice proteins were grouped within the Arabidopsis ‘glucosinolate biosynthesis’ clade (C19). A previous study indicated that this clade was derived from a β-type duplication event after Arabidopsis diverged from monocots but before diverging from Brassica (Chen et al., 2006), which may be the reason for its absence in wheat and rice.
Wheat MYB proteins respond to multiple abiotic stresses
For survival in unfavourable environmental conditions, plants have adopted different strategies to respond to various biotic and abiotic stresses (Ahuja et al., 2010). Many studies have indicated that the MYB TFs play essential roles in the regulation of gene expression to cope with environmental changes (Singh et al., 2002; Ahuja et al., 2010; Hirayama and Shinozaki, 2010). In the model plants, Arabidopsis and rice, a number of MYB genes were characterized to function as key factors in the signalling pathways for plant resistance to abiotic stresses (Vannini et al., 2004; Dai et al., 2007; Dubos et al., 2010). Although some of the wheat MYB gene sequences were present in the GenBank database, functional studies of this gene family were lacking. The expression patterns of all 60 wheat MYB genes under different abiotic stress conditions were studied. Thirty-two genes that responded to different stress treatments were discovered, of which 20 genes responded to multiple stress treatments, indicating that they were major factors involving cross-talk among different signal transduction pathways in response to abiotic stresses. However, multiple wheat MYB genes appeared to participate in responding to one stress stimulus, suggesting that there are multiple signalling pathways involving the response to abiotic stress treatment. Additionally, some genes exhibited opposite expression patterns under different stress conditions. However, for the most part, the gene expression profiles that respond to different stress treatments tend to be different, implying that the signalling pathways in the plant abiotic stress response are very complicated systems.
ABA is an important factor that participates in plant gene regulation networks involving abiotic response and tolerance. Stress response signalling transduction pathways are divided into two forms: ABA dependent and ABA independent (Shinozaki and Yamaguchi-Shinozaki, 2007). Under treatment with exogenous ABA, 15 wheat MYB genes changed their expression level: six inducible and nine repressible members. In those 15 genes, 14 of them also responded to one or more other stresses and might play roles in ABA-dependent signalling transduction pathways of abiotic stress responses.
Overexpression of the wheat R2R3-MYB gene TaMYB32 enhanced tolerance to salt stress
The functions of the MYB gene family have been characterized in Arabidopsis and rice (Du et al., 2009; Dubos et al., 2010; Feller et al., 2011). The overexpression of some MYB genes increased the tolerance of transgenic plants to biotic or abiotic stresses (Dai et al., 2007; Ma et al., 2009; Dubos et al., 2010). To date, only one wheat MYB gene, TaMYB1, had been characterized in wheat. The expression of TaMYB1 was enhanced by light under hypoxia, and gradually increased under treatment with NaCl. Furthermore, its transcript was slightly enhanced during the early stages of exogenous treatment with both ABA and PEG (Lee et al., 2007). However, a transgenic study of TaMYB1 is lacking, and its detailed function remains unknown. In the present study, TaMYB32 encoded an R2R3-MYB-type protein. Its expression was strongly induced during the early stages of treatment with NaCl (Fig. 4A). The TaMYB32 transgenic Arabidopsis showed improved tolerance to high salt. These results suggest that TaMYB32 was involved in the salt stress response and tolerance. The mechanism by which TaMYB32 conferred tolerance when subjected to salt stress and the effect of overexpression in wheat need further investigation.
In conclusion, 60 full-length cDNA sequences of wheat MYB genes have been isolated and their expression patterns have been analysed in different tissues and under different abiotic stress and exogenous ABA treatments. The phylogenetic comparison of wheat, rice, and Arabidopsis MYB members revealed the putative function of some wheat MYB proteins. The ectopic expression of TaMYB32 in Arabidopsis indicated that it was a stress response and tolerance gene. The results of this work provide significant information for improving the stress tolerance of crops through molecular breeding.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Specific primers for wheat MYB genes used in semi-quantitative RT-PCR.
Table S2. Characteristics of wheat MYB genes.
Table S3. Information on the wheat MYBs annotated in the NCBI database.
Table S4. Expression patterns of the wheat abiotic stress-responsive MYB genes.
Acknowledgments
We are grateful to Professor Yongfu Fu (Institute of Crop Sciences, Chinese Academy of Agricultural Sciences) for kindly providing the plant expression vector pLEELA. This work was supported by the National Transgenic Research Project (2008ZX08009-001 and 2011ZX08009-001).
