Abstract
Thioredoxins (Trxs) are ubiquitous disulphide reductases that play important roles in the redox regulation of many cellular processes. However, some redox-independent functions, such as chaperone activity, have also been attributed to Trxs in recent years. The focus of our study is on the putative chaperone function of the well-described plastid Trxs f and m. To that end, the cDNA of both Trxs, designated as NtTrxf and NtTrxm, was isolated from Nicotiana tabacum plants. It was found that bacterially expressed tobacco Trx f and Trx m, in addition to their disulphide reductase activity, possessed chaperone-like properties. In vitro, Trx f and Trx m could both facilitate the reactivation of the cysteine-free form of chemically denatured glucose-6 phosphate dehydrogenase (foldase chaperone activity) and prevent heat-induced malate dehydrogenase aggregation (holdase chaperone activity). Our results led us to infer that the disulphide reductase and foldase chaperone functions prevail when the proteins occur as monomers and the well-conserved non-active cysteine present in Trx f is critical for both functions. By contrast, the holdase chaperone activity of both Trxs depended on their oligomeric status: the proteins were functional only when they were associated with high molecular mass protein complexes. Because the oligomeric status of both Trxs was induced by salt and temperature, our data suggest that plastid Trxs could operate as molecular holdase chaperones upon oxidative stress, acting as a type of small stress protein.
Keywords: Chaperone, folding, oligomerization, plastid, thioredoxin, tobacco
Introduction
Thioredoxins (Trxs) are small ubiquitous thiol-disulphide oxidoreductases originally discovered in plants within the context of photosynthesis (Buchanan, 1980). All of the Trxs possess the so-called Trx fold, consisting of five β-strands surrounded by four short α-helices (Jeng et al., 1994), and contain a highly conserved active-site sequence with an extremely reactive disulphide bridge (WCG/PPC) (Holmgren, 1968, 1979). The major role of Trxs is to exert post-translational redox modifications on target proteins, which have been implicated in nearly all cellular processes, shifting them to an active or inactive state upon disulphide reduction. Plants, unlike bacteria and animals, contain several nuclear genes encoding Trxs that appear to be linked to almost 300 potential target proteins (Lemaire et al., 2007). Plant Trxs are located in various subcellular compartments, including the chloroplast, mitochondria, and cytosol. Trxs are reduced by a different electron donor system, depending on their location in the cell. Both cytosolic and mitochondrial Trxs are reduced by compartment-specific NADPH/Trx reductases, whereas chloroplast Trxs are reduced by ferredoxin/Trx reductase (FTR) with electrons provided by the photosynthetic electron transport chain (Lemaire et al., 2007; Schurmann and Buchanan, 2008). Besides the FTR, there is an interesting type of NADPH-dependent thioredoxin reductase (NTR) in the chloroplasts called NTRC. This protein contains a joined Trx module at the C-terminus (Serrato et al., 2004), and is able to act as a NTR/Trx system in a single polypeptide (Perez-Ruiz and Cejudo, 2009).
The chloroplast Trx system is particularly complex and includes five different types: f, m, x, y, and z (Collin et al., 2004; Arsova et al., 2010). Although all of these Trxs are encoded by nuclear genes, phylogenetic studies and structural comparisons have demonstrated that Trx m, x, y, and z have a prokaryotic origin (Meyer et al., 2005; Arsova et al., 2010). Conversely, Trx f shares an intron with cytosolic Trx h, and both seem to have a common eukaryotic ancestor (Sahrawy et al., 1996). Trx f and Trx m were initially identified as light-dependent regulators of the key enzymes of photosynthetic metabolism in chloroplasts (Buchanan, 1980), but new functions have been reported for these Trxs in recent years (Issakidis-Bourguet et al., 2001; Geigenberger et al., 2005; Lemaire et al., 2007; Benitez-Alfonso et al., 2009; Marri et al., 2009). Comparative genomic analyses have displayed the presence of gene families encoding Trxs in higher plants and, to date, these analyses have indicated that Trx m is present in multiple isoforms whereas Trx f is usually encoded by one or two genes (Chibani et al., 2009). Recently, the circadian regulation of some Trx f and m isoforms has been demonstrated in pea and Arabidopsis plants at both the transcriptional and the protein level (Barajas-Lopez et al., 2011). The presence of several Trx types and isoforms within the chloroplast that appear to be regulated differentially suggests the involvement of these Trxs in many processes. Therefore, Trxs may have redundant functions, act on different protein targets (an updated list of chloroplast targets is presented in Montrichard et al., 2009) or even exhibit organ specificity. Chloroplast Trxs, originally considered unique to photosynthetic tissues, were first reported in a non-photosynthetic organ some years ago (Pagano et al., 2000). Since then, a Trx m isoform and the complete ferredoxin/Trx system have been identified in amyloplasts from wheat endosperm, and new putative protein targets have been isolated using proteomic approaches (Balmer et al., 2006). Strong evidence that some plastid Trxs are present in both photosynthetic and heterotrophic tissues has also been reported in Arabidopsis (Collin et al., 2004) and pea (Barajas-Lopez et al., 2007; Traverso et al., 2008). Taken together, these findings open the possibility of additional roles for plastid Trxs, which are different from their classical function in photosynthetic enzyme regulation.
Several observations have shown that Trxs may have a second regulatory mechanism, independent from their oxidoreductase activity, that depends on their ability to interact with other proteins to form functional protein complexes, for instance, the interaction of E. coli Trx1 with phage T7 DNA polymerase (Mark and Richardson, 1976) or the protein–protein interaction between human Trx and apoptosis signalling kinase-1 (Saitoh et al., 1998). Trxs have also been shown to promote the folding of proteins in a redox-independent way, either by directly promoting protein folding or by enhancing the refolding activity of other molecular chaperones (reviewed in Berndt et al., 2008). Protein folding in vivo is mediated by a number of proteins that act either as ‘foldases’ or ‘molecular chaperones’. Foldases or folding catalysts, such as protein disulphide isomerase (PDI) and peptidylpropyl isomerase, accelerate the rate of target polypeptide folding (Miernyk, 1999). Molecular chaperones are a diverse group of proteins, but they share the property of binding to substrate proteins that are in unstable, non-native structural states (Boston et al., 1996). Arguments that Trxs might assist in the protein-folding pathway, analogously to a PDI, were proposed some years ago (Pigiet and Schuster, 1986). Similarly, new insights into the molecular chaperone activity of some Trxs have recently been revealed. E. coli Trx1 has been reported to interact with unfolded and denatured proteins in vitro, in a manner similar to that of molecular chaperones (Kern et al., 2003). Furthermore, the in vivo chaperone function of the E. coli Trx-like protein, YbbN, has also been reported (Kthiri et al., 2008). In plants, the chaperone function of an h-type Trx (AtTrxh3) (Park et al., 2009) and a Trx-like protein (AtTDX) (Lee et al., 2009) has recently been described. Other proteins harbouring the Trx fold, such as 2-Cys peroxiredoxin (Prx), have also been shown to function as molecular chaperones (Jang et al., 2004; Kim et al., 2009). Recently, four putative co-chaperones of Hsp90/Hsp70 that contain an additional Trx domain have been identified in Arabidopsis and rice using an in silico approach (Prasad et al., 2010). Therefore, it can be expected that other plant Trxs may function as facilitators and regulators of protein folding. In vivo, either the fusion or co-expression of E. coli Trx1 to a protein of interest has become a widely used tool to improve protein folding and solubility during recombinant protein expression (Yasukawa et al., 1995; LaVallie et al., 2000, 2003; Yuan et al., 2004). Similar to the bacterial expression system, the chloroplast transformation platform has emerged as a powerful tool for the expression of recombinant proteins in plants (Daniell et al., 2009; Bock and Warzecha, 2010). Recently, it has been shown that the co-expression of either tobacco Trx f or Trx m avoided inclusion body formation of recombinant human serum albumin (HSA) within the chloroplast (Sanz-Barrio et al., 2011), suggesting that both plastid Trxs may have acted as molecular chaperones and prevented the aggregation of HSA.
The present study aimed to investigate the putative chaperone activity of two plastid Trxs isolated from Nicotiana tabacum, with both eukaryotic (Trx f) and prokaryotic origin (Trx m). Biochemical evidence that both plastid tobacco Trxs may be directly involved in the folding of proteins through a redox-independent manner is presented. The isolated Trx f and Trx m proteins showed extensive similarity to previously reported genes, but contained additional surface-exposed cysteine (Cys) residues, which were subjected to site-directed mutagenesis to address their possible implication in the structure and functions of the Trxs.
Materials and methods
Cloning of the cDNAs for tobacco Trx f and Trx m
Well-developed tobacco (Nicotiana tabacum cv. Petite Havana SR1) leaves were dissected, frozen in liquid nitrogen, and ground in a Mikro-Dismembrator U (B Braun Biotech International, Melsungen, Germany). Total RNA was extracted using the Ultraspec RNA Isolation System (Biotecx Laboratories, Houston, TX), and samples were subjected to RNA ligase-mediated rapid amplification of cDNA ends (RLM-RACE) using the FirstChoice RLM-RACE kit (Ambion, Austin, TX), according to the manufacturer’s instructions. RACE PCR was done in two steps with nested gene-specific inner and outer primers, F1 (5′-GTTGCAATCAAGCTTCAGAAAGAC-3′) and F2 (5′-GTCTTTCTGAAGCTTGATTGCAAC-3′) for NtTrxf and M1 (5′-GAGGGGCTTTCATCTGTATTCAG-3′) and M2 (5′-CTGAATACAGATGAAAGCCCCTC-3′) for NtTrxm, and the nested primers provided in the kit. Fragments were cloned into the pGEM-T Easy vector (Promega, Madison, WI) and both strands were sequenced. The tobacco thioredoxin gene sequences (NtTrxf and NtTrxm) were deposited in the GenBank sequence database with the accession numbers HQ338526 and HQ338525, respectively.
3D structure modelling
Nicotiana tabacum Trxs were modelled by using the protein structure homology-modelling server, SWISS-MODEL (Arnold et al., 2006; Kiefer et al., 2009), with the representative structure of Trxs from Spinacea oleracea (Protein Data Bank entry 1F9M for Trx f and 1FB6A for Trx m; Capitani and Schurmann, 2004) as templates. The schematic representations and surfaces were obtained by using the PyMOL program.
