Abstract
(1,3;1,4)-β-D-glucans (mixed-linkage glucans) are found in tissues of members of the Poaceae (grasses), and are particularly high in barley (Hordeum vulgare) grains. The present study describes the isolation of three independent (1,3;1,4)-β-D-glucanless (betaglucanless; bgl) mutants of barley which completely lack (1,3;1,4)-β-D-glucan in all the tissues tested. The bgl phenotype cosegregates with the cellulose synthase like HvCslF6 gene on chromosome arm 7HL. Each of the bgl mutants has a single nucleotide substitution in the coding region of the HvCslF6 gene resulting in a change of a highly conserved amino acid residue of the HvCslF6 protein. Microsomal membranes isolated from developing endosperm of the bgl mutants lack detectable (1,3;1,4)-β-D-glucan synthase activity indicating that the HvCslF6 protein is inactive. This was confirmed by transient expression of the HvCslF6 cDNAs in Nicotiana benthamiana leaves. The wild-type HvCslF6 gene directed the synthesis of high levels of (1,3;1,4)-β-D-glucans, whereas the mutant HvCslF6 proteins completely lack the ability to synthesize (1,3;1,4)-β-D-glucans. The fine structure of the (1,3;1,4)-β-D-glucan produced in the tobacco leaf was also very different from that found in cereals having an extremely low DP3/DP4 ratio. These results demonstrate that, among the seven CslF and one CslH genes present in the barley genome, HvCslF6 has a unique role and is the key determinant controlling the biosynthesis of (1,3;1,4)-β-D-glucans. Natural allelic variation in the HvCslF6 gene was found predominantly within introns among 29 barley accessions studied. Genetic manipulation of the HvCslF6 gene could enable control of (1,3;1,4)-β-D-glucans in accordance with the purposes of use.
Keywords: Cell wall, grasses, Hordeum vulgare, mixed-linkage glucans, polysaccharide
Introduction
The non-starch cell-wall polysaccharides of cereal grains include cellulose, (1,3;1,4)-β-D-glucans (mixed-linkage glucans), and arabino-xylans as major components. Of these, (1,3;1,4)-β-D-glucans are found in tissues of members of the Poaceae (grasses). Barley (Hordeum vulgare) and oat (Avena sativa) grains are rich in (1,3;1,4)-β-D-glucans, while wheat (Triticum aestivum), rice (Oryza sativa), and maize (Zea mays) have much lower amounts. (1,3;1,4)-β-D-Glucans are linear, unbranched molecules, which contain both (1,3)- and (1,4)-β-D-glucosidic linkages. (1,3;1,4)-β-D-Glucans consist primarily of cellotriosyl and cellotetraosyl units linked by single (1,3)-β-D-glucosidic linkages, but they also contain a small proportion of blocks of 5–11 contiguous (1,4)-linked β-D-glucosyl residues. Adjacent (1,3)-β-D-glucosyl residues are absent in the case of the barley (1,3;1,4)-β-D-glucans (Fincher, 2009).
High (1,3;1,4)-β-D-glucan content in barley grains negatively affects the processes of malting and brewing because it prolongs endosperm modification and filtration of wort and beer, lowering the production efficiency and leaving beer with an unpleasant haze (von Wettstein, 2007). Therefore, a low (1,3;1,4)-β-D-glucan content is one of the desirable characteristics in the breeding of malting barley. On the other hand, (1,3;1,4)-β-D-glucan are dietary fibres that have beneficial effects in the prevention of various human diseases, including high serum cholesterol and cardiovascular disease (Brennan and Cleary, 2005). Thus, (1,3;1,4)-β-D-glucan is considered an important functional ingredient when barley is consumed as food. Although (1,3;1,4)-β-D-glucan content in ordinary barley cultivars is in the range of 2.9–6.4% (Kato et al., 1995), genotypes containing as high as 11–20% have been bred for food barley (Fujita et al., 1999; Munck et al., 2004). Several quantitative trait loci (QTLs) associated with (1,3;1,4)-β-D-glucan content are known (Han et al., 1995; Meyer et al., 2000; Molina-Cano et al., 2007; Li et al., 2008; Emebiri, 2009).
Recent studies have advanced our understanding of the genes involved in the synthesis of (1,3;1,4)-β-D-glucans (Burton et al., 2006, 2008; Doblin et al., 2009). CslF genes have been implicated in the biosynthesis of (1,3;1,4)-β-D-glucans (Burton et al., 2006). This was shown by inserting several genes from a cluster of six rice OsCslF genes on chromosome 7 into Arabidopsis thaliana, which does not have CslF genes or (1,3;1,4)-β-D-glucans in its walls; a small amount of (1,3;1,4)-β-D-glucans in the cell walls of transgenic plants was detected using a specific monoclonal antibody and enzymatic analysis. Comparative genomic studies have shown that, in contrast to the eight CslF genes in rice, barley has seven CslF family members; HvCslF3, HvCslF4, HvCslF8, and HvCslF10 are clustered on chromosome 2H, HvCslF9 is located on 1H, HvCslF7 is located on 5H, and HvCslF6 is located on 7H (Burton et al., 2008). By similar transgenic approaches, it was shown that the single CslH gene of barley can mediate (1,3;1,4)-β-D-glucan synthesis in Arabidopsis (Doblin et al., 2009). A recent study employing a (1,3;1,4)-β-D-glucanless (beta-glucanless hereafter abbreviated as bgl) mutant in barley indicated that HvCslF6 could be a key determinant controlling (1,3;1,4)-β-D-glucan synthesis as the bgl locus mapped close to the HvCslF6 gene and this gene had a single nucleotide change in the coding region (Tonooka et al., 2009). This finding was supported by the observation of reduced (1,3;1,4)-β-D-glucan of wholemeal flours from RNAi inhibition of CslF6 in wheat grain (Nemeth et al., 2010).
In this study, molecular and biochemical approaches were applied to prove that bgl mutants are mutations within the coding regions of the HvCslF6 gene that are essential for its enzymatic activity. The loss of function of this gene results in the complete absence of (1,3;1,4)-β-D-glucan both in vegetative and reproductive organs in barley suggesting a unique role for the HvCslF6 gene in the control of (1,3;1,4)-β-D-glucan biosynthesis.
Materials and methods
Plant materials
The barley (1,3;1,4)-β-D-glucanless mutant OUM125 was induced in the genetic background of ‘Akashinriki’ and carries the bgl gene (Tonooka et al., 2009). Backcross derivatives of OUM125 carrying bgl in the genetic background of other varieties were also used. For screening of new (1,3;1,4)-β-D-glucanless mutants, a sodium azide mutagenized population consisting of about 2000 M4 lines of cv. ‘Sachiho Golden’ were used. For allelism tests, putative (1,3;1,4)-β-D-glucanless mutants were test-crossed with the near isogenic line of ‘Nishinohoshi’ (Ni) with the introduced bgl gene after four backcrosses (abbreviated as bgl-Ni, Tonooka et al., 2009). At least three F1 grains from test cross were individually measured for (1,3;1,4)-β-D-glucan content as described below.
To test the dosage effect of the bgl gene, Ni and bgl-Ni were reciprocally crossed, and 12 grains each of the F1 hybrids and parental lines were individually measured for (1,3;1,4)-β-D-glucan content as described below. These materials are from harvests in the same growing season grown under the same greenhouse environment. Ni and bgl-Ni were also compared for expression of the HvCslF6 gene through qRT-PCR analyses.