References
- Agarwal M, Hao Y, Kapoor A, Dong CH, Fujii H, Zheng X, Zhu JK. A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. Journal of Biological Chemistry. 2006;281:37636–37645. doi: 10.1074/jbc.M605895200. [DOI] [PubMed] [Google Scholar]
- Ahuja I, de Vos RC, Bones AM, Hall RD. Plant molecular stress responses face climate change. Trends in Plant Science. 2010;15:664–674. doi: 10.1016/j.tplants.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Arumuganathan K, Earle ED. Nuclear DNA content of some important plant species. Plant Molecular Biology Reporter. 1991;9:208–218. [Google Scholar]
- Bang WY, Kim SW, Jeong IS, Koiwa H, Bahk JD. The C-terminal region (640–967) of Arabidopsis CPL1 interacts with the abiotic stress- and ABA-responsive transcription factors. Biochemical and Biophysical Research Communications. 2008;372:907–912. doi: 10.1016/j.bbrc.2008.05.161. [DOI] [PubMed] [Google Scholar]
- Baumann K, Perez-Rodriguez M, Bradley D, Venail J, Bailey P, Jin H, Koes R, Roberts K, Martin C. Control of cell and petal morphogenesis by R2R3 MYB transcription factors. Development. 2007;134:1691–1701. doi: 10.1242/dev.02836. [DOI] [PubMed] [Google Scholar]
- Byrt CS, Munns R. Living with salinity. New Phytologist. 2008;179:903–905. doi: 10.1111/j.1469-8137.2008.02596.x. [DOI] [PubMed] [Google Scholar]
- Chen YH, Yang XY, He K, et al. The MYB transcription factor superfamily of Arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Molecular Biology. 2006;60:107–124. doi: 10.1007/s11103-005-2910-y. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip:a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Coggill P, Finn RD, Bateman A. Identifying protein domains with the Pfam database. Current Protocols in Bioinformatics Chapter 2, 2008 doi: 10.1002/0471250953.bi0205s23. Unit 2 5. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Research. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Xu Y, Ma Q, Xu W, Wang T, Xue Y, Chong K. Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiology. 2007;143:1739–1751. doi: 10.1104/pp.106.094532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du H, Zhang L, Liu L, Tang XF, Yang WJ, Wu YM, Huang YB, Tang YX. Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry. 2009;74:1–11. doi: 10.1134/s0006297909010015. [DOI] [PubMed] [Google Scholar]
- Dubos C, Stracke R, Grotewold E, Weisshaar B, Martin C, Lepiniec L. MYB transcription factors in Arabidopsis. Trends in Plant Science. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- Feller A, Machemer K, Braun EL, Grotewold E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. The Plant Journal. 2011;66:94–116. doi: 10.1111/j.1365-313X.2010.04459.x. [DOI] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, et al. The Pfam protein families database. Nucleic Acids Research. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga N, Kato K, Murase M, Araki S, Kubo M, Demura T, Suzuki K, Muller I, Voss U, Jurgens G, Ito M. R1R2R3-Myb proteins positively regulate cytokinesis through activation of KNOLLE transcription in Arabidopsis thaliana. Development. 2007;134:1101–1110. doi: 10.1242/dev.02801. [DOI] [PubMed] [Google Scholar]
- He K, Guo AY, Gao G, Zhu QH, Liu XC, Zhang H, Chen X, Gu X, Luo J. Computational identification of plant transcription factors and the construction of the PlantTFDB database. Methods in Molecular Biology. 2010;674:351–368. doi: 10.1007/978-1-60761-854-6_21. [DOI] [PubMed] [Google Scholar]
- Hemm MR, Herrmann KM, Chapple C. AtMYB4: a transcription factor general in the battle against UV. Trends in Plant Science. 2001;6:135–136. doi: 10.1016/s1360-1385(01)01915-x. [DOI] [PubMed] [Google Scholar]
- Hirayama T, Shinozaki K. Research on plant abiotic stress responses in the post-genome era: past, present and future. The Plant Journal. 2010;61:1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x. [DOI] [PubMed] [Google Scholar]
- Hong SH, Kim HJ, Ryu JS, Choi H, Jeong S, Shin J, Choi G, Nam HG. CRY1 inhibits COP1-mediated degradation of BIT1, a MYB transcription factor, to activate blue light-dependent gene expression in Arabidopsis. The Plant Journal. 2008;55:361–371. doi: 10.1111/j.1365-313X.2008.03508.x. [DOI] [PubMed] [Google Scholar]
- International Brachypodium Initiative Genome sequencing and analysis of the model grass Brachypodium distachyon. Nature. 2010;463:763–768. doi: 10.1038/nature08747. [DOI] [PubMed] [Google Scholar]
