Abstract
Genetic disorders of amino acid and fatty acid metabolism can be detected with tandem mass spectrometry (MS/MS). MS/MS screening of mice subjected to chemical mutagenesis defined a new disorder of branched-chain amino acid metabolism resembling human maple syrup urine disease. This approach has general application to the discovery of gene function in developmental and metabolic disorders.
The post-genomic era has arrived and has created an exciting environment for the discovery of genetic disorders, multigenic etiologies of disease, and pharmacogenomics, and the delineation of gene function, with unusual approaches (1). In this issue of the JCI, Y.-T. Chen, an accomplished biochemical geneticist, and coworkers use an intriguing process with potential general application to mammalian disease and congenital anomalies to demonstrate the phenotype of a new metabolic disorder (Figure 1). The results document the usefulness of studying the “metabolome,” a term that refers to the low-molecular-weight compounds present in organisms that are substrates, products, and regulatory molecules in macromolecular metabolic pathways. This commentary projects three major conclusions. First, screening of mutagenized animals with clinically relevant approaches will define phenotypes similar to critical human disorders. Second, the study of the metabolome will likely define critical compounds, relevant genetic disorders, and modifying genes in animal models that will be revealing in our understanding of human disease. Third, highly sensitive technologies applied to both animal models and humans will result in the discovery of new disorders; the aberrant, causative genes; and new treatments.
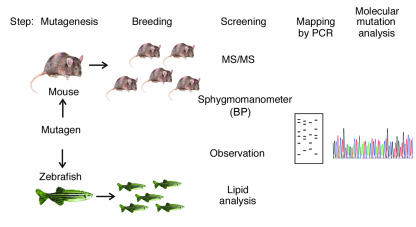
Figure 1.
Clinical observation and clinically relevant screening of mutated mouse and zebrafish populations in gene discovery. The common approach that has allowed identification of novel genes essential in clinically relevant disorders or congenital malformations is illustrated. The first step is chemical mutagenesis, likely to produce point mutations, which is achieved by injection of a mutagen into a substantial number of animals. The second step is breeding of the offspring to search for either recessive or dominant phenotypes. The key third step is screening for clinically relevant phenotypes, illustrated here as either tandem mass spectrometry (MS/MS) for disorders of amino acid or fatty acid metabolism (1); measurement of blood pressure (in mice by tail cuff assessment) to find mutations causing hypertension; lipid analysis, either of plasma samples from mice or other whole animals or embryos in fish to assess lipoprotein or membrane lipid abnormalities; and clinical observation for congenital anomalies by noting abnormalities of organ development. The latter has been accomplished using the transparent zebrafish embryo to detect heart and vascular anomalies and aberrant gut or eye development and can also be assessed in mice by magnetic resonance imaging, echocardiography, or other clinically used imaging modalities. The subsequent steps of mapping of the mutated gene using polymorphisms and delineation of mutations by DNA sequencing of mapped regions or candidate genes provide proof that the mutated gene causes the observed phenotype.
Metabolomics, screening, and tandem mass spectrometry
Following chemical mutagenesis of mice using N-ethyl-N-nitrosourea and three generations of breeding designed to assess recessively inherited disorders, the authors screened families of mutant mice for disorders of amino acid and fatty acid metabolism using tandem mass spectroscopy (MS/MS). MS/MS analysis of body fluids, including plasma, urine, bile, and tissues has become a powerful tool to discern some 40 metabolic disorders in humans and is now the method of choice for newborn screening of these disorders in many American states and European countries (2). In this instance (1), elevations of branched-chain amino acids in one family of mutagenized mice suggested a disorder similar to human maple syrup urine disease (MSUD), a disorder that is secondary to mutations in the branched-chain α-keto-acid dehydrogenase complex. However, in the murine family, a missense mutation in another enzyme in this amino acid pathway, the mitochondrial branched-chain aminotransferase, was discovered. Thus, through the use of mutagenesis of mice and metabolome screening of offspring with MS/MS, a novel disorder with a phenotype similar to a known human metabolic disease was discovered. Because mouse and human metabolism is similar, it is very likely that humans with a similar clinical and biochemical phenotype will turn out to have mutations in this gene. Thus, use of the highly sophisticated methodology of MS/MS, currently applied to human newborn screening, in mice led to the discovery of a murine genetic disorder that in all probability will subsequently be found in people.
Murine models of metabolic disorders and treatment
Chen and coworkers (1) utilized their newly discovered model of human MSUD to assess the efficacy of dietary management used in humans. The mutant mice exhibited failure to thrive, muscular weakness, hair loss, and premature death. All of these features were reversed by feeding the animals a diet low in branched-chain amino acids. This result demonstrates the important general principle that murine mammalian models of human disorders can be used to determine the natural history of such disorders and quickly assess outcomes of treatment, studies that are often impossible to complete in humans and, even if feasible, take years to perform. Although gene ablation mouse models have allowed similar studies, knockout mice that are null for gene expression often are embryonic lethal or have a much more severe phenotype than those of monogenic human disorders that are secondary to missense or splice site mutations with partial expression or activities of protein. Thus, as illustrated by this manuscript (1), a milder phenotype more typical of many human genetic disorders is often more relevant to human disease than a complete knockout. Another obvious advantage of these animal models is the ability to study outcomes in the whole organism, not just in cell lines derived from human patients.
Animal model screening
The application of typical medically relevant clinical and laboratory assessment of large populations of mutagenized animals, especially those such as mice and zebrafish that have relatively short reproductive cycles, gestations, and life spans, and adequately characterized genomes, opens up an entirely new era for the characterization of human anomalies and disease. In zebrafish, large-scale screens for developmental anomalies of the cardiovascular (3) and other organ systems through visual observation have allowed delineation and characterization of genes essential for organogenesis that have human homologs and are mutated in human disease. In mice, screening for phenotypes highly relevant to common human disorders, including such examples as hypertension, diabetes, hyperlipidemias, congenital anomalies such as congenital heart disease, and behavioral or movement disorders, is now possible with newer or microtechnologies for the measurement of blood pressure, blood clinical chemistries, echocardiography, and behavioral screening.
Conclusion
More widespread application of the mutagenesis approach reported (1) with screening by clinical observational and sophisticated laboratory studies will allow delineation of a broader spectrum of genetically determined or modified disorders directly relevant to human disease.
Footnotes
See the related article beginning on page 434.
Conflict of interest: The author has declared that no conflict of interest exists.
Nonstandard abbreviations used: tandem mass spectrometry (MS/MS); maple syrup urine disease (MSUD).
References
- 1.Wu J-Y, et al. ENU mutagenesis identifies mice with mitochondrial branched-chain aminotransferase deficiency resembling human maple syrup urine disease. J. Clin. Invest. 2004;113:434–440. doi:10.1172/JCI200419574. doi: 10.1172/JCI19574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zytkovicz TH, et al. Tandem mass spectrometric analysis for amino, organic, and fatty acid disorders in newborn dried blood spots: a two-year summary from the New England newborn screening program. Clin. Chem. 2001;47:1945–1955. [PubMed] [Google Scholar]
- 3.Fishman MD, Olson EN. Parsing the heart: genetic modules for organ assembly. Cell. 1997;91:153–156. doi: 10.1016/s0092-8674(00)80397-9. [DOI] [PubMed] [Google Scholar]