For the assay of (1,3;1,4)-β-D-glucan synthase activity, two sets of field-grown materials were used; one is Shikoku Hadaka 84 (SH84) and its isogenic line with the introduced bgl gene of OUM125 by recurrent backcrosses (abbreviated as bgl-SH84), and the other is the (1,3;1,4)-β-D-glucanless mutant KM27 and its parental cv. ‘Sachiho Golden’.
For quantification of the (1,3;1,4)-β-D-glucan content in field-grown leaves, three sets of isogenic lines were used, i.e. ‘Akashinriki’ and OUM125, Ni and bgl-Ni, and SH84 and bgl-SH84.
Genetic mapping of bgl and HvCslF6
For mapping, 104 F2 plants derived from a cross between ‘Bowman’ (two-rowed, covered caryopsis) and (1,3;1,4)-β-D-glucanless mutant OUM125 (six-rowed, naked caryopsis) was used. Parental accessions with contrasting morphology were selected for the cross to maximize genetic polymorphisms. Individual F2 grains were cut in half, and half grains minus the embryo were used for (1,3;1,4)-β-D-glucan quantification as described below, and the remaining embryo-containing half grains were germinated for DNA extraction from leaves. DNA was isolated by the modified methods of Edwards et al. (1991) with an additional extraction step with phenol–chloroform–isoamyl alcohol (25:24:1 by vol.) after initial extraction with the buffer. The following DNA markers were used for mapping: the CDO673 primer is from Heun et al. (1991) and detected according to Kikuchi et al. (2003), nud for naked caryopsis is detected according to Taketa et al. (2008), the MWG511 primer is from Künzel et al. (2000), and the SSR markers are from publicly available sources (Ramsay et al., 2000; Varshney et al., 2007). Molecular markers for waxy and beta-glucanase II (Waxy and HvGlb2, respectively) were originally developed in this study, as shown in Supplementary Table S1 at JXB online. Molecular markers were assigned to specific chromosome arms by using wheat–barley disomic and ditelosomic chromosome addition lines (Islam et al., 1981; Islam 1983).
Analysis of (1,3;1,4)-β-D-glucan content in barley sample
For grain samples, half grains minus the embryo were individually squashed with pliers and then ground into a fine powder with a mortar and pestle in the presence of liquid nitrogen. During grinding, efforts were made to remove hulls as much as possible. The flours were weighed and about 18 mg of flours per grain were subjected to (1,3;1,4)-β-D-glucan quantification by an enzymatic method (McCleary and Codd, 1991) using a Mixed linkage beta-Glucan Assay Kit (Megazyme International Ireland, Wicklow, Ireland). The percentages of (1,3;1,4)-β-D-glucan are shown on a dry-weight basis.
For (1,3;1,4)-β-D-glucan quantification in leaves, the three sets of materials described above were planted in late October 2008 in a field of the National Agriculture Research Center in Tsukuba, Japan. Leaf blades and leaf sheaths were collected from three plants of each line in early March and immediately frozen in liquid nitrogen. The samples were finely milled by a hi-speed vibration sample mill (TI-100, CMT Corp., Iwaki) after being freeze-dried. Following a treatment with 80% ethanol, (1,3;1,4)-β-D-glucan content were measured using a Mixed linkage beta-Glucan Assay Kit.
Expression analysis of HvCslF6 by quantitative RT-PCR (qRT-PCR)
qRT-PCR was conducted according to the procedures described in Yuo et al. (2009). Briefly, total RNAs were extracted from various tissues of plants at the vegetative stage (10 d after germination) and the reproductive stages [5 d and 10 d after flowering (DAF)] using an Isogen kit (Nippon Gene, Tokyo, Japan). RNA samples were treated with DNaseI (Promega, Madison, WI). For qRT-PCR, a first strand cDNA was synthesized with PrimeScript RT reagent Kit (TaKaRa, Otsu, Japan). Quantitative analyses were carried out on Thermal cycle dice TP800 (TaKaRa) using SYBR Premix Ex Taq and SYBR green detection kit (TaKaRa) according to the manufacturer's instructions. Normalization was carried out using the GAPDH gene. Primers used for expression analyses of HvCslF6 and GAPDH are the same as those described in Burton et al. (2008).
Assay of activity of (1,3;1,4)-β-D-glucan synthase
Seeds of the two lines (SH84 and bgl-SH84) were taken at different developmental stages from 7 to 35 DAF. Seeds of KM 27 and its parental cv. ‘Sachiho Golden’, were also harvested at 19 DAF. The seeds were stored at –80 °C until use. The microsomal fraction of the endosperms was prepared as described previously (Tsuchiya et al., 2005). The microsomal fraction was used for the enzyme assays immediately after preparation.
Enzyme activity of (1,3;1,4)-β-D-glucan synthase was determined according to our previous procedures (Tsuchiya et al., 2005) with a slight modification. A reaction mixture (60 μl) consisted of 2 mM UDP-Glc including 2.96 kBq UDP-[14C]Glc (Perkin Elmer Life Sciences, Boston, MA), 20 mM MgCl2, 200 mM sucrose, 50 mM TRIS-HCl buffer (pH 9.0), and the microsomal fraction (protein, 200–400 μg) at 25 °C for 30 min. Protein was determined by the method of Bradford (1976) using BSA as the standard. The reaction was terminated by dipping in a boiling water bath for 5 min, and the products containing radiolabelled (1,3;1,4)-β-D-glucans were precipitated by methanol (final 55% concentration). The products were digested with lichenase (1 unit; Megazyme), and the hydrolysate was analysed by paper chromatography. Radioactive spots on paper chromatograms were detected by a fluoroimage analyser FLA-7000 (Fujifilm), while spots of reducing sugars were visualized with alkaline AgNO3 (Trevelyan et al., 1950). The radioactive spots corresponding to trisaccharide (G4G3G) and tetrasaccharide (G4G4G3G) were cut off, and radioactivity (dpm) was counted with a liquid scintillation counter (Ishikawa et al., 2000) to estimate enzyme activity.
Analyses of carbohydrates in developing endosperm cell walls
The sugar constituents in cell walls of developing endosperms of barley were analysed essentially according to previous method (Urahara et al., 2004) and described briefly as follows. Barley endosperm specimens (about 10 mg) were separated from the seed coats and embryos by hand. The samples were homogenized to powder in liquid nitrogen, dissolved in 1 ml of 17.5% NaOH containing 0.04% NaBH4 by mixing in a boiling water bath. The mixtures were neutralized with acetic acid, dialysed against 5 mM MOPS-KOH buffer (pH 7.0), and then incubated at 37 °C with α-amylase (80 units, type I-A; Sigma-Aldrich, Tokyo, Japan) to remove starch. The resulting cell wall polysaccharides were hydrolysed by the 72%–8% sulphuric acid method of Bouveng et al. (1965). Separation of monosaccharide constituents was carried out by high-performance anion-exchange chromatography (HPAEC) using a Dionex DX-500 liquid chromatograph (Dionex Japan, Osaka, Japan) fitted with the CarboPac PA-1 column and a pulsed amperometric detector as described previously (Ishikawa et al., 2000).