- Jakoby MJ, Falkenhan D, Mader MT, Brininstool G, Wischnitzki E, Platz N, Hudson A, Hulskamp M, Larkin J, Schnittger A. Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiology. 2008;148:1583–1602. doi: 10.1104/pp.108.126979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Cominelli E, Bailey P, Parr A, Mehrtens F, Jones J, Tonelli C, Weisshaar B, Martin C. Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO Journal. 2000;19:6150–6161. doi: 10.1093/emboj/19.22.6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Martin C. Multifunctionality and diversity within the plant MYB-gene family. Plant Molecular Biology. 1999;41:577–585. doi: 10.1023/a:1006319732410. [DOI] [PubMed] [Google Scholar]
- Keller T, Abbott J, Moritz T, Doerner P. Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. The Plant Cell. 2006;18:598–611. doi: 10.1105/tpc.105.038588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik V, Kolle K, Wohlfarth T, Misera S, Baumlein H. Ectopic expression of a novel MYB gene modifies the architecture of the Arabidopsis inflorescence. The Plant Journal. 1998;13:729–742. doi: 10.1046/j.1365-313x.1998.00072.x. [DOI] [PubMed] [Google Scholar]
- Kirik V, Simon M, Huelskamp M, Schiefelbein J. The ENHANCER OF TRY and CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Developmental Biology. 2004;268:506–513. doi: 10.1016/j.ydbio.2003.12.037. [DOI] [PubMed] [Google Scholar]
- Lai LB, Nadeau JA, Lucas J, Lee EK, Nakagawa T, Zhao L, Geisler M, Sack FD. The Arabidopsis R2R3 MYB proteins FOUR LIPS and MYB88 restrict divisions late in the stomatal cell lineage. The Plant Cell. 2005;17:2754–2767. doi: 10.1105/tpc.105.034116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TG, Jang CS, Kim JY, Kim DS, Park JH, Kim DY, Seo YW. A Myb transcription factor (TaMyb1) from wheat roots is expressed during hypoxia: roles in response to the oxygen concentration in root environment and abiotic stresses. Physiologia Plantarum. 2007;129:375–385. [Google Scholar]
- Li L, Yu X, Thompson A, Guo M, Yoshida S, Asami T, Chory J, Yin Y. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. The Plant Journal. 2009;58:275–286. doi: 10.1111/j.1365-313X.2008.03778.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Zhang P, Fellers JP, Friebe B, Gill BS. Sequence composition, organization, and evolution of the core Triticeae genome. The Plant Journal. 2004;40:500–511. doi: 10.1111/j.1365-313X.2004.02228.x. [DOI] [PubMed] [Google Scholar]
- Ma Q, Dai X, Xu Y, Guo J, Liu Y, Chen N, Xiao J, Zhang D, Xu Z, Zhang X, Chong K. Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiology. 2009;150:244–256. doi: 10.1104/pp.108.133454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar A, Browse J. MYB108 acts together with MYB24 to regulate jasmonate-mediated stamen maturation in Arabidopsis. Plant Physiology. 2009;149:851–862. doi: 10.1104/pp.108.132597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandaokar A, Thines B, Shin B, Lange BM, Choi G, Koo YJ, Yoo YJ, Choi YD, Browse J. Transcriptional regulators of stamen development in Arabidopsis identified by transcriptional profiling. The Plant Journal. 2006;46:984–1008. doi: 10.1111/j.1365-313X.2006.02756.x. [DOI] [PubMed] [Google Scholar]
- Manickavelu A, Kawaura K, Oishi K, Shin IT, Kohara Y, Yahiaoui N, Keller B, Suzuki A, Yano K, Ogihara Y. Comparative gene expression analysis of susceptible and resistant near-isogenic lines in common wheat infected by. Puccinia triticina. DNA Research. 2010;17:211–222. doi: 10.1093/dnares/dsq009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matus JT, Aquea F, Arce-Johnson P. Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biology. 2008;8:83. doi: 10.1186/1471-2229-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Gubler F. The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. The Plant Cell. 2005;17:705–721. doi: 10.1105/tpc.104.027920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitsuda N, Ohme-Takagi M. Functional analysis of transcription factors in Arabidopsis. Plant and Cell Physiology. 2009;50:1232–1248. doi: 10.1093/pcp/pcp075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munns R, Tester M. Mechanisms of salinity tolerance. Annual Review of Plant Biology. 2008;59:651–681. doi: 10.1146/annurev.arplant.59.032607.092911. [DOI] [PubMed] [Google Scholar]
- Nilsson L, Muller R, Nielsen TH. Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in. Arabidopsis thaliana. Plant, Cell and Environment. 2007;30:1499–1512. doi: 10.1111/j.1365-3040.2007.01734.x. [DOI] [PubMed] [Google Scholar]
- Ogata K. Structure and dynamics of the transcription factor, Myb, in DNA-sequence recognition. Seikagaku. 1998;70:1233–1250. [PubMed] [Google Scholar]
- Ogata K, Nishimura Y. Specific DNA recognition by Myb protein. Tanpakushitsu Kakusan Koso. 1995;40:1592–1597. [PubMed] [Google Scholar]