Expression and purification of recombinant tobacco Trx f and Trx m in E. coli
Tobacco Trx f and Trx m were expressed in E. coli as histidine (His)-tagged polypeptides. The coding sequences, excluding those for the putative transit peptide (55 residues at the N-terminus in Trx f and 70 residues in Trx m), were amplified from the full-length cDNA with gene-specific primers, which included a poly(His)-tag (italics) and an NcoI site (underlined) in the forward primer and a NotI site (underlined) in the reverse primer. The sequence of the gene-specific primers was as follows: Trxf-F, 5′-CCATGGGTCACCATCACCATCACCATAGCTCCGATGCTACTG-3′; Trxf-R, 5′-GCGGCCGCT TAACTTGACCGCACATCCTCAATTG-3′; Trxm-F, 5′-CCATGG GTCACCATCACCATCACCATGAAGCGCAAA-3′; and Trxm-R, 5′-GCGGCCGCTTACAAGAATTTCTCTATGCAGGTGG-3′. The PCR fragments were digested with NcoI and NotI and cloned into a pET 28a(+) vector (Novagen, Merck KGaA, Darmstadt, Germany). The resulting plasmids, termed pET Trxf and pET Trxm, were introduced into E. coli (BLR DE3, Novagen). Over-expressed proteins were purified by chromatography on affinity columns packed with Ni-NTA agarose (Qiagen, Hilden, Germany). The protein concentration was measured using the RC-DC protein assay (Bio-Rad, Hercules, CA), according to the manufacturer’s instructions.
Activity assays
Insulin reduction assay:
The disulphide oxidoreductase activity of the His-tagged Trxs was determined according to the DTT-dependent insulin reduction assay (Holmgren, 1979). Reactions were assayed at 25 °C and initiated by the addition of 0.5 mM DTT to the reaction mixture, which contained 100 mM TRIS-HCl (pH 7.5), 2 mM EDTA, 1 mg ml−1 insulin (Sigma-Aldrich, St Louis, MO) and 2 or 4 μM of purified Trx. The oxidoreductase activity was determined by monitoring the increase in turbidity at 650 nm using a Hitachi U-2000 spectrophotometer (Hitachi High Technologies America Inc., Schaumburg, IL).
Trx-dependent fructose 1,6-bisphosphatase (FBPase) activity:
The Trx-dependent FBPase activity was measured as described previously (Serrato et al., 2009), in the presence of pea chloroplast FBPase (a gift from Dr M Sahrawy, Estación Experimental del Zaidín, Granada, Spain). The reaction mixture contained the following in a final volume of 0.2 ml: 100 mM TRIS-HCl (pH 8.8), 0.25 mM DTT, 1 μg FBPase, 15 mM MgCl2, 0.3 mM NADP, 0.7 U ml−1 G6PDH (Roche Diagnostics, Rotkreuz, Switzerland), 1.4 U ml−1 PGI (Roche Diagnostics), 0.6 mM fructose-1,6-bisphosphate (Sigma-Aldrich) and increasing concentrations of Trx f or Trx m. FBPase activity was measured by monitoring the increase in absorbance at 340 nm using a Multiskan MS spectrophotometer (Labsystems, Helsinki, Finland).
Refolding of glucose 6-phosphate dehydrogenase (G6PDH):
Chaperone foldase activity was analysed by using guanidine-denatured G6PDH as a substrate. Denaturation, renaturation, and activity measurements were carried out according to a method previously described (Hansen and Gafni, 1993) in a 62.5 mM TRIS buffer (pH 8.0). The denaturation of 20 μM G6PDH from Leuconostoc mesenteroides (Sigma-Aldrich) was carried out by incubation with 1.8 M guanidine HCl at 20 °C. The refolding of denatured G6PDH was initiated by 37-fold dilution at 20 °C for 60 min in TRIS buffer containing different concentrations (2.5, 5, 10 or 20 μM) of Trxs or BSA as a negative control. Spontaneous refolding was also monitored in the absence of proteins. The activity recovery was determined after dilution, and the reactivation yield was expressed as a percentage of the activity of the native enzyme, which showed full activity. The reaction mixture contained 1.6 nM G6PDH, 7.5 mM MgCl2, 2 mM glucose 6-phosphate, and 1 mM NADP. The activity was assayed by following the rate of change of optical density at 340 nm with a U-2000 spectrophotometer (Hitachi High Technologies America Inc.).
Thermal aggregation of malate dehydrogenase (MDH):
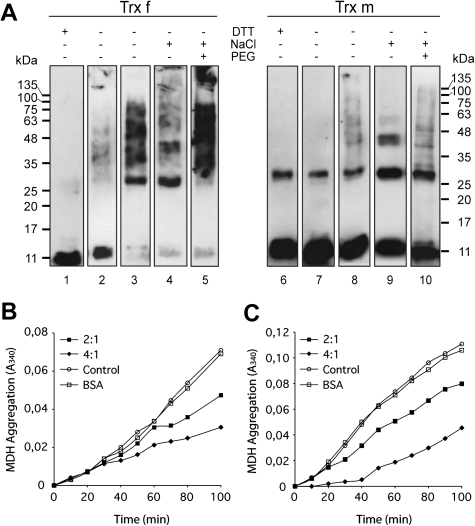
Chaperone holdase activity was assayed by measuring the thermal aggregation of MDH from porcine heart (Sigma-Aldrich) following the Park et al. (2009) procedure. MDH at 2 μM was incubated in 50 mM HEPES-KOH (pH 8.0) buffer at 43 °C with various Trx concentrations (2:1 and 4:1, Trx:MDH molar ratio). The thermal aggregation of MDH was determined in a microtitre plate by monitoring the increase in turbidity at 340 nm in a temperature-controlled spectrophotometer (Spectra Max 340PC; Molecular Devices, Sunnyvale, CA). To study the effects of salt and macromolecular crowding on the Trx holdase activity, Trx f or Trx m was incubated in the presence of 150 mM NaCl either alone or in combination with 100 mg ml−1 PEG 20 000 as a crowding agent. The aggregation of MDH in the absence of Trxs or in the presence of 8 μM BSA were used as controls. The per cent protection of aggregation was calculated relative to the MDH controls.
Electron microscopy and immunogold labelling
Leaf samples of well-developed tobacco plants grown in a greenhouse were fixed in 4% formaldehyde in PBS and dehydrated in methanol by the ‘Progressive Lowering of Temperature’ (PLT) method in a Leica AFS automated system (Leica, Wetzlar, Germany). The samples were infiltrated in Lowicryl K4M resin and polymerized at –30 °C under ultraviolet light. For immunolabelling, the Lowicryl sections (thickness, ∼80 nm) were deposited on Formvar- and carbon-coated nickel grids, and the immunogold labelling was performed as described previously (Segui-Simarro et al., 2003), with some modifications. Briefly, sections were hydrated, floated on PBS, blocked with 5% BSA in PBS, and incubated for 3 h at 25 °C with specific tobacco anti-Trx f and anti-Trx m antibodies (Sanz-Barrio et al., 2011), both diluted 1:2. The sections were then incubated for 1 h at 25 °C with an anti-rabbit IgG antibody conjugated to 10 nm gold particles (British Biocell International, Cardiff, UK), diluted 1:25 in 1% BSA. Lastly, the sections were washed, air-dried, counterstained with uranyl acetate and lead citrate, and observed under a Philips CM10 transmission electron microscope operating at 100 kV. Controls were performed by excluding the corresponding primary antibody from the incubation buffer.
Gene expression analysis
Total RNA (1.4 μg per sample), extracted from mature leaves, apical and basal stems, roots, flowers, and seeds from 60-d-old tobacco plants grown in a greenhouse, was used to perform random hexamer-primed reverse transcription with the reagents contained in a SYBR® GreenER™ Two-Step qRT-PCR Kit (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. Aliquots (0.2 μl) from each reverse transcription reaction were subjected to real-time quantitative PCR in an ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Foster City, CA). Amplifications were performed in a final volume of 25 μl. The PCR conditions included 1 cycle at 50 °C for 2 min, 1 cycle at 95 °C for 15 min, and 40 cycles at 94 °C for 15 s, 63 °C for 30 s, and 72 °C for 30 s. Agarose gel (2%) electrophoresis confirmed the homogeneity of the DNA products. Gene-specific primers were designed with Primer Express Software (Applied Biosystems), as follows: Trx f, qRTf_F (5′-AGTGATCGCTCCAAAGTTTCAAG-3′) and qRTf_R (5′-TT GTCCTGGTTACAGTCCAGCTT-3′); Trx m, qRTm_F (5′-AA TTTTGGGCTCCGTGGTG-3′) and qRTm_R (5′-GCTTGCCAAC ATATTCCTTTGC-3′); and 18S, 18S_F (5′-ACAAACCCCGACTTCTGGAA-3′) and 18S_R (5′-CATGAATCATCGCAGCAACG-3′). Relative quantification for a given transcript was calculated with the 2−ΔΔCT method (Livak and Schmittgen, 2001), and the relative amount of each NtTrxf or NtTrxm transcript was quantified using the 18S rRNA transcript as reference gene.
Western-blot analysis
Tobacco tissues (mature leaf, apical and basal stem, root, flower, and seed) were frozen in liquid nitrogen, ground in a Mickro-Dismembrator U (B Braun Biotech), resuspended in sample buffer (250 mM TRIS-HCl, pH 6.8, 4% SDS, 10% glycerol, and 10% β-mercaptoethanol) and boiled for 5 min. After centrifugation at 20 000 g for 5 min, the supernatant protein content was measured using the RC-DC protein assay (Bio-Rad) with BSA as a standard. Extracted proteins (50 μg) were separated by electrophoresis on 15% SDS-PAGE and electrophoretically transferred to a nitrocellulose membrane (Hybond C, GE Healthcare, Fairfield, CT). Trx detection was performed with a primary antibody, anti-Trx f or anti-Trx m, at 1:7500 or 1:5000 dilution, respectively, and a secondary antibody (anti-rabbit peroxidase conjugated, Sigma-Aldrich) at 1:20 000 dilution. Immunoblot detection was performed by chemiluminescence (ECL Western blotting, GE Healthcare).
For non-reducing inmunoblot analyses of Trx f and Trx m in young tobacco leaves, 100 μg of plant tissue was homogenized in 3 vols of extraction buffer (100 mM HEPES, pH 7.5, and 2 mM EDTA) and incubated for 30 min on ice. After centrifugation at 20 000 g for 10 min, 10 μl of supernatant was subjected to a 15% non-reducing or reducing SDS-PAGE and blotted onto nitrocellulose membrane. Trx detection by Western blotting was performed as described above.
For inmunoblot analyses of purified recombinant His-tagged Trxs and mutants, 0.25 μg of protein was subjected to 13% non-reducing or reducing SDS-PAGE and blotted onto nitrocellulose membrane. Anti-poly(His) peroxidase conjugated antibody (Sigma-Aldrich) was used at 1:2000 dilution. Detection was performed as described above.