Functional characterization of HvCslF6 genes in Nicotiana benthamiana
Full-length HvCslF6 cDNAs from wild-type ‘Akashinriki’ and mutant lines (OUM125 and KM27) were amplified from leaf cDNA using primers shown in Supplementary Table S1 at JXB online, and cloned into pCR Blunt (Invitrogen, Carlsbad, CA). For unknown reasons it has not been possible to clone the full-length cDNA from the KM30 mutant. The full-length cDNAs were excised as EcoRI fragments and cloned in the sense orientation into the same site of a modified pORE02 (Coutu et al., 2007) Agrobacterium binary vector containing a cauliflower mosaic virus 35S promoter was inserted at the SfoI site (Wood et al., 2009). The expression constructs were transformed into Agrobacterium tumefaciens strain AGL1 and transient expression of Agrobacterium-infilitrated Nicotiana benthamiana leaves was performed as described in Wood et al. (2009) except that the Agrobacterium density had an OD600 of 0.4. Leaves were harvested 5 d after infiltration and freeze-dried. The (1,3;1,4)-β-D-glucan content of the leaves was assayed as follows. Firstly, a crude cell wall preparation was made from 20 mg of ground leaf material by heating for 30 min at 80 °C in 1.8 ml of 80% ethanol in a 2 ml Eppendorf tube with mixing. The supernatant was removed after centrifugation at 10 000 rpm for 5 min and the residue was re-extracted in the same volume of 80% ethanol at 80 °C for 10 min. After centrifugation, the pellet was washed at room temperature for 10 min in 50% ethanol with a final wash in 20 mM sodium phosphate buffer pH 6.5. The pellet was resuspended in 0.5 ml of the same buffer and the material was solubilized by heating at 90 °C for 30 min. The sample was cooled to 50 °C and incubated for 2 h with 20 μl (1 U) lichenase (Megazyme) to digest the (1,3;1,4)-β-D-glucan. Following centrifugation, 100 μl of sample was dried in a Speedvac and the oligosaccharides were fluorescently labelled by reductive amination with 8-amino-1,3,6-pyrenetrisulphonic acid (APTS) and separation by fluorophore-assisted-capillary electrophoresis (FACE) with laser-induced fluorescence detection as described (O'Shea et al., 1998). Similarly, FACE analysis was also applied to lichenase digests of cell wall extracts from various barley seedling tissues of ‘Akashinriki’ and OUM125.
Molecular characterization and diversity analysis of the HvCslF6 gene
On the basis of reported cDNA sequence of HvCslF6 (Burton et al., 2008), sequencing primers for genomic DNA and cDNA were designed (see Supplementary Table S1 at JXB online). About a 5.2 kb genomic fragment was PCR-amplified and sequenced using internal primers.
A set of 29 barley accessions with diverse origins were sequenced for the HvCslF6 gene. These accessions included five H. vulgare ssp. spontaneum, and 15 two-rowed and seven six-rowed domesticated barley accessions. Barley accessions used for QTL analysis of (1,3;1,4)-β-D-glucan content in the previous studies (Han et al., 1995; Meyer et al., 2000; Li et al., 2008; Molina-Cano et al., 2007) were included. The 29 HvCslF6 sequences reported in this article have been deposited in the EMBL/GenBank/DDBJ databases under accession numbers AB621305–AB621333. Multiple alignments of genomic sequences were performed using ClustalW with default options (Thompson et al., 1994). Phylogenetic analyses were conducted in MEGA v4.0 (Tamura et al., 2007) based on the bootstrap Neighbor–Joining (NJ) method (Saitou and Nei, 1987). Multiple alignments of amino acid sequences of seven HvCslF family genes of barley and CslF6 orthologues from five Poaceae species (wheat, rice, Brachypodium distachyon, sorghum, and oats) were also prepared.
Results
Genetic mapping of bgl
A previous study showed that bgl co-segregated with the HvCslF6 gene in the centromeric region of 7H using 227 F2 plants from the Ni×bgl-Ni cross (Tonooka et al., 2009). In this study, bgl was more precisely mapped using a more polymorphic mapping population of 104 F2 plants from the ‘Bowman’×OUM125 cross (see Supplementary Fig. S1 at JXB online). The Megazyme kit assay of half grains clearly distinguished bgl homozygous plants. The bgl gene perfectly co-segregated with HvCslF6, and HvCslF6 was assigned to the 7HL arm using wheat–barley chromosome addition lines. bgl–HvCslF6 was flanked by MWG511, 2.9 cM distal on the 7HS arm and Bmac0162, 1 cM distal on the 7HL arm. The centromere of chromosome 7H lies between MWG511 and HvCslF6. As Burton et al. (2008) reported, HvCslF6 showed a close linkage with CDO673 with a distance of 4.9 cM. The present mapping results again strongly indicate that bgl represents a mutation of HvCslF6.
Dosage effects of bgl
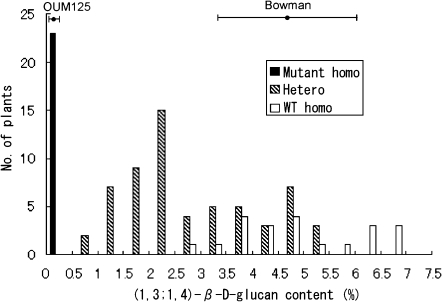
A frequency distribution of (1,3;1,4)-β-D-glucan content of the grain in the F2 mapping population is shown in Fig. 1. F2 plants were assigned to three classes according to their genotypes of HvCslF6, either mutant homozygous, heterozygous, or wild-type homozygous. The results show that heterozygous plants had intermediate levels of (1,3;1,4)-β-D-glucan content, being distributed between mutant homozygote class and wild-type homozygote class. Moreover, about half of the heterozygotes overlapped with the range of the wild-type homozygote (‘Bowman’) with a broad distribution. The wider distribution of the heterozygotes probably reflects a dosage effect from the triploid nature of the endosperm. For heterozygotes, two genotypes are expected; namely Bglbglbgl and BglBglbgl, probably the former showing a slightly lower (1,3;1,4)-β-D-glucan content and the latter showing a slightly higher level.
Fig. 1.
Frequency distribution of (1,3;1,4)-β-D-glucan content in 104 F2 plants from the cross between ‘Bowman’ and OUM125. F2 plants were classified into three classes (mutant homozygous, heterozygous, and wild-type homozygous) according to the genotypes of the HvCslF6 gene.
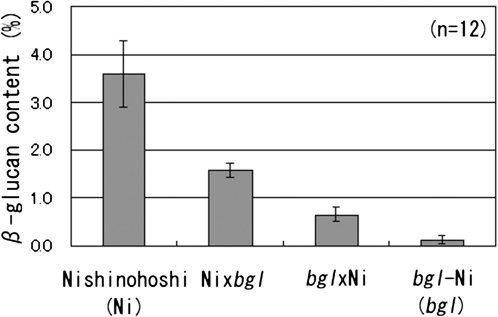
To confirm a dosage effects of the bgl gene, (1,3;1,4)-β-D-glucan content in grain obtained from reciprocal crosses was analysed. The results showed that two types of heterozygotes were intermediate between wild-type homozygotes (BglBglBgl) and mutant homozygotes (bglbglbgl) and that BglBglbgl is higher than Bglbglbgl (Fig. 2). Significant differences among four genotypic classes in (1,3;1,4)-β-D-glucan content were detected by t test.
Fig. 2.
(1,3;1,4)-β-D-Glucan content in the grains of wild-type ‘Nishinohoshi’ (Ni) and mutant bgl-Ni and their reciprocal F1 hybrids. Bars indicate standard deviations.
qRT-PCR analysis
Expression of the HvCslF6 gene was compared between Ni and bgl-Ni. Expression of the HvCslF6 gene was detected in all tissues studied, but the expression was highest in 10-d-old caryopsis and lowest in the 10-d-old leaf blade. There were no apparent differences between Ni and bgl-Ni (see Supplementary Fig. S2 at JXB online). Expression patterns generally confirmed the report by Burton et al. (2008).