- Pabo CO, Sauer RT. Transcription factors: structural families and principles of DNA recognition. Annual Review of Biochemistry. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- Paz-Ares J, Ghosal D, Wienand U, Peterson PA, Saedler H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO Journal. 1987;6:3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Vailleau F, Leger A, Joubes J, Miersch O, Huard C, Blee E, Mongrand S, Domergue F, Roby D. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. The Plant Cell. 2008;20:752–767. doi: 10.1105/tpc.107.054858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JL, Chua NH. ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. The Plant Journal. 2007;49:592–606. doi: 10.1111/j.1365-313X.2006.02980.x. [DOI] [PubMed] [Google Scholar]
- Riechmann JL, Heard J, Martin G, et al. Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science. 2000;290:2105–2110. doi: 10.1126/science.290.5499.2105. [DOI] [PubMed] [Google Scholar]
- Rosinski JA, Atchley WR. Molecular evolution of the Myb family of transcription factors: evidence for polyphyletic origin. Journal of Molecular Evolution. 1998;46:74–83. doi: 10.1007/pl00006285. [DOI] [PubMed] [Google Scholar]
- Schrumpfova PP, Kuchar M, Palecek J, Fajkus J. Mapping of interaction domains of putative telomere-binding proteins AtTRB1 and AtPOT1b from. Arabidopsis thaliana. FEBS Letters. 2008;582:1400–1406. doi: 10.1016/j.febslet.2008.03.034. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Park CM. MYB96-mediated abscisic acid signals induce pathogen resistance response by promoting salicylic acid biosynthesis in Arabidopsis. New Phytologist. 2010;186:471–483. doi: 10.1111/j.1469-8137.2010.03183.x. [DOI] [PubMed] [Google Scholar]
- Seo PJ, Xiang F, Qiao M, Park JY, Lee YN, Kim SG, Lee YH, Park WJ, Park CM. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiology. 2009;151:275–289. doi: 10.1104/pp.109.144220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K. Gene networks involved in drought stress response and tolerance. Journal of Experimental Botany. 2007;58:221–227. doi: 10.1093/jxb/erl164. [DOI] [PubMed] [Google Scholar]
- Singh K, Foley RC, Onate-Sanchez L. Transcription factors in plant defense and stress responses. Current Opinion in Plant Biology. 2002;5:430–436. doi: 10.1016/s1369-5266(02)00289-3. [DOI] [PubMed] [Google Scholar]
- Stracke R, Werber M, Weisshaar B. The R2R3-MYB gene family in. Arabidopsis thaliana. Current Opinion in Plant Biology. 2001;4:447–456. doi: 10.1016/s1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Vailleau F, Daniel X, Tronchet M, Montillet JL, Triantaphylides C, Roby D. A R2R3-MYB gene, AtMYB30, acts as a positive regulator of the hypersensitive cell death program in plants in response to pathogen attack. Proceedings of the National Academy of Sciences, USA. 2002;99:10179–10184. doi: 10.1073/pnas.152047199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vannini C, Locatelli F, Bracale M, Magnani E, Marsoni M, Osnato M, Mattana M, Baldoni E, Coraggio I. Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. The Plant Journal. 2004;37:115–127. doi: 10.1046/j.1365-313x.2003.01938.x. [DOI] [PubMed] [Google Scholar]
- Velasco R, Zharkikh A, Troggio M, et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS One. 2007;2:e1326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins O, Nahal H, Foong J, Provart NJ, Campbell MM. Expansion and diversification of the populus R2R3-MYB family of transcription factors. Plant Physiology. 2009;149:981–993. doi: 10.1104/pp.108.132795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Lee E, Lucas JR, Morohashi K, Li D, Murray JA, Sack FD, Grotewold E. Regulation of cell proliferation in the stomatal lineage by the Arabidopsis MYB FOUR LIPS via direct targeting of core cell cycle genes. The Plant Cell. 2010;22:2306–2321. doi: 10.1105/tpc.110.074609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Zhang W, Zhao Y, Gong X, Guo L, Zhu G, Wang X, Gong Z, Schumaker KS, Guo Y. SAD2, an importin-like protein, is required for UV-B response in Arabidopsis by mediating MYB4 nuclear trafficking. The Plant Cell. 2007;19:3805–3818. doi: 10.1105/tpc.106.048900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Verslues PE, Zheng X, et al. HOS10 encodes an R2R3-type MYB transcription factor essential for cold acclimation in plants. Proceedings of the National Academy of Sciences, USA. 2005;102:9966–9971. doi: 10.1073/pnas.0503960102. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhu JK. Salt and drought stress signal transduction in plants. Annual Review of Plant Biology. 2002;53:247–273. doi: 10.1146/annurev.arplant.53.091401.143329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.