Site-directed mutagenesis of tobacco Trx f and Trx m
Site-directed mutagenesis was performed by PCR using the His-tagged NtTrxf or NtTrxm cDNA as templates. The TrxfC19S mutant was produced with oligonucleotides C19S-f (5′-GTGACTGAAGTTAGTAAAGATACC-3′) and C19S-r (5′-GGTATCTTTACTAACTTCAGTCAC-3′), which included a single change (underlined) to replace Cys19 with Ser. The TrxfC71S mutant was produced with oligonucleotides C71S-f (5′-CTGAAGCTTGATAGCAACCAGG-3′) and C71S-r (5′- CCTGGTTGCTATCAAGCTTCAG-3′), which included a single change (underlined) to replace Cys71 with Ser. To produce the double mutant, TrxfC19/71S, the same pair of oligonucleotides (C71S-f and C71S-r) was used with the mutant TrxfC19S construct as a template. The TrxmC107S mutant was produced with oligonucleotides Trxm-F (described above) and C107S-r (5′-GCGGCCGCTTACAAGAATTTCTCTATGCTGGT GG-3′), which included a single change (underlined) to replace Cys107 with Ser. The PCR products were then cloned in the pET 28a(+) vector (Novagen). The resulting cDNAs were sequenced to check for the correct introduction of the mutations and that no additional mutations occurred during the process. The resulting plasmids, termed pET TrxfC19S, pET TrxfC71S, pET TrxfC19/71S, and pET TrxmC107S were introduced into E. coli (BLR DE3, Novagen). Over-expressed mutant proteins were purified and quantified as described before.
Results
Isolation and characterization of cDNAs encoding NtTrxf and NtTrxm from tobacco
Two coding sequences, which encode a plastid Trx f and Trx m, were isolated from Nicotiana tabacum cv. Petite Havana by RACE and named NtTrxf and NtTrxm, respectively. The nucleotide sequences of both cDNAs revealed an open reading frame (ORF) of 528 and 549 nucleotides, respectively (GenBank accession numbers HQ338526 and HQ338525). According to the nucleotide similarity, the isolated NtTrxm belongs to cluster I, which includes Trx m1, m2 and m4 orthologues of dicot species (Chibani et al., 2009). With the exception of Arabidopsis, the vast majority of sequenced plant genomes contain a single Trx f gene (Chibani et al., 2009), suggesting that the cloned NtTrxf could also be the single Trx f gene in tobacco. These Trx ORFs encoded deduced polypeptides of 175 and 182 residues for Trx f and m, respectively. Subcellular prediction programs (iPSORT, ChloroP), modified according to Zybailov et al. (2008), predicted a chloroplast transit peptide (TP) of 55 amino acids that would be cleaved upon import to produce a mature Trx f with an estimated mass of 13.2 kDa and a pI of 6. For the mature Trx m protein (TP length of 70 residues), the deduced molecular mass was 12.4 kDa with a pI of 5.4. Both Trxs contain the conserved WCGPC motif (Fig. 1A) and showed high identity with the extensively studied Trx f and m in other plant species (see Supplementary Figs S1 and S2, respectively, at JXB online). The highest similarities for Trx f were found with Arabidopsis Trx f1 and f2 (80% and 81%, respectively; GenBank accessions NP_186922 and NP_197144) and pea Trx f (80%; GenBank accession AAC49357). Trx m shows the highest sequence similarity with poplar Trx m4 and m2 (78% and 71%, respectively; GenBank accessions EEE97513 and EEF00430) and pea Trx m2 (74%; GenBank accession CAC69854).
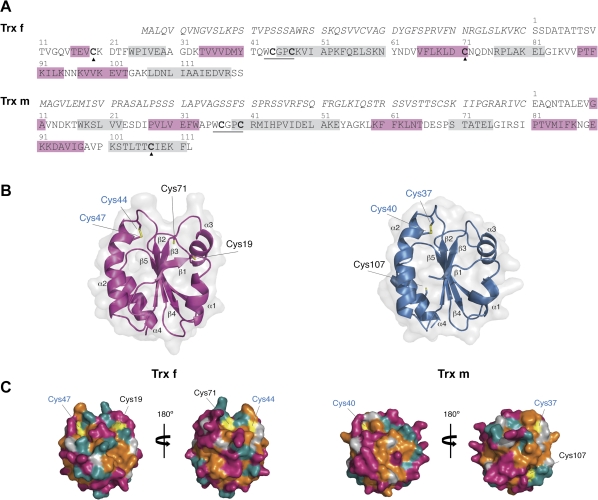
Fig. 1.
Amino acid sequences and model structures of chloroplast Trxs displaying additional Cys residues. (A) Protein sequences deduced from Nicotiana tabacum cDNAs. The predicted transit peptide is given in italics and the mature protein numbered above the sequence. The secondary structural elements appear boxed in pink (β-strands) or gray (α-helixes). The Trx conserved WCGPC motif is underlined and additional Cys residues are indicated with black triangles. (B) Cartoon representation of the Trx f (left) and Trx m (right) structure displaying the active-site Cys in blue and the additional Cys in black. Secondary structural elements are labelled according to the Trx convention. (C) Surface representation of Trxs, viewing from both sides of the molecule (rotated by 180°) and mapped by residue type. Parts of the surface contributed by polar residues appear in cyan, by charged residues in magenta, by hydrophobic residues in orange, and by Cys residues in yellow. The active-site Cys are given in blue and the additional Cys in black. The molecules were prepared using PyMOL.
In addition to the two active site Cys residues, the mature Trx f enzyme contains two additional Cys, one of them in the C-terminal region (Cys71; Fig. 1A), which is well-conserved in all known Trx f structures (see Supplementary Fig. S1 at JXB online). According to the Trx f predicted structural model (Fig. 1B, C, left panel), this residue was surface exposed close to the active site and surrounded by a highly hydrophobic region (Fig. 1C, left panel). The other additional Cys residue (Cys19; Fig. 1A), though not being a common feature on f-type Trxs, was predicted to be solvent-exposed on a rather flexible part of the molecule, in the loop connecting strand β1 and helix α1 (Fig. 1B, left panel). The mature Trx m protein also has an additional Cys (Cys107, Fig. 1A) not conserved among the m-type Trxs (see Supplementary Fig. S2 at JXB online), which was remote from the active site and located in a secondary structure of the modelled tobacco Trx m (Fig. 1B, right panel). However, surface plots showed that the Cys107 residue was near a solvent-exposed hydrophobic surface area (Fig. 1C, right panel), indicating that it might be important for protein-protein interactions.
Enzymatic characterization and subcellular localization of Trx f and Trx m
The predicted mature proteins of tobacco Trx f and Trx m were expressed in E.coli as His-tagged recombinant polypeptides (see Supplementary Fig. S3 at JXB online). To confirm the disulphide reductase activity of these recombinant Trxs, their ability to reduce the insulin β-chain was first tested using the turbidimetric insulin precipitation assay (Holmgren, 1979). Both Trxs accelerated the reduction of insulin in the presence of DTT in a concentration-dependent way, as shown by the increase in turbidity at 650 nm (Fig. 2A). The specificity of each protein in the activation of fructose 1,6-bisphosphatase, a target plastidial enzyme, was also tested. Under our assay conditions, Trx f was able to activate the FBPase enzyme in a concentration-dependent way, whereas FBPase remained inactive when Trx m was assayed at analogous concentrations (Fig. 2B). However, a slight FBPase activity was observed when higher amounts of Trx m (up to 5 μM) were used (data not shown). Therefore, both proteins were able to reduce insulin in a DTT-dependent way, although Trx f showed a higher affinity for FBPase than that displayed by Trx m, providing evidence that the cloned tobacco Trxs are bioactive disulphide oxidoreductases.
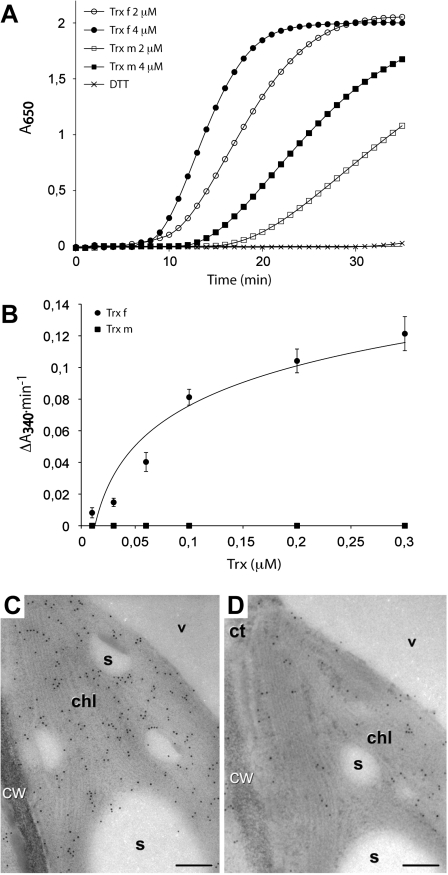
Fig. 2.
Activity assays and immunolocalization of chloroplastic Nicotiana tabacum Trx f and Trx m. (A) DTT-dependent insulin reduction assay of Trx f and Trx m. The assay was determined according to Holmgren (1979) in an incubation mixture containing 2 μM or 4 μM of Trx as indicated, supplemented with 0.5 mM DTT. Assays in the absence of Trx showing no activity were used as controls. (B) FBPase activation by Trx f and Trx m. FBPase activity was carried out according to Serrato et al. (2009). The incubation mixture contained a final concentration of FBPase 0.1 μM and increasing concentrations of Trxs as indicated. Each value is the mean ±SE (bars) of three independent determinations. (C, D) Immunogold labelling with anti- Trx f (C) and anti- Trx m (D) over different sections of a same chloroplast. Both figures exhibit specific labelling within the chloroplast (chl), in the form of dispersed particles. ct, Cytoplasm; cw, cell wall; s, starch; v, vacuole. Bars: 200 nm.
The subcellular localization of tobacco Trx f and Trx m was investigated further by immunocytochemical analysis. For that purpose, specific tobacco anti-Trx f and anti-Trx m antibodies were used (Sanz-Barrio et al., 2011), which did not show cross-reaction with either m- or f-type Trxs (see Supplementary Fig. S4 at JXB online). Immunogold labelling showed specific labelling within the chloroplast for both Trxs (Fig. 2C and D), although labelling with anti-Trx f was more abundant than with anti-Trx m. Such differences can easily be explained in terms of a different affinity of the antibodies for their antigens, as confirmed by the antibodies manufactured through titration analysis (data not shown). No signal above background was observed when the sections were probed with the pre-immune serum (data not shown).