Mutant screening and analysis
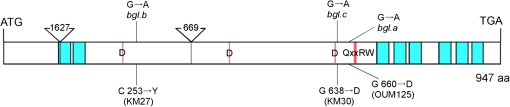
For screening of new (1,3;1,4)-β-D-glucanless mutants from sodium azide mutagenized M4 lines of cv. ‘Sachiho Golden’, chilling injury was used as a selection criterion. This is based on the observation that OUM125 and its derivatives with the bgl gene show chilling injury when sown in autumn and exposed to a cold climate in Tochigi, Japan (Fig. 3). It is likely that (1,3;1,4)-β-D-glucanless mutants are more chilling sensitive probably because thinner cell walls would provide weaker protection. Out of 11 chilling-susceptible lines, two were confirmed to be (1,3;1,4)-β-D-glucanless mutants by the Megazyme grain assay. The two new betaglucanless mutants (KM27 and KM30) were confirmed to be allelic to bgl of OUM125 by test-crossing as F1 hybrid grains showed a (1,3;1,4)-β-D-glucanless phenotype by the Megazyme assay method. Thus, allele names are assigned as follows: bgl.a for OUM125, bgl.b for KM27, and bgl.c for KM30. Analysis of the genomic sequences of the HvCslF6 gene determined that each of them carried a SNP within the coding sequence (CDS) region. Figure 4 summarizes the mutation points and alteration in predicted amino acid sequences. OUM125 had a G to A change at the 4275th position (1979th in CDS), which is predicted to cause an amino acid change at the 660th position from glycine (G) to aspartic acid (D). KM27 had a G to A change at the 2385th position (758th in CDS), which is predicted to cause an amino acid change at the 253th position from cysteine (C) to tyrosine (Y). KM30 had a G to A change at 4209th position (1913th in CDS), which is predicted to cause an amino acid change at the 638th position from G to D. Multiple alignments showed that the single amino acid substitutions in all three bgl mutants affected amino acids that are highly conserved among the seven HvCslF family proteins of barley (see Supplementary Fig. S3 at JXB online) and orthologous CslF6 proteins from five other Poaceae species (data not shown). A CAPS marker that distinguishes the mutation site of OUM125 was reported (Tonooka et al., 2009). Similarly, this study developed CAPS markers that specifically detect mutation sites of KM27 and KM30 (see Supplementary Table S1 at JXB online).
Fig. 3.
Plant phenotype after exposure to winter chilling in the field. Left is ‘Nishinohoshi’ (Ni), and right is bgl-Ni.
Fig. 4.
Structure of the HvCslF6 gene, which is predicted to encode 947 amino acid (aa) residues. Mutation points of three mutants (bgl.a, bgl.b, and bgl.c) are indicated. The positions of introns are indicated by the triangles and the lengths of the introns (in base pairs) are indicated within each triangle. The blue boxes show the positions of eight times of trans-membrane helices. The red bars indicate the positions of the D, D, D, QxxRW motifs.
(1,3;1,4)-β-D-glucan synthase activity during development of endosperms
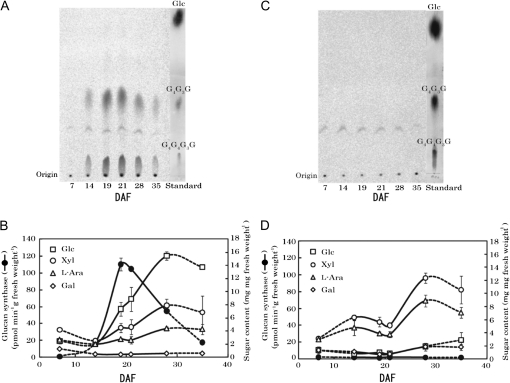
Enzyme activity and sugar composition of cell walls were measured for endosperms harvested at various developmental stages of SH84 and bgl-SH84 from 7–35 DAF, in order to examine the physiological role of the HvCslF6 gene in the synthesis of (1,3;1,4)-β-D-glucan in vivo. It appeared that enzyme activity (expressed based on a fresh weight of endosperms) of SH84 was low at 7 DAF. The activity then rapidly increased during the initial developmental stages, reached its maximal level around 19 DAF, and then fell with the maturation of the seeds (Fig. 5A, B). The activity at the maximal level was 110 pmol Glc transferred min−1 g−1 fresh weight. Similar activity profiles were observed when activities were expressed based on protein content of microsomal fractions (specific activity), although the maximal level shifted to from 19 DAF to 21 DAF: 230 and 260 pmol min−1 mg−1 protein at 19 DAF and 21 DAF, respectively. The major sugars detected in the endosperm cell walls were Glc, Xyl, and L-Ara together with a small amount of Gal throughout the development of endosperms (Fig. 5B). The content of Glc increased continuously during the development of the endosperms, and the initial deposition of (1,3;1,4)-β-D-glucan was accompanied by an increase in (1,3;1,4)-β-D-glucan synthase activity. The presence of (1,3;1,4)-β-D-glucan in SH84 endosperms was confirmed by incubation of the α-amylase-treated cell wall polysaccharides with lichenase, which resulted in a 94% reduction of the Glc content.
Fig. 5.
Changes of (1,3;1,4)-β-D-glucan synthase activity and the sugar composition of cell walls during development of endosperms of SH84 and bgl-SH84. (A, C) Enzyme activity was determined with microsomes prepared from endosperms of SH84 (A) or bgl-SH84 (C) at different developing stages for 7–35 d after flowering (DAF). The [14C]Glc transfer products were digested with lichenase, separated by paper chromatography, and analysed with a fluoroimage analyser. Note that the spots detected between tri- (G4G3G) and tetrasaccharides (G4G4G3G) are a contaminant contained in the commercial UDP-[14C]Glc specimen. (B, D) Enzyme activities are expressed based on the fresh weight of endosperms. The sugar composition of the cell walls was analysed as described in the Materials and methods. Endosperms at 7 and 35 DAF were tightly attached to pericarps. Hence, the values shown reflect the involvement of pericarps and are connected to other data by dotted lines. Data for activity and sugar composition are averages of duplicate or triplicate assays.
By contrast, bgl-SH84 showed extremely low levels of enzyme activity throughout the development of endosperms (Fig. 5C, D). Concurrently, the content of Glc in the endosperm cell walls of bgl-SH84 was much lower in comparison with SH84, indicating that no or little synthesis of (1,3;1,4)-β-D-glucan occurred (Fig. 5D). The amounts of Xyl and L-Ara increased to 1.7–2.0-fold at the late developmental stages when compared with those of SH84, suggesting that the deposition of arabinoxylan partly compensates for the lack of (1,3;1,4)-β-D-glucan in endosperm cell walls of bgl-SH84. However, the profiles of increases in the amounts of Xyl and L-Ara during development of endosperms were somewhat different from those of SH84. The reason for the temporal decreases of Xyl and L-Ara contents around 21 DAF is unclear. These results indicate that the HvCslF6 gene regulates the synthesis of (1,3;1,4)-β-D-glucans during the development of barely endosperms, and that the bgl.a mutation causes a loss of gene function. This conclusion was supported by another observation using endosperms prepared from KM27 (bgl.b) and ‘Sachiho Golden’ at 19 DAF. ‘Sachiho Golden’ showed high (1,3;1,4)-β-D-glucan synthase activity (126 pmol min−1 g−1 fresh weight), while that of KM27 was almost entirely depleted (1.5 pmol min−1 g−1 fresh weight).