Expression pattern of Trx f and Trx m in different tobacco tissues
The expression levels of NtTrxf and NtTrxm were investigated by quantitative PCR in different tobacco tissues, including mature leaves, stem sections, roots, flowers, and mature seeds. Both genes displayed maximal expression in photosynthetic tissues, such as mature leaf (ML) and apical stem (AS), with NtTrxm mRNA as the most abundant transcript (Fig. 3A). A significant level of mRNA expression was also detected in flowers (Fl; Fig. 3A). The expression of both genes was also detected in roots (R) and basal stem (BS), but the transcript levels dropped significantly in these tissues; in addition, no transcripts were detected in seeds (S; Fig. 3A).
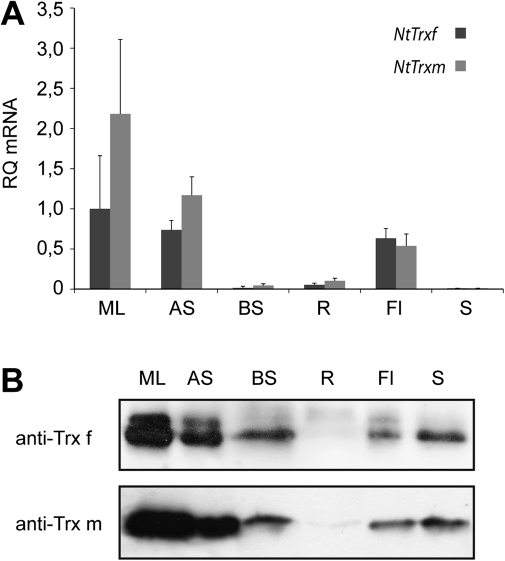
Fig. 3.
Tissue distribution of tobacco plastid Trx f and Trx m. (A) RT-PCR determination of specific mRNA for NtTrxf and NtTrxm in mature leaf (ML), apical stem (AS), basal stem (BS), root (R), flower (Fl), and seed (S) of 60-d-old tobacco plants grown in a greenhouse. Bars represent arbitrary units relative to the respective levels of NtTrxf in the mature leaves set to 1.0. Mean values (±SE) of four determinations are represented. (B) Western blot analysis of protein extracts from the above-described tobacco tissues. Fifty micrograms of protein were loaded in each lane. Anti-Trx f, antibody against tobacco Trx f; anti-Trx m, antibody against tobacco Trx m.
Immunoblot analyses were performed with specific antibodies against tobacco Trx f and Trx m to confirm the presence of both Trxs in different tobacco tissues (Fig. 3B). As stated before for Trx transcripts, the highest content of Trx f and Trx m proteins was detected in mature leaves and apical stems of greenhouse-grown plants, which is consistent with their well-established role in the activation of Calvin cycle enzymes and other photosynthetic processes; to a lower extent, Trx f and m were also detected in basal stems, flowers, and seeds. According to a previous study (Traverso et al., 2008), the localisation of both f and m Trxs in the basal stem could mainly be associated with the vascular stem tissues. In the case of flowers and seeds, these Trxs could be associated with functions related to cell division, germination, and plant reproduction (Barajas-Lopez et al., 2007). Interestingly, the high amounts of Trxs found in seeds are in contrast to an undetectable level of the transcripts. Because dry tobacco seeds were collected for the mRNA and protein extraction, it can be concluded that no expression of the NtTrxf and NtTrxm genes occurs in seeds at this maturation stage, and that Trx f and m accumulate in this tissue as a result of the Trx stability in dry seeds. Moreover, as has been reported to occur in pea (Pagano et al., 2000; Barajas-Lopez et al., 2007), a faintly specific band was visualized in roots for both Trx f and Trx m (Fig. 3B). These results confirm that Trx f and Trx m are ubiquitous proteins present in all tobacco tissues and leads us to expand the range of their potential roles in planta.
Trx f and Trx m increase the amount of correctly folded glucose 6-phosphate dehydrogenase acting as molecular chaperones
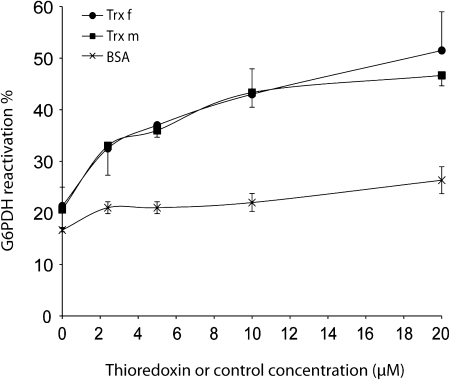
Molecular chaperones are largely classified into two groups: foldase chaperones, which support the folding of denatured proteins into their native state, and holdase chaperones, which bind to folding intermediates, thereby preventing their non-specific aggregation (Beissinger and Buchner, 1998). It was first investigated whether f- and m-type Trxs act as foldase chaperones, assisting the proper folding of proteins. Glucose 6-phosphate dehydrogenase (G6PDH), for which refolding is facilitated by several chaperones (Hansen and Gafni, 1993; Lee et al., 2009), was chosen as the substrate for this reaction. The renaturation degree was determined by measuring G6PDH activity. To distinguish the foldase chaperone function of Trxs from its disulphide reductase activity, the Cys-free form of G6PDH was used (Hansen and Gafni, 1993). G6PDH was denatured in the presence of guanidine hydrochloride and allowed to refold upon dilution of the denaturant, in the presence or absence of Trx. Under our experimental conditions, the refolding yield of G6PDH was raised from approximately 20% (spontaneous refolding) to 47% or 52% in the presence of Trx f or Trx m, respectively, within 1 h (Fig. 4), which represents more than a 2-fold G6PDH refolding increase. This effect was significantly higher than that of bovine serum albumin (BSA) when used as a control protein (up to 26% of G6PDH renaturation; Fig. 4). Maximal G6PDH renaturation was attained in the presence of 20 μM of either Trx f or Trx m, and a half-maximal reactivation occurred at 3–4 μM Trx, concentrations significantly lower than those found in the chloroplast (Scheibe and Anderson, 1981). These results suggest that, like molecular chaperones, Trx f and Trx m specifically interact with unfolded proteins and are able to increase their productive folding.
Fig. 4.
Chaperone foldase activity of chloroplastic tobacco Trx f and Trx m. The Cys-free form of G6PDH (20 μM) was denatured in guanidine-HCl and subsequently refolded in a renaturation buffer in the presence of increasing concentrations of Trx f (closed circles) or Trx m (closed squares) as indicated. The refolding yield of G6PDH was determined by measuring G6PDH activity and expressed as a percentage of native G6PDH activity set to 100%. Reactions using analogous concentrations of BSA (crosses) instead of Trx were used as controls. Each value is the mean ±SE (bars) of three independent determinations.
Protection of malate dehydrogenase (MDH) from thermal aggregation by plastid Trxs depends on their oligomeric status
The holdase chaperone activity of f- and m-type tobacco Trxs was analysed by assessing their ability to inhibit the thermal aggregation of MDH, a model substrate extensively used with other chaperones (Jang et al., 2004; Cheng et al., 2008; Lee et al., 2009; Park et al., 2009). Purified recombinant tobacco Trx f and Trx m mainly appeared as monomers under non-reducing SDS-PAGE (Fig. 5A, lanes 2 and 7). In this monomeric state, both Trxs exhibited a very weak holdase chaperone activity (data not shown). However, the ability of self-associated tobacco Trx f and Trx m to inhibit the thermal aggregation of MDH efficiently has been demonstrated (Sanz-Barrio et al., 2011). Thus, in this work, we speculate on the relationship between the oligomeric state and the holdase-like activity of both tobacco Trxs and investigate in detail the process of oligomerization of tobacco Trx f and Trx m.
Fig. 5.
Structural changes of chloroplastic tobacco Trx f and Trx m in vitro lead chaperone holdase activity. (A) Structural changes of Trx f and Trx m, incubated in the presence or absence of DTT, NaCl or PEG 20 000, were analysed by Western blot on 13% non-reducing SDS-PAGE gels. Reduced Trxs (lanes 1 and 6), oxidized Trxs (lanes 2 and 7), oxidized Trxs incubated for 3 h at 50 °C (lanes 3 and 8), oxidized Trxs in the presence of 1 M NaCl (lanes 4 and 9), and oxidized Trxs in the presence of 150 mM NaCl and 100 mg ml−1 PEG 20 000 (lanes 5 and 10). (B, C) Holdase chaperone activity of Trx f and Trx m under different oligomeric conditions. Thermal aggregation of 2 μM MDH was examined at 43 °C in the absence (open circles) or presence of Trxs. Trx to MDH molar ratio of 2:1 (closed squares) or 4:1 (closed diamonds) were used. Bovine serum albumin (BSA, 8 μM) was used as the protein control (open squares). (B) 2 μM MDH in HEPES-KOH pH 7.5 was incubated at 43 °C in the presence of 150 mM NaCl. (C) 2 μM MDH in HEPES-KOH pH 7.5 was incubated at 43 °C in the presence of 150 mM NaCl and 100 mg ml−1 PEG 20 000. A representative plot of three independent experiments is shown.
The oligomeric status of tobacco Trxs was analysed under non-reducing SDS-PAGE. In addition to Trx f monomers, there is also evidence of a population of self-associated SDS-resistant species (Fig. 5A, lane 2). These multimeric protein structures disappeared completely in the presence of DTT (Fig. 5A, lane 1), which suggested that Trx f polymeric structures were associated with intermolecular redox-dependent disulphide bonds involving Cys residues. In the case of Trx m, both monomers and dimers were detected in the absence of DTT, with monomers being the dominant species (Fig. 5A, lane 7). However, dimers still remained under reducing conditions (Fig. 5A, lane 6), suggesting that monomers probably associated into dimers by hydrophobic forces. Changes in the protein structure, from low to high molecular weight complexes, could be caused not only by redox status but also by heat-shock (Lee et al., 2009; Park et al., 2009) or hydrophobic interactions (Akhtar et al., 2004). When Trx f was incubated at 50 °C for 3 h, the monomeric species almost disappeared and appeared mainly as high molecular complexes (Fig. 5A, lane 3). Despite the fact that Trx m has been reported to be more heat stable than Trx f (Wolosiuk et al., 1979), some oligomeric species were also observed (Fig. 5A, lane 8). Because hydrophobic interactions are enhanced in the presence of salt, the effect of NaCl on the structure of the Trxs was also studied and it was found that the addition of at least 1 M NaCl resulted in the formation of dimers, trimers, and higher oligomers, especially for Trx f (Fig. 5A, lanes 4 and 9). In the presence of a more physiological salt concentration (150 mM NaCl), the oligomerization of Trx f slightly increased, but no higher order changes were observed for Trx m (data not shown). Inside the cell, high concentrations of macromolecules physically occupy a substantial portion of the volume (crowding conditions), and, therefore, the effective concentration of each molecule increases significantly (Ellis, 2001). To simulate molecular crowding in vitro, crowding agents, such as polyethylene glycol (PEG), Ficoll, and dextran, are added to the solution (Chebotareva et al., 2004). Accordingly, the oligomerization status of tobacco plastid Trxs was investigated at physiological salt concentrations in the presence of PEG as a crowding agent. Figure 5A (lanes 5 and 10) shows that both Trxs displayed multimeric protein structures in the presence of 150 mM NaCl when PEG 20 000 was added, though oligomers were more abundant in the Trx f sample than in the Trx m sample.
In addition, the ability of those Trx oligomeric species to inhibit the thermal aggregation of MDH was also studied. As shown in Fig. 5B, increasing amounts of self-associated Trx f (in the presence of 150 mM NaCl) gradually prevented MDH thermal aggregation, with ∼63% inhibition at a Trx to MDH molar ratio of 4:1. By contrast, Trx m incubated at the same physiological salt concentration did not prevent MDH aggregation (data not shown), which is consistent with its monomeric status in this condition. Interestingly, Trx m oligomers formed in the presence of 150 mM NaCl and 100 mg ml−1 of PEG 20 000 as crowding agent offered ∼60% protection against the thermal aggregation of MDH at a molar ratio of 4:1 (Fig. 5C). When Trxs were replaced in the reaction mixture with 8 μM BSA as a control, no holdase activity was detected (Fig. 5B, C). These results show that only the oligomeric species of Trx f and Trx m exhibit holdase chaperone activity, and this structural and functional switching may be caused by changes in redox status and stress conditions.
Oligomerization status of Trxs f and m in planta
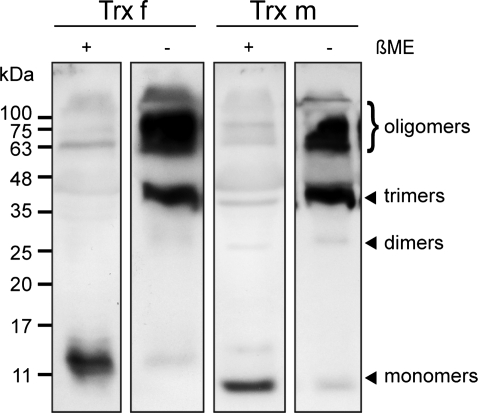
On the basis of the observation that both Trxs tend to self-oligomerize in vitro, it was investigated whether high molecular mass protein complexes were also present in planta. For that purpose, tobacco leaf proteins were analysed by SDS-PAGE under reducing and non-reducing conditions with specific anti-Trx f and anti-Trx m antibodies. Although both Trxs were detected as monomers in the absence of a reducing agent, they predominantly associated into trimers (∼40 kDa) and higher oligomeric complexes (Fig. 6) that may correspond either to Trx homo-oligomers or to heterocomplexes with other target proteins. When leaf protein extracts were treated with β-mercaptoethanol prior to SDS-PAGE, both Trxs migrated mainly as monomers (Fig. 6), but some SDS-resistant polymeric structures still remained detectable in these reductive conditions, suggesting that hydrophobic forces might also play an important role in the protein–protein interaction.
Fig. 6.
Oligomerization status of Trx f and Trx m in planta. Inmunoblot analysis of proteins extracts from young tobacco leaves of plants grown in a greenhouse was performed with specific anti-Trx f or anti-Trx m antibodies. Protein samples were extracted in the presence or absence of 2.5% β-mercaptoethanol (+ or –) and loaded onto a 15% SDS-PAGE gel. βME, β-mercaptoethanol.
Effect of additional Cys residues on the structure and function of Trx f and Trx m
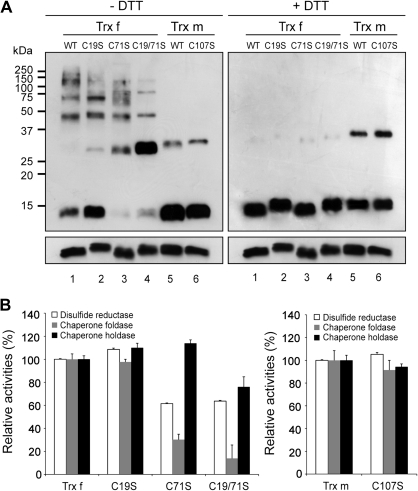
Given that the oligomeric status of Trxs depends, to some extent, on the formation of redox-dependent disulphide bonds, the role of Cys residues in Trx f and Trx m structure and function was investigated. It has been reported that the mutation of the Trx active Cys residues did not create a pronounced effect on its chaperone function (Kern et al., 2003; Park et al., 2009). In addition to the two active-site Cys residues, both tobacco Trxs possess additional solvent-exposed cysteines (Fig. 1C), which were subjected to site-directed mutagenesis. Thus, Cys19 and Cys71 of Trx f (Fig. 1A) were replaced by serine (C19S and C71S mutants), and the double mutant C19/71S was also generated. Similarly, Cys107 of Trx m (Fig. 1A) were replaced by a serine to generate the C107S mutant. All of the mutant proteins were produced in E. coli and purified to homogeneity by Ni-affinity chromatography (see Supplementary Fig. S3 at JXB online). To investigate the redox-dependent structural changes in the Trx mutants, SDS-PAGE was performed under reducing and non-reducing conditions (Fig. 7A). In the absence of DTT, oligomeric complexes were visible in wild-type and Trx f mutants (Fig. 7A, left panel, lanes 1 to 4) and disappeared completely under reducing conditions (Fig. 7A, right panel, lanes 1 to 4). Interestingly, the monomers virtually disappear when the well-conserved Cys71 was mutated, with dimers prevailing over monomers for the C71S and C19/71S mutants (Fig. 7A, left panel, lanes 3 and 4), which directly implicated this Cys in the Trx f monomeric state. In addition, the abundance of the higher-order oligomers dropped significantly in the C19/71S double mutant (Fig. 7A, left panel, lane 4). Conversely, the C107S mutant of tobacco Trx m did not show any significant structural change when compared with the wild-type protein (Fig. 7A, lanes 5 and 6). As occurred in planta, the prevalence of trimers rather than dimers was also observed in vitro for Trx f (Fig. 7A, right panel, lane 1).
Fig. 7.
Effect of additional Cys residues on function and protein structure of chloroplastic tobacco Trx f and Trx m. (A) Redox-dependent structural changes of wild-type (WT) and mutant Trxs containing the site-specific replacement of additional Cys residues by Ser (Trx f mutants: C19S, C71S, and C19/71S; Trx m mutant: C107S). Proteins were analysed by Western blot on 13% non-reducing SDS-PAGE in the absence (left panel) or presence (right panel) of DTT. (B) Relative disulphide reductase (white bar), chaperone foldase (grey bar) and chaperone holdase (black bar) activities of the wild-type and Cys-mutant proteins. Activities of the mutants were compared with those of wild-type Trxs. The wild-type Trx f (left panel) or Trx m (right panel) activities were measured at A650 (DTT-dependent insulin reduction activity) and A340 (chaperone foldase and holdase activities) under the assay conditions described above and set to 100%, respectively. Each value is the mean ±SE (bars) of three independent determinations.
The disulphide reductase and chaperone functions of the mutants generated were investigated using the above-described DTT-dependent insulin reduction, G6PDH reactivation, and MDH thermal aggregation assays. As shown in Fig. 7B, the mutation of the Trx f non-conserved Cys19 and the Trx m Cys107 did not alter the disulphide reduction and chaperone activities, when compared with the wild-type Trxs, indicating that none of these additional Cys residues are required for those functions. By contrast, the replacement of Trx f Cys71, either alone (C71S mutant) or in combination with Cys19 (C19/71S mutant), significantly reduced both the disulphide reductase and foldase activities (a 40% and 80% decrease, respectively; Fig. 7B). The effect of replacing the extra Cys residues on Trx f activity was also analysed by measuring the kinetic behaviour of these mutants in the activation of FBPase. Mutation of the well-conserved Cys residue (C71S and C19/71S mutants) completely abolished the reactivity with FBPase when assayed at similar concentrations as wild-type Trx f, whereas the reactivity was partially retained in the C19S mutant (data not shown). Regarding the holdase-chaperone function, only the double mutant, C19/71S, showed an impaired activity (30% decrease; Fig. 7B). Altogether, our results strengthen the hypothesis that the well-conserved Cys residue among the f-type Trxs is involved in protein–protein interactions; thus, mutation of this residue impairs the capacity of Trx f to interact and reduce target enzymes, and seems to be equally important for the Trx f chaperone function.
Discussion
Much progress has been made in recent years in discovering specific protein targets and functions mainly associated with intrinsic redox activity of chloroplast Trxs (Motohashi et al., 2001, 2003; Balmer et al., 2003, 2006; Marchand et al., 2010). In all of these reports, the involvement of plastid Trxs in the redox control of targets implicated in protein folding and assembly has been widely stated. However, in the last few years, a direct redox-independent control over protein folding has been identified in some proteins belonging to the Trx-fold class, providing new insights into their physiological roles (Jang et al., 2004; Berndt et al., 2008; Kthiri et al., 2008; Kim et al., 2009; Lee et al., 2009; Prasad et al., 2010). In particular, chaperone-like properties have been reported for Trxs in E. coli and Arabidopsis (Kern et al., 2003; Park et al., 2009). However, the potential functions of plastid Trxs as facilitators of protein folding remain unexplored.
As yet, only three h-type Trxs have been characterised in Nicotiana tabacum (Brugidou et al., 1993; Sun et al., 2010). In the present study, two plastid Trxs, named NtTrxf and NtTrxm, were isolated from tobacco plants. The full-length cDNAs displayed a high sequence similarity to both Trx f and Trx m from other plant species. In addition to the catalytic cysteines, Trx f possesses two extra Cys residues at positions 19 and 71 of the mature peptide (Fig. 1A). The Cys71 residue, located near the active site, is well conserved among f-type Trxs, and has been reported to be essential for protein–protein interaction (del Val et al., 1999) and responsible for Trx f glutathionylation as a redox signalling mechanism in plants (Michelet et al., 2005). By contrast, the Cys19 residue was, to date, only detected in a Trx f sequence from wheat (GenBank accession CBH32529). The analysis of ESTs from a variety of plants has identified cDNAs encoding Trx f from other solanaceous species carrying this extra Cys19 residue, suggesting that it could be well conserved within the Solanaceae family. The mature Trx m from tobacco also shows a non-conserved additional Cys residue at position 107. The modelled structure of both proteins showed that all of these Cys residues were solvent exposed (Fig. 1C), which might allow for their implication in both the structure and function of tobacco Trx f and Trx m.