Functional characterization of HvCslF6 genes in Nicotiana benthamiana
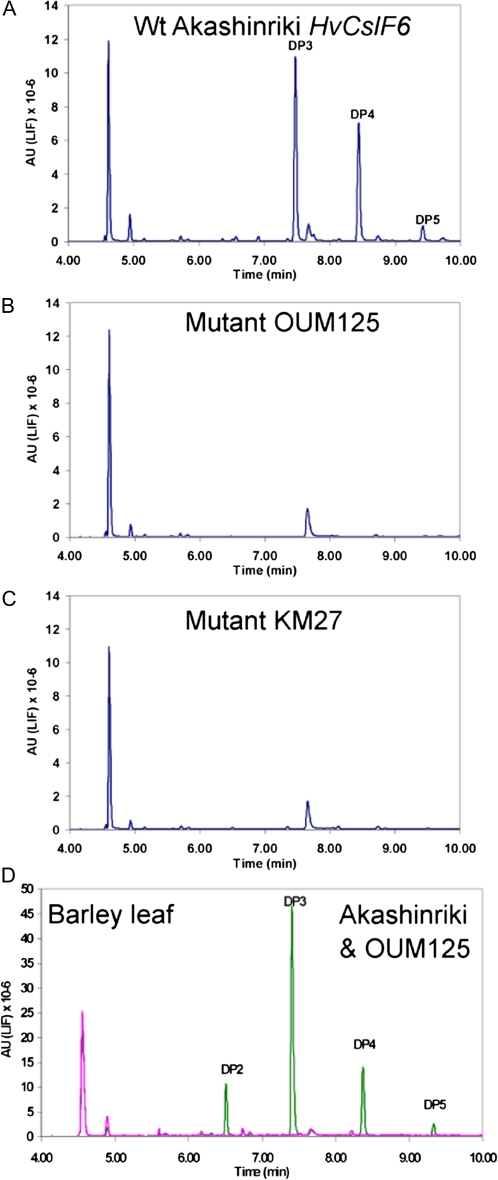
To determine if the changes in the amino acid sequence of the HvCslF6 protein of bgl mutants result in a loss of function, the ability of the genes to direct synthesis of (1,3;1,4)-β-D-glucan was tested in a N. benthamiana transient leaf expression system. Agrobacterium tumefaciens cultures containing binary vector plasmids driving expression of the wild-type HvCslF6, or mutant (OUM125 and KM27) HvCslF6 genes with the strong CaMV 35S promoter were infiltrated into N. benthamiana leaves. After 5 d, the presence of (1,3;1,4)-β-D-glucan was assayed by lichenase digestion of a crude cell wall preparation and detection of the released oligosaccharides by FACE (O'Shea et al., 1998). Lichenase specifically cleaves only (1,3;1,4)-β-D-glucan at a (1,4)-β-D-glucosidic linkage following a (1,3)-β-D-glucosidic linkage releasing oligosaccharides with a degree of polymerizsation (DP) of mainly DP3 and DP4 (G4G3G and G4G4G3G), respectively (Lazaridou and Biliaderis, 2007). The wild-type HvCslF6 gene directed the synthesis of appreciable quantities of (1,3;1,4)-β-D-glucan in the leaves as indicated by the large DP3 and DP4 peaks with lesser amounts of DP5 (Fig. 6A). The DP3/DP4 ratio was 1.35 compared with 2.6 from barley grain (Megazyme standard, data not shown). Very similar results have been obtained in more than four separate experiments and quantification using the Megayzme assay kit, routinely gave levels of (1,3;1,4)-β-D-glucan between 1.0% and 3.5% of the dry weight of the leaves. Both mutant HvCslF6 synthases, however, showed no evidence of synthesis of (1,3;1,4)-β-D-glucan in any of the experiments as indicated by the absence of the DP3 and DP4 peaks (Fig. 6B, C).
Fig. 6.
FACE analysis of oligosaccharides released from lichenase-digested cell walls. Wild-type ‘Akashinriki’ (A), mutant OUM125 (B), and KM27 (C) HvCSlF6 genes were transiently expressed in Nicotiana benthamiana leaves and oligosaccharides released from cell wall preparations after lichenase digestion were analysed by 8-amino-1,3,6-pyrenetrisulphonic acid (APTS) fluorescence labelling and separation by capillary electrophoresis. Lichenase digests of barley leaf cell walls from the wild type (‘Akashinriki’, green line) and mutant (OUM125, pink line) are shown for comparison in (D). The degree of polymerization (DP) of the oligosaccharides is indicated. The large peak at the beginning of the trace (4.5 min) and smaller peak at approximately 7.75 min are unlabelled APTS and a non-specific labelled product as they appear in minus lichenase controls (data not shown).
Characterization of (1,3;1,4)-β-D-glucan structure in wild-type and mutant barley leaves
Previously, analysis of the (1,3;1,4)-β-D-glucan composition of the bgl mutant was confined to grain tissues using the Megazyme assay kit which can only give information on the amount of (1,3;1,4)-β-D-glucan. The structure of (1,3;1,4)-β-D-glucan in leaf tissue was therefore analysed by FACE as described above. Tissue from the tips of mature leaves was chosen as the bgl mutant was morphologically different in this region. Compared with the wild type ‘Akashinriki’, the bgl.a mutant (OUM125) showed an obvious constriction below the leaf tip which also appeared distinctly chlorotic (see Supplementary Fig. S4 at JXB online). Lichenase digestion released the characteristic DP3 and DP4 as the major oligosaccharides from wild-type (1,3;1,4)-β-D-glucan in a ratio of 3.2 (Fig. 6D). A small amount of DP5 was also detected as was a significant amount of DP2. No oligosaccharides were detected in the bgl leaves confirming the complete absence of (1,3;1,4)-β-D-glucan in the mutant. A similar analysis of 10-d-old seedling tissues also showed the complete absence of (1,3;1,4)-β-D-glucan in the leaf tip, the leaf base, the stem (including the coleoptile) as well as in root tissues (see Supplementary Fig. S5 at JXB online).
In addition, the (1,3;1,4)-β-D-glucan content in leaves was analysed in three sets of field-grown plants using a Megazyme assay kit. (1,3;1,4)-β-D-Glucan was not detected in the leaves of OUM125, bgl-Ni or bgl-SH84, whereas those of ‘Akashinriki’, Ni, and SH84 contained 13.5–14.1 mg g−1 of (1,3;1,4)-β-D-(1,3;1,4)-β-D-glucan (see Supplementary Fig. S6 at JXB online).
Phylogenetic analysis of natural variation in the HvCslF6 gene
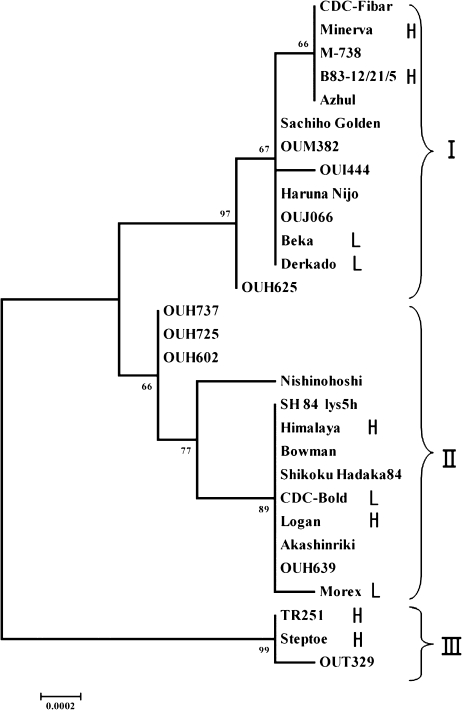
Sequencing of the HvCslF6 gene from 29 (five wild and 24 domesticated) barley accessions identified 30 polymorphisms (24 SNPs, five indels, and one SSR; see Supplementary Table S2 at JXB online). Twenty-seven polymorphisms were located within introns 1 and 2, and only three polymorphisms were localized within exons; only SNP23 produces a change in the translated protein [590th alanine (A) to threonine (T)], but this amino acid residue is not conserved among members of the HvCslF gene family (see Supplementary Fig. S3 at JXB online). An unrooted phylogenetic tree was constructed by using the Neighbor–Joining method (Fig. 7). Barley accessions were classified into three major groups (clades I, II, and III). Steptoe, OUT329, and TR251 were distinctly separated from other accessions (clade III). Three H. vulgare ssp. spontaneum accessions (OUH602, OUH725, and OUH737) formed a subclade sister to other members of the second major group because of an insertion of a miniature inverted-repeat transposable (MITE) in intron 2. The present study did not measure (1,3;1,4)-β-D-glucan content of the accessions used for phylogenetic analysis. However, limited information available from the literature suggests that there seems to be no apparent relationship between HvCslF6 polymorphisms and (1,3;1,4)-β-D-glucan contents in 29 accessions representing natural variation, because the two largest clades included both high and low (1,3;1,4)-β-D-glucan accessions. From haplotype analysis of approximately 40 barley accessions, Burton et al. (2010) identified just four SNPs in the HvCslF6 gene. This sharply contrasts with the present results where as many as 30 polymorphisms were detected in 29 barley accessions analysed. This discrepancy could be attributable to the wider range of germplasms used for natural variation analysis in the present study.