Recently, the solubilization of recombinant HSA, which was produced largely as protein bodies in chloroplasts, has been demonstrated by the co-expression with tobacco plastid Trxs (Sanz-Barrio et al., 2011), suggesting that Trxs may act as molecular chaperones preventing the aggregation of HSA. To shed light on the potential chaperone properties of plastid Trxs, the ability of tobacco Trx f and Trx m to promote the in vitro functional folding of the chemically-denatured Cys-free form of G6PDH was investigated. As shown in Fig. 4, both tobacco Trxs increased (up to ∼2.5-fold) the yield of active G6PDH renaturation, and this reactivation was regulated by molar ratios of Trxs to G6PDH. This reactivation yield did not differ significantly from those that have been reported for other molecular chaperones, such as GroEL (Hansen and Gafni, 1993) and AtTDX (Lee et al., 2009). Thus, our data show that Trx f and Trx m exhibit a foldase chaperone-like activity with the Cys-free form of G6PDH, which is partially independent of their active site cysteines. The holdase chaperone function of the plastid Trxs has also been explored by studying their ability to protect MDH against thermodenaturation. As stated previously for E. coli Trx1 (Kern et al., 2003), neither Trx f nor Trx m were able to prevent efficiently the thermal aggregation of MDH when they were present as monomers. However, both Trxs exhibited holdase chaperone-like activity when they were associated with high molecular mass complexes in the presence of physiological salt concentrations or molecular crowding agents (Fig. 5). Enhancement of chaperone activity upon oligomer formation has also been reported under molecular crowding conditions for other well-described bacterial molecular chaperones, such as Hsp33 and GroEL (Ren et al., 2003; Akhtar et al., 2004). Moreover, the Trx oligomers detected in leaf extracts (Fig. 6) might indicate that Trx f or m oligomerization is of a functional significance in vivo. Oligomerization-dependent functional changes have also been observed for other plant chaperones, such as AtTDX and AtTrxh3, which change their functions from disulphide reductases to holdase chaperones, depending on their oligomeric status (Lee et al., 2009; Park et al., 2009). Similarly, structural and functional switching has been reported for some plant peroxiredoxins (Barranco-Medina et al., 2008; Kim et al., 2009). Because chaperone activity is in direct proportion to the degree of hydrophobicity (Akhtar et al., 2004; Park et al., 2009), it might be assumed that an increased hydrophobic surface in the Trx f and m oligomers, compared with the monomers, allows the protection of MDH against thermal denaturation.
Trxs are known to be redox active in their monomeric form. However, biochemical and crystal structural studies on human Trx have suggested that it has a tendency to associate into dimers (Weichsel et al., 1996; Gronenborn et al., 1999) or even higher-order oligomeric complexes (Andersen et al., 1997). In plants, the dimerization of Trxh1 from Hordeum vulgare has been reported to be involved in target recognition (Maeda et al., 2008). A relationship between Trx f dimer formation and its proper binding to chloroplastic FBPase has also been reported in pea (Soulie et al., 1985). Recently, evidence of a population of self-associated species has been demonstrated by analytical ultracentrifugation for PsTrxh1 and PsTrxf (Aguado-Llera et al., 2011). In all of these cases, the interaction between the Trxs is a result of the formation of an intermolecular disulphide bond through a well-conserved non-active Cys residue. Because Trx f oligomers disappear under reducing conditions (Fig. 7A), the Trx f polymeric structures might be associated with redox-dependent disulphide bonds. Thus, the extra Cys residues present in tobacco Trx f could be responsible for its oligomeric behaviour. However, none of the mutations created a pronounced change on the oligomerization ability of Trx f (Fig. 7A, –DTT), suggesting that the Trx active site Cys residues could also be involved in intermolecular disulphide bonds. On the other hand, our results showed that the self-association of either Trx f or Trx m into polymeric protein structures takes place in the presence of salt (Fig. 5A), which indicates that the higher-order species are held together by hydrophobic interactions. Several hydrophobic regions, including the area surrounding the Trx active site, have been proposed to be important in the interaction of Trxs with other proteins (Holmgren, 1995; Lennon et al., 2000), which could account, in part, for the oligomer formation of both tobacco Trxs. However, the ability of Trx f to self-associate was significantly higher than that of Trx m, which required a molecular crowding agent to bring about its own in vitro oligomerization at physiological salt concentrations. By using a protein pocket prediction algorithm (http://fpocket.sourceforge.net/), it was possible to identify the potential binding sites in the modelled tobacco Trxs. The program predicted five binding pockets for tobacco Trx m and six for Trx f. The major difference between both Trxs was a binding pocket that was situated around the well-conserved Cys71, which has also been reported for PsTrxf (Aguado-Llera et al., 2011). Therefore, we suggest that besides the redox-dependent Trx f polymerization, the area around the Cys71 residue may also be important for Trx f oligomer formation. In addition, hydrogen bonds and salt bridges could potentially operate in the oligomerization of both Trxs (Qin et al., 1996; Nordstrand et al., 1999).
It was also studied whether mutation of the non-active site Cys residues could alter the reported Trx functions. Regarding the Trx chaperone holdase-like function, only a slight decrease in activity was observed for the Trx f double mutant (C19/71S). This finding is consistent with the lower ability of this mutant to assemble into higher-order oligomer complexes in the absence of DTT (Fig. 7A, lane 4) and strengthens the direct relationship between holdase activity and oligomeric status. However, mutation of the Trx f well-conserved Cys71 residue, either alone or with Cys19, had a strong influence on the disulphide reductase (insulin reduction and FBPase activation) and chaperone foldase functions. The decrease in both activities and the virtual absence of monomers with those mutant proteins (Fig. 7A, –DTT, lanes 3 and 4) implies that these functions predominate in the monomeric species, as has been reported previously (Lee et al., 2009). The loss of disulphide reductase activity might be due to the close proximity of this Cys71 residue to the Trx active site, thus, making it critical for the efficient binding of Trx f to target proteins. In agreement, the activation of various target enzymes was strongly decreased when this residue was glutathionylated (Michelet et al., 2005), showing that the status of this Cys probably plays a key role in modulating Trx f efficiency. Similarly, the Trx f foldase chaperone activity was severely diminished in both Cys71 mutants, indicating that this activity may operate by means of protein interactions involving the Cys71 residue and, most likely, the binding pocket around it. These findings reinforce the well-established role of this Cys in promoting protein–protein interactions (Soulie et al., 1985; del Val et al., 1999; Aguado-Llera et al., 2011). However, as stated previously for E. coli Trx (Kern et al., 2003), an involvement of the moderately hydrophobic surface around the Trx active site on the foldase function of both Trx f and Trx m cannot be discounted. The absence of reactivity of C19S and C107S mutants could be because of the fact that their additional cysteines are considerably distant from their active site.
In conclusion, our studies revealed a novel chaperone function of plastid Trx f and Trx m as a result of their oligomeric status. It appears that the oligomerization of both plastid Trxs brings about a functional switch. The disulphide reductase and foldase activities of Trx f and Trx m occur when the proteins are monomeric, whereas the holdase chaperone function occurs predominantly when high molecular mass complexes form. This behaviour strongly resembles that of small stress family proteins in plants (Miernyk, 1999). However, the regulatory mechanism that promotes Trx f and Trx m structural and functional changes remains unclear. The reported oligomerization of AtTrxh3, with a concomitant switch in activity, seems to be regulated by heat-shock and redox status (Park et al., 2009). Other chaperones become active under heat and oxidative stress, a state in which they exist predominantly as oligomeric species (Akhtar et al., 2004; Lee et al., 2009). In this study, it was shown that plastid Trx oligomerization was induced by salt and temperature. Thus, it is hypothesized that, under reduced conditions, Trx f and Trx m are redox active in their monomeric form, and are also able to operate as molecular foldase chaperones. However, upon oxidative stress, Trx oligomers could interact with unfolded proteins preventing their aggregation (as holdase chaperones) and protecting chloroplast structures from oxidative damage. Nevertheless, the occurrence of multiple species (low and high molecular mass) in the chloroplast that display different functions cannot be ruled out.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Alignment of the amino acid sequence of tobacco Trx f with other Trx f proteins from plant sources, using the ClustalW software.
Supplementary Fig. S2. Alignment of the amino acid sequence of tobacco Trx m with other Trx m proteins from plant sources, using the ClustalW software.
Supplementary Fig. S3. SDS-PAGE analysis of purified tobacco Trx f, Trx m and mutant proteins expressed in E. coli.
Supplementary Fig. S4. Cross-reactivity of anti-Trx f and anti-Trx m antibodies.
Acknowledgments
The authors appreciate the assistance of Dr Santiago Mora (Instituto Leloir, Argentine) for his collaboration in the first steps of Trxs cDNA isolation. We gratefully acknowledge the provision of the FBPase enzyme by the laboratory of Dr Mariam Sahrawy (Estación Experimental del Zaidín, Spain) and the unconditional help of Dr César Santiago (Centro Nacional de Biotecnología, CSIC, Spain) in protein modelling. This work was supported by grants from Gobierno de Navarra (Spain; Res.17/2004) and Gobierno de Navarra-European Union funding (Project EuroInnova IIM10865.RI1-EP12). RSB and PCM were supported by pre-doctoral fellowships from Consejo Superior de Investigaciones Cientificas (Spain) and Generalitat Valenciana (Spain), respectively. AFS post-doctoral fellowship was granted by Public University of Navarra (Spain).
References
- Aguado-Llera D, Martinez-Gomez AI, Prieto J, Marenchino M, Traverso JA, Gomez J, Chueca A, Neira JL. The conformational stability and biophysical properties of the eukaryotic thioredoxins of Pisum sativum are not family-conserved. PLoS One. 2011;6 doi: 10.1371/journal.pone.0017068. e17068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akhtar MW, Srinivas V, Raman B, Ramakrishna T, Inobe T, Maki K, Arai M, Kuwajima K, Rao Ch M. Oligomeric Hsp33 with enhanced chaperone activity: gel filtration, cross-linking, and small angle X-ray scattering (SAXS) analysis. Journal of Biological Chemistry. 2004;279:55760–55769. doi: 10.1074/jbc.M406333200. [DOI] [PubMed] [Google Scholar]
- Andersen JF, Sanders DA, Gasdaska JR, Weichsel A, Powis G, Montfort WR. Human thioredoxin homodimers: regulation by pH, role of aspartate 60, and crystal structure of the aspartate 60 → asparagine mutant. Biochemistry. 1997;36:13979–13988. doi: 10.1021/bi971004s. [DOI] [PubMed] [Google Scholar]
- Arnold K, Bordoli L, Kopp J, Schwede T. The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- Arsova B, Hoja U, Wimmelbacher M, Greiner E, Ustun S, Melzer M, Petersen K, Lein W, Bornke F. Plastidial thioredoxin z interacts with two fructokinase-like proteins in a thiol-dependent manner: evidence for an essential role in chloroplast development in Arabidopsis and Nicotiana benthamiana. The Plant Cell. 2010;22:1498–1515. doi: 10.1105/tpc.109.071001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Koller A, del Val G, Manieri W, Schurmann P, Buchanan BB. Proteomics gives insight into the regulatory function of chloroplast thioredoxins. Proceedings of the National Academy of Sciences, USA. 2003;100:370–375. doi: 10.1073/pnas.232703799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balmer Y, Vensel WH, Cai N, Manieri W, Schurmann P, Hurkman WJ, Buchanan BB. A complete ferredoxin/thioredoxin system regulates fundamental processes in amyloplasts. Proceedings of the National Academy of Sciences, USA. 2006;103:2988–2993. doi: 10.1073/pnas.0511040103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas-Lopez JD, Serrato AJ, Cazalis R, Meyer Y, Chueca A, Reichheld JP, Sahrawy M. Circadian regulation of chloroplastic f and m thioredoxins through control of the CCA1 transcription factor. Journal of Experimental Botany. 2011;62:2039–2051. doi: 10.1093/jxb/erq394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barajas-Lopez JD, Serrato AJ, Olmedilla A, Chueca A, Sahrawy M. Localization in roots and flowers of pea chloroplastic thioredoxin f and thioredoxin m proteins reveals new roles in nonphotosynthetic organs. Plant Physiology. 2007;145:946–960. doi: 10.1104/pp.107.105593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barranco-Medina S, Krell T, Bernier-Villamor L, Sevilla F, Lazaro JJ, Dietz KJ. Hexameric oligomerization of mitochondrial peroxiredoxin PrxIIF and formation of an ultrahigh affinity complex with its electron donor thioredoxin Trx-o. Journal of Experimental Botany. 2008;59:3259–3269. doi: 10.1093/jxb/ern177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beissinger M, Buchner J. How chaperones fold proteins. Biological Chemistry. 1998;379:245–259. [PubMed] [Google Scholar]
- Benitez-Alfonso Y, Cilia M, San Roman A, Thomas C, Maule A, Hearn S, Jackson D. Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proceedings of the National Academy of Sciences, USA. 2009;106:3615–3620. doi: 10.1073/pnas.0808717106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt C, Lillig CH, Holmgren A. Thioredoxins and glutaredoxins as facilitators of protein folding. Biochimica et Biophysica Acta. 2008;1783:641–650. doi: 10.1016/j.bbamcr.2008.02.003. [DOI] [PubMed] [Google Scholar]
- Bock R, Warzecha H. Solar-powered factories for new vaccines and antibiotics. Trends in Biotechnology. 2010;28:246–252. doi: 10.1016/j.tibtech.2010.01.006. [DOI] [PubMed] [Google Scholar]
- Boston RS, Viitanen PV, Vierling E. Molecular chaperones and protein folding in plants. Plant Molecular Biology. 1996;32:191–222. doi: 10.1007/BF00039383. [DOI] [PubMed] [Google Scholar]
- Brugidou C, Marty I, Chartier Y, Meyer Y. The Nicotiana tabacum genome encodes two cytoplasmic thioredoxin genes which are differently expressed. Moecular and General Genetics. 1993;238:285–293. doi: 10.1007/BF00279557. [DOI] [PubMed] [Google Scholar]
- Buchanan BB. Role of light in the regulation of chloroplast enzymes. Annual Review of Plant Physiology. 1980;31:341–374. [Google Scholar]
- Capitani G, Schurmann P. On the quaternary assembly of spinach chloroplast thioredoxin m. Photosynthesis Research. 2004;79:281–285. doi: 10.1023/B:PRES.0000017197.03467.81. [DOI] [PubMed] [Google Scholar]
- Chebotareva NA, Kurganov BI, Livanova NB. Biochemical effects of molecular crowding. Biochemistry (Moscow) 2004;69:1239–1251. doi: 10.1007/s10541-005-0070-y. [DOI] [PubMed] [Google Scholar]
- Cheng G, Basha E, Wysocki VH, Vierling E. Insights into small heat shock protein and substrate structure during chaperone action derived from hydrogen/deuterium exchange and mass spectrometry. Journal of Biological Chemistry. 2008;283:26634–26642. doi: 10.1074/jbc.M802946200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibani K, Wingsle G, Jacquot JP, Gelhaye E, Rouhier N. Comparative genomic study of the thioredoxin family in photosynthetic organisms with emphasis on. Populus trichocarpa. Molecular Plant. 2009;2:308–322. doi: 10.1093/mp/ssn076. [DOI] [PubMed] [Google Scholar]
- Collin V, Lamkemeyer P, Miginiac-Maslow M, Hirasawa M, Knaff DB, Dietz KJ, Issakidis-Bourguet E. Characterization of plastidial thioredoxins from Arabidopsis belonging to the new y-type. Plant Physiology. 2004;136:4088–4095. doi: 10.1104/pp.104.052233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Singh ND, Mason H, Streatfield SJ. Plant-made vaccine antigens and biopharmaceuticals. Trends in Plant Science. 2009;14:669–679. doi: 10.1016/j.tplants.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Val G, Maurer F, Stutz E, Schurmann P. Modification of the reactivity of spinach chloroplast thioredoxin f by site-directed mutagenesis. Plant Science. 1999;149:183–190. [Google Scholar]
- Ellis RJ. Macromolecular crowding: an important but neglected aspect of the intracellular environment. Current Opinion in Structural Biology. 2001;11:114–119. doi: 10.1016/s0959-440x(00)00172-x. [DOI] [PubMed] [Google Scholar]
- Geigenberger P, Kolbe A, Tiessen A. Redox regulation of carbon storage and partitioning in response to light and sugars. Journal of Experimental Botany. 2005;56:1469–1479. doi: 10.1093/jxb/eri178. [DOI] [PubMed] [Google Scholar]
- Gronenborn AM, Clore GM, Louis JM, Wingfield PT. Is human thioredoxin monomeric or dimeric? Protein Science. 1999;8:426–429. doi: 10.1110/ps.8.2.426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JE, Gafni A. Thermal switching between enhanced and arrested reactivation of bacterial glucose-6-phosphate dehydrogenase assisted by GroEL in the absence of ATP. Journal of Biological Chemistry. 1993;268:21632–21636. [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin. 6. The amino acid sequence of the protein from Escherichia coli B. European Journal of Biochemistry. 1968;6:475–484. doi: 10.1111/j.1432-1033.1968.tb00470.x. [DOI] [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin catalyzes the reduction of insulin disulfides by dithiothreitol and dihydrolipoamide. Journal of Biological Chemistry. 1979;254:9627–9632. [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin structure and mechanism: conformational changes on oxidation of the active-site sulfhydryls to a disulfide. Structure. 1995;3:239–243. doi: 10.1016/s0969-2126(01)00153-8. [DOI] [PubMed] [Google Scholar]
- Issakidis-Bourguet E, Mouaheb N, Meyer Y, Miginiac-Maslow M. Heterologous complementation of yeast reveals a new putative function for chloroplast m-type thioredoxin. The Plant Journal. 2001;25:127–135. doi: 10.1046/j.0960-7412.2000.00943.x. [DOI] [PubMed] [Google Scholar]
- Jang HH, Lee KO, Chi YH, et al. Two enzymes in one; two yeast peroxiredoxins display oxidative stress-dependent switching from a peroxidase to a molecular chaperone function. Cell. 2004;117:625–635. doi: 10.1016/j.cell.2004.05.002. [DOI] [PubMed] [Google Scholar]
- Jeng MF, Campbell AP, Begley T, Holmgren A, Case DA, Wright PE, Dyson HJ. High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin. Structure. 1994;2:853–868. doi: 10.1016/s0969-2126(94)00086-7. [DOI] [PubMed] [Google Scholar]
- Kern R, Malki A, Holmgren A, Richarme G. Chaperone properties of Escherichia coli thioredoxin and thioredoxin reductase. Biochemical Journal. 2003;371:965–972. doi: 10.1042/BJ20030093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer F, Arnold K, Kunzli M, Bordoli L, Schwede T. The SWISS-MODEL Repository and associated resources. Nucleic Acids Research. 2009;37:D387–392. doi: 10.1093/nar/gkn750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SY, Jang HH, Lee JR, et al. Oligomerization and chaperone activity of a plant 2-Cys peroxiredoxin in response to oxidative stress. Plant Science. 2009;177:227–232. [Google Scholar]
- Kthiri F, Le HT, Tagourti J, Kern R, Malki A, Caldas T, Abdallah J, Landoulsi A, Richarme G. The thioredoxin homolog YbbN functions as a chaperone rather than as an oxidoreductase. Biochemical and Biophysical Research Communications. 2008;374:668–672. doi: 10.1016/j.bbrc.2008.07.080. [DOI] [PubMed] [Google Scholar]
- LaVallie ER, DiBlasio-Smith EA, Collins-Racie LA, Lu Z, McCoy JM. Thioredoxin and related proteins as multifunctional fusion tags for soluble expression in. E. coli. Methods in Molecular Biology. 2003;205:119–140. doi: 10.1385/1-59259-301-1:119. [DOI] [PubMed] [Google Scholar]
- LaVallie ER, Lu Z, Diblasio-Smith EA, Collins-Racie LA, McCoy JM. Thioredoxin as a fusion partner for production of soluble recombinant proteins in. Escherichia coli. Methods in Enzymology. 2000;326:322–340. doi: 10.1016/s0076-6879(00)26063-1. [DOI] [PubMed] [Google Scholar]