Fig. 7.
Phylogenetic analysis of the natural variation of the HvCslF6 gene in 29 barley accessions. The tree is generated through genomic sequences. The materials are grouped into three clades (I, II, and III). Numbers indicate bootstrap values. Accessions with (1,3;1,4)-β-D-glucan content information from the literature are denoted either by L (low content) or H (high content) after the cultivar name.
Discussion
A previous report revealed that the bgl.a mutant was a possible loss-of-function mutation of the HvCslF6 gene on the basis of cosegregation of the bgl phenotype (i.e. lack of (1,3;1,4)-β-D-glucan) and the mutant HvCslF6 allele (Tonooka et al., 2009). The lack of (1,3;1,4)-β-D-glucan was only assayed by the Megazyme assay kit, therefore, a more detailed biochemical and functional characterization of the mutant has now been performed for the unequivocal verification that a mutation in the HvCslF6 gene causes the loss of (1,3;1,4)-β-D-glucan from all the tissues tested. In the present study, two additional allelic mutants at the bgl locus were isolated (KM27 and KM30), and were confirmed to harbour a mutation within the coding region of the HvCslF6 gene, that results in the substitution of highly conserved amino acid residues. Isolation of three independent mutants at the HvCslF6 locus is strong evidence that bgl represents a loss-of-function mutation of HvCslF6. Here, more critical biochemical evidence for the absence of functionality of the mutant genes was provided on the basis of measurement of isolated (1,3;1,4)-β-D-glucan synthase enzyme activity and heterologous expression of the mutant and wild-type HvCslF6 alleles in N. benthamiana.
The bgl mutants lack the activity of (1,3;1,4)-β-D-glucan synthase and contain extremely low levels of (1,3;1,4)-β-D-glucan throughout endosperm development. The amount of (1,3;1,4)-β-D-glucan in developing endosperms of SH84 increased essentially linearly from 14 DAF to the late stages, as did the amount of arabinoxylan, consistent with the initial increase in the activity of (1,3;1,4)-β-D-glucan synthase. However, the deposition of (1,3;1,4)-β-D-glucan continued with the maturation of the seeds, even after the enzyme activity gradually decreased. This is also the case of our previous observation for Shikoku Hadaka 97 (Tsuchiya et al., 2005), and the reason is not yet clearly explained. It was also observed that the amount of arabinoxylan increased to about 2-fold in developing endosperms of bgl-SH84 when compared with its isogenic line, SH84, as observed similarly for the bgl-Ni and Ni pair (Tonooka et al., 2009). Based on the metabolic pathway of nucleotide sugars in plants (Bar-Peled and O'Neill, 2011), it is probable that the bgl mutation decreases the consumption of UDP-Glc, leading to alteration of the carbon flow to other pathways such as the UDP-Xyl and -L-Ara formation. This would increase the substrates available for the synthesis of arabinoxylan, resulting in the increased deposition of the polysaccharide in endosperm cell walls.
Previous heterologous expression studies characterizing the function of CslF and CslH genes from barley and rice have used stable transformation of Arabidopsis plants and have resulted in only very low levels of (1,3;1,4)-β-D-glucan (Burton et al., 2006; Doblin et al., 2009). By contrast, the N. benthamiana transient expression system used here results in significant levels of (1,3;1,4)-β-D-glucan synthesis, in the order of 1–3% dry wt of leaf tissue. One reason for this could be that the N. benthamiana transient expression system is well known to give high levels of expression of heterologous genes through the use of the viral suppressor protein P19 which shuts down the RNAi-mediated host response allowing high expression over prolonged periods compared with transgenic plants (Wood et al., 2009). Another possibility is that the HvCslF6 gene encodes a protein that is much more active than the other HvCslF and HvCslH genes that have been expressed so far. In fact, the HvCslF6 gene appears to encode the major (1,3;1,4)-β-D-glucan synthase as it is constitutively expressed at much higher levels than all the other HvCslF and HvCslH genes which generally show low expression only in specific tissues (Burton et al., 2008).
The hypothesis that the CslF6 protein is the core subunit of the (1,3;1,4)-β-D-glucan synthase in the structure model proposed by Buckeridge et al. (2004), is supported by the demonstration that loss-of-function mutations of the HvCslF6 gene cause the complete absence of (1,3;1,4)-β-D-glucan not only in grains but also in vegetative tissues. The structure of the (1,3;1,4)-β-D-glucan produced in the tobacco leaves by expression of the HvCslF6 gene is also different from that found in the barley grain as it has a much lower DP3/DP4 ratio (1.35 compared to 2.6 in barley grain and 3.6 in barley leaves) (Doblin et al., 2009). This agrees with the findings of Burton et al. (2011) where over-expression of the HvCslF6 gene in barley grain reduced the DP3/DP4 ratio and this also altered the solubility of the (1,3;1,4)-β-D-glucan. Heterologous expression of the HvCslH gene in Arabidopsis leaves produced a (1,3;1,4)-β-D-glucan with a DP3/DP4 ratio of 3.6 (Doblin et al., 2009) similar to that found in barley leaves (3.2, Fig. 6D). It is possible is that the HvCslF6 gene encodes the core synthase and the other HvCslF and HvCslH genes act like modifier genes to produce enzymes that produce (1,3;1,4)-β-D-glucan of different structures in different tissues depending on the properties of the cell walls needed in those tissues. These results suggest that the HvCslF6 gene encodes an enzyme that has unique properties that are not complemented by other HvCslF or HvCslH genes. In this respect, it is interesting to note that the HvCslF6 protein differs from all other HvCslF and HvCslH proteins in having an extended loop at about amino acid 535 (Burton et al., 2008) and that this may be the reason for the unique properties of the (1,3;1,4)-β-D-glucan synthase encoded by the HvCslF6 gene.
(1,3;1,4)-β-d-Glucan synthase activity is reported to reside in the Golgi membrane (Gibeaut and Carpita, 1993; Carpita and McCann, 2010), however, little is known about the structure of the (1,3;1,4)-β-D-glucan synthase other than that the CslF and CslH proteins are predicted to be integral membrane proteins and that there is a large central catalytic domain probably facing the cytoplasm. In the three bgl mutants identified here, the affected amino acid residues are located close to one of the D, D, D, QxxRW motifs within the central catalytic domain (Fig. 4). These motifs are believed to be involved in nucleotide sugar binding and catalytic activity of the enzyme (Charnock et al., 2001). Of these, two are substitutions of G to D (KM30 and OUM125) near the C-terminus of the central domain, whereas the other mutant (KM27) is a C to Y change near the N-terminal conserved D residue. Sequencing of the HvCslF6 gene from 29 barley accessions did not identify any changes in conserved amino acids indicating that the identified amino acid changes in the mutants are significant. Using the N. bethamiana expression system described here it should be relatively easily to use mutagenesis to determine other amino acids necessary for activity or even make changes that would alter the structure of the (1,3;1,4)-β-D-glucan.