- Lee JR, Lee SS, Jang HH, et al. Heat-shock dependent oligomeric status alters the function of a plant-specific thioredoxin-like protein, AtTDX. Proceedings of the National Academy of Sciences, USA. 2009;106:5978–5983. doi: 10.1073/pnas.0811231106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaire SD, Michelet L, Zaffagnini M, Massot V, Issakidis-Bourguet E. Thioredoxins in chloroplasts. Curent Genetics. 2007;51:343–365. doi: 10.1007/s00294-007-0128-z. [DOI] [PubMed] [Google Scholar]
- Lennon BW, Williams CH, Jr, Ludwig ML. Twists in catalysis: alternating conformations of Escherichia coli thioredoxin reductase. Science. 2000;289:1190–1194. doi: 10.1126/science.289.5482.1190. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maeda K, Hagglund P, Finnie C, Svensson B, Henriksen A. Crystal structures of barley thioredoxin h isoforms HvTrxh1 and HvTrxh2 reveal features involved in protein recognition and possibly in discriminating the isoform specificity. Protein Science. 2008;17:1015–1024. doi: 10.1110/ps.083460308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchand CH, Vanacker H, Collin V, Issakidis-Bourguet E, Marechal PL, Decottignies P. Thioredoxin targets in Arabidopsis roots. Proteomics. 2010;10:2418–2428. doi: 10.1002/pmic.200900835. [DOI] [PubMed] [Google Scholar]
- Mark DF, Richardson CC. Escherichia coli thioredoxin: a subunit of bacteriophage T7 DNA polymerase. Proceedings of the National Academy of Sciences, USA. 1976;73:780–784. doi: 10.1073/pnas.73.3.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marri L, Zaffagnini M, Collin V, Issakidis-Bourguet E, Lemaire SD, Pupillo P, Sparla F, Miginiac-Maslow M, Trost P. Prompt and easy activation by specific thioredoxins of calvin cycle enzymes of Arabidopsis thaliana associated in the GAPDH/CP12/PRK supramolecular complex. Molecular Plant. 2009;2:259–269. doi: 10.1093/mp/ssn061. [DOI] [PubMed] [Google Scholar]
- Meyer Y, Reichheld JP, Vignols F. Thioredoxins in Arabidopsis and other plants. Photosynthesis Research. 2005;86:419–433. doi: 10.1007/s11120-005-5220-y. [DOI] [PubMed] [Google Scholar]
- Michelet L, Zaffagnini M, Marchand C, et al. Glutathionylation of chloroplast thioredoxin f is a redox signaling mechanism in plants. Proceedings of the National Academy of Sciences, USA. 2005;102:16478–16483. doi: 10.1073/pnas.0507498102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miernyk JA. Protein folding in the plant cell. Plant Physiology. 1999;121:695–703. doi: 10.1104/pp.121.3.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montrichard F, Alkhalfioui F, Yano H, Vensel WH, Hurkman WJ, Buchanan BB. Thioredoxin targets in plants: the first 30 years. Journal of Proteomics. 2009;72:452–474. doi: 10.1016/j.jprot.2008.12.002. [DOI] [PubMed] [Google Scholar]
- Motohashi K, Kondoh A, Stumpp MT, Hisabori T. Comprehensive survey of proteins targeted by chloroplast thioredoxin. Proceedings of the National Academy of Sciences, USA. 2001;98:11224–11229. doi: 10.1073/pnas.191282098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motohashi K, Koyama F, Nakanishi Y, Ueoka-Nakanishi H, Hisabori T. Chloroplast cyclophilin is a target protein of thioredoxin. Thiol modulation of the peptidyl-prolyl cis-trans isomerase activity. Journal of Biological Chemistry. 2003;278:31848–31852. doi: 10.1074/jbc.M304258200. [DOI] [PubMed] [Google Scholar]
- Nordstrand K, slund F, Holmgren A, Otting G, Berndt KD. NMR structure of Escherichia coli glutaredoxin 3-glutathione mixed disulfide complex: implications for the enzymatic mechanism. Journal of Molecular Biology. 1999;286:541–552. doi: 10.1006/jmbi.1998.2444. [DOI] [PubMed] [Google Scholar]
- Pagano EA, Chueca A, Lopez-Gorge J. Expression of thioredoxins f and m, and of their targets fructose-1,6-bisphosphatase and NADP-malate dehydrogenase, in pea plants grown under normal and light/temperature stress conditions. Journal of Experimental Botany. 2000;51:1299–1307. [PubMed] [Google Scholar]
- Park SK, Jung YJ, Lee JR, et al. Heat-shock and redox-dependent functional switching of an h-type Arabidopsis thioredoxin from a disulfide reductase to a molecular chaperone. Plant Physiology. 2009;150:552–561. doi: 10.1104/pp.109.135426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Ruiz JM, Cejudo FJ. A proposed reaction mechanism for rice NADPH thioredoxin reductase C, an enzyme with protein disulfide reductase activity. FEBS Leters. 2009;583:1399–1402. doi: 10.1016/j.febslet.2009.03.067. [DOI] [PubMed] [Google Scholar]
- Pigiet VP, Schuster BJ. Thioredoxin-catalyzed refolding of disulfide-containing proteins. Proceedings of the National Academy of Sciences, USA. 1986;83:7643–7647. doi: 10.1073/pnas.83.20.7643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad BD, Goel S, Krishna P. In silico identification of carboxylate clamp type tetratricopeptide repeat proteins in Arabidopsis and rice as putative co-chaperones of Hsp90/Hsp70. PLoS One. 2010 doi: 10.1371/journal.pone.0012761. 5, e12761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Clore GM, Kennedy WP, Kuszewski J, Gronenborn AM. The solution structure of human thioredoxin complexed with its target from Ref-1 reveals peptide chain reversal. Structure. 1996;4:613–620. doi: 10.1016/s0969-2126(96)00065-2. [DOI] [PubMed] [Google Scholar]
- Ren G, Lin Z, Tsou CL, Wang CC. Effects of macromolecular crowding on the unfolding and the refolding of D-glyceraldehyde-3-phosophospate dehydrogenase. Journal of Protein Chemistry. 2003;22:431–439. doi: 10.1023/b:jopc.0000005458.08802.11. [DOI] [PubMed] [Google Scholar]
- Sahrawy M, Hecht V, Lopez-Jaramillo J, Chueca A, Chartier Y, Meyer Y. Intron position as an evolutionary marker of thioredoxins and thioredoxin domains. Journal of Molecular Evolution. 1996;42:422–431. doi: 10.1007/BF02498636. [DOI] [PubMed] [Google Scholar]
- Saitoh M, Nishitoh H, Fujii M, Takeda K, Tobiume K, Sawada Y, Kawabata M, Miyazono K, Ichijo H. Mammalian thioredoxin is a direct inhibitor of apoptosis signal-regulating kinase (ASK) 1. EMBO Journal. 1998;17:2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Barrio R, Fernandez-San Millan A, Corral-Martinez P, Segui-Simarro JM, Farran I. Tobacco plastidial thioredoxins as modulators of recombinant protein production in transgenic chloroplasts. Plant Biotechnology Journal. 2011;9:639–650. doi: 10.1111/j.1467-7652.2011.00608.x. [DOI] [PubMed] [Google Scholar]
- Scheibe R, Anderson LE. Dark modulation of NADP-dependent malate dehydrogenase and glucose-6-phosphate dehydrogenase in the chloroplast. Biochimica et Biophysica Acta. 1981;636:58–64. doi: 10.1016/0005-2728(81)90075-x. [DOI] [PubMed] [Google Scholar]
- Schurmann P, Buchanan BB. The ferredoxin/thioredoxin system of oxygenic photosynthesis. Antioxidant Redox Signalling. 2008;10:1235–1274. doi: 10.1089/ars.2007.1931. [DOI] [PubMed] [Google Scholar]
- Segui-Simarro JM, Testillano PS, Risueno MC. Hsp70 and Hsp90 change their expression and subcellular localization after microspore embryogenesis induction in Brassica napus L. Journal of Structural Biology. 2003;142:379–391. doi: 10.1016/s1047-8477(03)00067-4. [DOI] [PubMed] [Google Scholar]
- Serrato AJ, Perez-Ruiz JM, Spinola MC, Cejudo FJ. A novel NADPH thioredoxin reductase, localized in the chloroplast, which deficiency causes hypersensitivity to abiotic stress in Arabidopsis thaliana. Journal of Biological Chemistry. 2004;279:43821–43827. doi: 10.1074/jbc.M404696200. [DOI] [PubMed] [Google Scholar]
- Serrato AJ, Yubero-Serrano EM, Sandalio LM, Munoz-Blanco J, Chueca A, Caballero JL, Sahrawy M. cpFBPaseII, a novel redox-independent chloroplastic isoform of fructose-1,6-bisphosphatase. Plant, Cell and Environment. 2009;32:811–827. doi: 10.1111/j.1365-3040.2009.01960.x. [DOI] [PubMed] [Google Scholar]
- Soulie JM, Buc J, Riviere M, Ricard J. Equilibrium binding of thioredoxin fB to chloroplastic fructose bisphosphatase. Evidence for a thioredoxin site distinct from the active site. European Journal of Biochemistry. 1985;152:565–568. doi: 10.1111/j.1432-1033.1985.tb09232.x. [DOI] [PubMed] [Google Scholar]
- Sun L, Ren H, Liu R, Li B, Wu T, Sun F, Liu H, Wang X, Dong H. An h-type thioredoxin functions in tobacco defense responses to two species of viruses and an abiotic oxidative stress. Moecular Plant–Microbe Interactions. 2010;23:1470–1485. doi: 10.1094/MPMI-01-10-0029. [DOI] [PubMed] [Google Scholar]
- Traverso JA, Vignols F, Cazalis R, Serrato AJ, Pulido P, Sahrawy M, Meyer Y, Cejudo FJ, Chueca A. Immunocytochemical localization of Pisum sativum TRXs f and m in non-photosynthetic tissues. Journal of Experimental Botany. 2008;59:1267–1277. doi: 10.1093/jxb/ern037. [DOI] [PubMed] [Google Scholar]
- Weichsel A, Gasdaska JR, Powis G, Montfort WR. Crystal structures of reduced, oxidized, and mutated human thioredoxins: evidence for a regulatory homodimer. Structure. 1996;4:735–751. doi: 10.1016/s0969-2126(96)00079-2. [DOI] [PubMed] [Google Scholar]
- Wolosiuk RA, Crawford NA, Yee BC, Buchanan BB. Isolation of three thioredoxins from spinach leaves. Journal of Biological Chemistry. 1979;254:1627–1632. [PubMed] [Google Scholar]
- Yasukawa T, Kanei-Ishii C, Maekawa T, Fujimoto J, Yamamoto T, Ishii S. Increase of solubility of foreign proteins in Escherichia coli by coproduction of the bacterial thioredoxin. Journal of Biological Chemistry. 1995;270:25328–25331. doi: 10.1074/jbc.270.43.25328. [DOI] [PubMed] [Google Scholar]
- Yuan S, Duan H, Liu C, Liu X, Liu T, Tao H, Zhang Z. The role of thioredoxin and disulfide isomerase in the expression of the snake venom thrombin-like enzyme calobin in Escherichia coli BL21 (DE3) Protein Expression and Purification. 2004;38:51–60. doi: 10.1016/j.pep.2004.08.004. [DOI] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One. 2008;3:e1994. doi: 10.1371/journal.pone.0001994. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.