Successful identification of two new (1,3;1,4)-β-D-glucanless mutants using chilling injury as a selection criterion indicates that bgl mutants generally show a cold-sensitive phenotype. Besides, all the three bgl mutants identified in this study showed a reduction in plant height, plant vigour and yield (approximately 70% of the control, data not shown). Interestingly the present study noted that bgl mutants show some morphological changes and chlorosis of the leaf tip which is a region where a high level of expression of HvCslH has been reported (Doblin et al., 2009) possibly indicating that a (1,3;1,4)-β-D-glucan of different structure was synthesized in the absence of HvCslF6. However, no trace of (1,3;1,4)-β-D-glucan was detected in this region of the leaf or in any other tissues of the bgl mutant tested. It may be possible that (1,3;1,4)-β-D-glucan is present in extremely small amounts in this or other tissues but is not extractable under the conditions used; further immunolabelling microscopic studies may resolve this issue. As the agronomic characteristics are reduced, the utility of the bgl mutants in malting may not be good even though they have favourable characteristics such as softer grains in addition to a (1,3;1,4)-β-D-glucanless phenotype, however, the present mutants are useful materials for basic research such as the enzymatic investigation of the biosynthesis of (1,3;1,4)-β-D-glucans.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Table S1. Primer sequences used in this study.
Supplementary Table S2. Allelic variation in the genomic sequences of the HvCslF6 gene.
Supplementary Fig. S1. Molecular mapping of the bgl and HvCslF6 genes on chromosome 7H using 104 F2 plants from a cross between ‘Bowman’ and OUM125.
Supplementary Fig. S2. Quantitative RT-PCR analysis of expression of the HvCslF6 gene in various tissues and developmental stages.
Supplementary Fig. S3. Alignments of seven barley HvCslF proteins and rice OsCslF6 protein.
Supplementary Fig. S4. Difference in appearance of leaf tip between wild-type barley (‘Akashinriki’) and HvCslF6 mutant OUM125.
Supplementary Fig. S5. FACE analysis of lichenase digests of cell wall extracts from various barley seedling tissues of ‘Akashinriki’ (shown in blue line) and OUM125 (shown in red line) demonstrating the absence DP3 and DP4 peaks characteristic of (1,3;1,4)-β-D-glucan in all tissues of the mutant.
Supplementary Fig. S6. Content of (1,3;1,4)-β-D-glucan in leaves of three sets of isogenic lines.
Acknowledgments
The authors are grateful to Dr WTB Thomas, Dr BG Rossnagel, Dr WG Legge, Dr JL Molina-Cano, and Dr T Yanagisawa for supplying seed samples of the barley breeding lines. Barley landraces and wild accessions were supplied by Dr K Sato under the support of National Bioresource Project–Barley, Japan. Ms Y Ito, Ms Y Yamashita, Mr Y Tsujino, Ms N Shigematsu, and Ms Robin Chapple are acknowledged for technical assistance. The authors also thank Dr K Yoshida and Dr S Kidou for their useful suggestions. The research was partly supported by a grant in aid (no. 21580007) from the Ministry of Education, Culture, Sports, Science, and Technology, and grants from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation Grant TRC1007, Development of crop production technology for all-year-round multi-utilization of paddy fields), and funding from the CSIRO Food Futures Flagship.
References
- Bar-Peled M, O'Neill MA. Plant nucleotide sugar formation, interconversion, and salvage by sugar recycling. Annual Review of Plant Biology. 2011;62:127–155. doi: 10.1146/annurev-arplant-042110-103918. [DOI] [PubMed] [Google Scholar]
- Bouveng HS, Lindberg B. Hydrolysis of methylated polysaccharides. Methods in Carbohydrate Chemistry. 1965;5:269–276. [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brennan CS, Cleary LJ. The potential use of (1→3,1→4)-β-D-glucans as functional food ingredients. Journal of Cereal Science. 2005;42:1–13. [Google Scholar]
- Buckeridge MS, Rayon C, Urbanowicz B, Tine MAS, Carpita NC. Mixed linkage (1→3),(1→4)-β-D-glucans of grasses. Cereal Chemistry. 2004;81:115–127. [Google Scholar]
- Burton RA, Collins HM, Kibble NAJ, et al. Over-expression of specific HvCslF cellulose synthase-like genes in transgenic barley increases the levels of cell wall (1,3;1,4)-β-D-glucans and alters their fine structure. Plant Biotechnology Journal. 2011;9:117–135. doi: 10.1111/j.1467-7652.2010.00532.x. [DOI] [PubMed] [Google Scholar]
- Burton RA, Jobling SA, Harvey AJ, Shirley NJ, Mather DE, Bacic A, Fincher GB. The genetics and transcriptional profiles of the cellulose synthase-like HvCslF gene family in barley. Plant Physiology. 2008;146:1821–1833. doi: 10.1104/pp.107.114694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton RA, Wilson SM, Hrmova M, Harvey AJ, Shirley NJ, Medhurst A, Stone BA, Newbigin EJ, Bacic A, Fincher GB. Cellulose synthase-like CslF genes mediate the synthsis of cell wall (1,3;1,4)-β-D-glucans. Science. 2006;311:1940–1942. doi: 10.1126/science.1122975. [DOI] [PubMed] [Google Scholar]
- Burton RA, Ma G, Baumann U, et al. A customized gene expression microarray reveals that the brittle stem phenotype fs2 of barley is attributable to a retroelement in the HvCesA4 cellulose synthase gene. Plant Physiology. 2010;153:1716–1728. doi: 10.1104/pp.110.158329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita NC, McCann MC. The maize mixed-linkage (1→3),(1→4)-β-D-glucan polysaccharide is synthesized at the Golgi membrane. Plant Physiology. 2010;153:1362–1371. doi: 10.1104/pp.110.156158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charnock SJ, Henrissat B, Davies GJ. Three-dimensional structures of UDP-sugar glycosyltransferases illuminate the biosynthesis of plant polysaccharides. Plant Physiology. 2001;125:527–531. doi: 10.1104/pp.125.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutu C, Brandle JE, Brown DCW, Brown K, Simmonds JA, Miki BLA, Hegedus DD. pORE: a modular binary vector series suited for both monocot and dicot plant transformation. Transgenic Research. 2007;16:771–781. doi: 10.1007/s11248-007-9066-2. [DOI] [PubMed] [Google Scholar]
- Doblin MS, Pettolino FA, Wilson SM, Campbell R, Burton RA, Fincher GB, Newbigin Ed. Bacic A. 2009. A barley cellulose synthase-like CSLH gene mediates (1,3;1,4)-β-D-glucan synthesis in transgenic Arabidopsis. Proceedings of the National Academy of Sciences, USA. 106:5996–6001. doi: 10.1073/pnas.0902019106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards K, Jhonstone C, Thompson C. A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Research. 1991;19:1349. doi: 10.1093/nar/19.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emebiri LC. EST-SSR markers derived from an elite barley cultivar (Hordeum vulgare L. ‘Morex’): polymorphism and genetic marker potential. Genome. 2009;52:665–676. doi: 10.1139/g09-040. [DOI] [PubMed] [Google Scholar]
- Fincher GB. Revolutionary times in our understanding of cell wall biosynthesis and remodeling in the grasses. Plant Physiology. 2009;149:27–37. doi: 10.1104/pp.108.130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Domon E, Doi Y. Grain and starch characteristics of the double recessive lines for amylase-free and high amylase gene in barley. Breeding Science. 1999;49:217–219. [Google Scholar]
- Gibeaut DM, Carpita NC. Synthesis of (1→3),(1→4)-β-D-glucan in the Golgi apparatus of maize colepotiles. Proceedings of the National Academy of Sciences, USA. 1993;91:3850–3854. doi: 10.1073/pnas.90.9.3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han F, Ullrich SE, Chirat S, et al. Mapping of ß-glucan content and ß-glucanase activity loci in barley grain and malt. Theoretical and Applied Genetics. 1995;91:921–927. doi: 10.1007/BF00223901. [DOI] [PubMed] [Google Scholar]
- Heun M, Kennedy AE, Anderson JA, Lapitan NLV, Sorrells ME, Tanksley SD. Construction of a restriction fragment length polymorphism map for barley (Hordeum vulgare) Genome. 1991;34:437–447. [Google Scholar]
- Ishikawa M, Kuroyama H, Takeuchi Y, Tsumuraya Y. Characterization of pectin methyltransferase from soybean hypocotyls. Planta. 2000;210:782–791. doi: 10.1007/s004250050680. [DOI] [PubMed] [Google Scholar]
- Islam AKMR. Ditelosomic additions of barley chromosomes to wheat. In: Sakamoto S, editor. Proceedings of the 6th International Wheat Genetics Symposium. Kyoto: Maruzen; 1983. pp. 233–238. [Google Scholar]
- Islam AKMR, Shepherd KW, Sparrow DHB. Isolation and characterization of euplasmic wheat–barley chromosome addition lines. Heredity. 1981;46:161–174. [Google Scholar]
- Kato T, Sasaki A, Takeda G. Genetic variation of β-glucan contents and β-glucanase activities in barley, and their relationship to malting quality. Breeding Science. 1995;45:471–477. [Google Scholar]
- Kikuchi S, Taketa S, Ichii M, Kawasaki S. Efficient fine mapping of the naked caryopsis gene (nud) by HEGS (High efficiency genome scanning)/AFLP in barley. Theoretical and Applied Genetics. 2003;108:73–78. doi: 10.1007/s00122-003-1413-y. [DOI] [PubMed] [Google Scholar]
- Künzel G, Korzun L, Meister A. Cytologically integrated physical restriction fragment length polymorphism maps for the barley genome based on translocation breakpoints. Genetics. 2000;154:397–412. doi: 10.1093/genetics/154.1.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridou A, Biliaderis CG. Molecular aspects of cereal beta-glucan functionality: physical properties, technological applications and physiological effects. Journal of Cereal Science. 2007;46:101–118. [Google Scholar]
- Li J, Baga M, Rossnagel BG, Legge WG, Chibbar RN. Identification of quantitative trait loci for β-glucan concentration in barley grain. Journal of Cereal Science. 2008;48:647–655. [Google Scholar]
- McCleary BV, Codd R. Measurement of (1→3),(1→4)-β-D-glucan in barley and oats: a streamlined enzymatic procedure. Journal of Science, Food and Agriculture. 1991;55:303–312. [Google Scholar]
- Meyer RC, Swanston JS, Young GR, et al. Genetic approaches to improving distilling quality in barley. Barley Genetics. 2000;8:107–114. [Google Scholar]
- Molina-Cano JL, Moralejo M, Elia M, Munoz P, Russell JR, Perez-Vendrell AM, Ciudad F, Swanston JS. QTL analysis of a cross between European and North American malting barleys reveals a putative candidate gene for β-glucan content on chromosome 1H. Molecular Breeding. 2007;19:275–284. [Google Scholar]
- Munck L, Møller B, Jacobsen S, Søndergaard Near infrared spectra indicate specific mutant endosperm genes and reveal a new mechanism for substituting starch with (1→3, 1→4)-β-D-glucan in barley. Journal of Cereal Science. 2004;40:213–222. [Google Scholar]
- Nemeth C, Freeman J, Jones HD, et al. Down regulation of the CSLF6 gene results in decreased (1,3;1,4)-β-D-glucan in endosperm of wheat. Plant Physiology. 2010;152:1209–1218. doi: 10.1104/pp.109.151712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shea MG, Samuel MS, Konik CM, Morell MK. Fluorophore-assisted carbohydrate electrophoresis (FACE) of oligosaccharides: efficiency of labelling and high-resolution separation. Carbohydrate Research. 1998;307:1–12. [Google Scholar]
- Ramsay L, Macauley M, Ivanissevich DS, et al. A simple sequence repeat-based linkage map of barley. Genetics. 2000;156:1997–2005. doi: 10.1093/genetics/156.4.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou N, Nei M. The Neighbor–Joining method: a new method for reconstructing phylogenetic trees. Molecular Biology and Evolution. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Taketa S, Amano S, Tsujino Y, et al. Barley grain with adhering hulls is controlled by an ERF family transcription factor gene regulating a lipid biosynthesis pathway. Proceedings of the National Academy of Sciences, USA. 2008;105:4062–4067. doi: 10.1073/pnas.0711034105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonooka T, Aoki E, Yoshioka T, Taketa S. A novel mutant gene for (1,3;1,4)-β-D-glucanless grain on barley (Hordeum vulgare L.) chromosome 7H. Breeding Science. 2009;59:47–54. [Google Scholar]
- Trevelyan WE, Procter DP, Harrison JS. Detection of sugars on paper chromatograms. Nature. 1950;166:444–445. doi: 10.1038/166444b0. [DOI] [PubMed] [Google Scholar]
- Tsuchiya K, Urahara T, Konishi T, Kotake T, Tohno-oka T, Komae K, Kawada N, Tsumuraya Y. Biosynthesis of (1→3),(1→4)-β-glucan in developing endosperms of barley (Hordeum vulgare L. Physiologia Plantarum. 2005;125:181–191. [Google Scholar]
- Urahara T, Tsuchiya K, Kotake T, Tohno-oka T, Komae K, Kawada N, Tsumuraya Y. A β-(1→4)-xylosyltransferase involved in the synthesis of arabinoxylans in developing barley endosperms. Physiologia Plantarum. 2004;122:169–180. [Google Scholar]
- Varshney RK, Marcel TC, Ramsay L, Russel J, Röder MS, Stein N, Waugh R, Langridge P, Niks RE, Graner A. A high density barley microsatellite consensus map with 775 SSR loci. Theoretical and Applied Genetics. 2007;114:1091–1103. doi: 10.1007/s00122-007-0503-7. [DOI] [PubMed] [Google Scholar]
- von Wettstein D. From analysis of mutants to genetic engineering. Annual Review of Plant Biology. 2007;58:1–19. doi: 10.1146/annurev.arplant.58.032806.104003. [DOI] [PubMed] [Google Scholar]
- Yuo T, Toyota M, Ichii M, Taketa S. Molecular cloning of a root hairless gene rth1 in rice. Breeding Science. 2009;59:13–20. [Google Scholar]
- Wood CC, Petrie JR, Shrestha P, Mansour MP, Nichols PD, Green AG, Singh SP. A leaf-based assay using interchangeable design principles to rapidly assemble multistep recombinant pathways. Plant Biotechnology Journal. 2009;7:914–924. doi: 10.1111/j.1467-7652.2009.00453.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.