Abstract
COG0354 proteins have been implicated in synthesis or repair of iron/sulfur (Fe/S) clusters in all domains of life, and those of bacteria, animals, and protists have been shown to require a tetrahydrofolate to function. Two COG0354 proteins were identified in Arabidopsis and many other plants, one (At4g12130) related to those of α-proteobacteria and predicted to be mitochondrial, the other (At1g60990) related to those of cyanobacteria and predicted to be plastidial. Grasses and poplar appear to lack the latter. The predicted subcellular locations of the Arabidopsis proteins were validated by in vitro import assays with purified pea organelles and by targeting assays in Arabidopsis and tobacco protoplasts using green fluorescent protein fusions. The At4g12130 protein was shown to be expressed mainly in flowers, siliques, and seeds, whereas the At1g60990 protein was expressed mainly in young leaves. The folate dependence of both Arabidopsis proteins was established by functional complementation of an Escherichia coli COG0354 (ygfZ) deletant; both plant genes restored in vivo activity of the Fe/S enzyme MiaB but restoration was abrogated when folates were eliminated by deleting folP. Insertional inactivation of At4g12130 was embryo lethal; this phenotype was reversed by genetic complementation of the mutant. These data establish that COG0354 proteins have a folate-dependent function in mitochondria and plastids, and that the mitochondrial protein is essential. That plants retain mitochondrial and plastidial COG0354 proteins with distinct phylogenetic origins emphasizes how deeply the extant Fe/S cluster assembly machinery still reflects the ancient endosymbioses that gave rise to plants.
Keywords: COG0354, folate, iron–sulfur cluster, mitochondrion, plastid
Introduction
Iron/sulfur (Fe/S) clusters are highly versatile but labile cofactors of ancient origin that occur in all domains of life (Beinert, 2000). Fe/S cluster proteins play central roles in electron transfer, catalysis, and gene expression (Johnson et al., 2005). Plants have over a hundred such proteins, located in mitochondria, plastids, cytosol, and nuclei (Balk and Pilon, 2011).
Although Fe/S clusters are simple structures, their biogenesis and maintenance require a complex machinery. The grand lines of this machinery are now clear and are similar in bacteria and eukaryotes (Johnson et al., 2005; Lill, 2009). Thus, Fe/S cluster assembly begins with a cysteine desulfarase that transfers sulfur from free cysteine to a scaffold protein, which also receives Fe from an Fe-binding protein. The scaffold protein acts as a platform on which clusters are assembled and from which they are mobilized to target apoproteins, often with the help of chaperones. Fe/S cluster biogenesis systems are of several types. Bacteria such as Escherichia coli have two independent systems, ISC and SUF. Plants also have ISC and SUF systems, ISC being in mitochondria and SUF in plastids; these organelles thus have autonomous Fe/S assembly machinery that reflects their endosymbiotic origins (Xu and Møller, 2008; Balk and Pilon, 2011). Plants have in addition a cytosolic system, CIA, which depends in part on the mitochondrial ISC system (Balk and Pilon, 2011). Other eukaryotes have mitochondrial ISC and cytosolic CIA systems (Lill, 2009).
Despite the progress in dissecting Fe/S cluster biogenesis, much remains unclear, including the roles of proteins involved in the maturation of specific subsets of Fe/S enzymes (Sheftel et al., 2010). One such protein is COG0354, which occurs in all domains of life and is known to be mitochondrial in yeast and animals (Gelling et al., 2008; Pagliarini et al., 2008). Yeast COG0354 (Iba57p) is required for Fe/S cluster formation on aconitase and activation of two radical SAM (S-adenosylmethionine) Fe/S enzymes (Gelling et al., 2008). E. coli COG0354 (YgfZ) supports succinate dehydrogenase, fumarase, dimethylsulphoxide reductase, and the radical SAM enzyme MiaB, particularly during oxidative stress (Ote et al., 2006; Waller et al., 2010). These functions are crucial at the organism level: deleting iba57 causes loss of mitochondrial function and a petite phenotype (Gelling et al., 2008); deleting ygfZ impairs growth and increases oxidative stress sensitivity (Lin et al., 2010; Waller et al., 2010); and knockdown of zebrafish COG0354 (C1orf69) causes anaemia (Nilsson et al., 2009).
The biochemical action of COG0354 is not yet understood, but it appears to be universally conserved because the ΔygfZ growth defects are complemented by COG0354 genes from all domains of life, including plants (Waller et al., 2010), and the Δiba57 growth defects are complemented by human C1orf69 (Gelling et al., 2008). Major clues to the action of COG0354 have been the prediction of a folate-binding site on the E. coli protein (Teplyakov et al., 2004), the finding that this protein indeed binds a model folate (5-formyltetrahydrofolate) in vitro (Waller et al., 2010), and the genetic demonstration that E. coli, mammalian, and protistan COG0354 proteins depend on a tetrahydrofolate to function in vivo (Waller et al., 2010).
Nothing was known until now about plant COG0354 proteins except that there are two types, and that both types from Arabidopsis complement the E. coli ΔygfZ mutation (Waller et al., 2010). Here, it is shown that one plant COG0354 protein is mitochondrial and that the other is plastidial, that they are respectively predicted to derive from α-proteobacteria and cyanobacteria, that they both require folate for in vivo activity, and that the mitochondrial protein is essential for embryo development.
Materials and methods
Bioinformatics
COG0354 protein sequences (identified by the KGC-[F/Y]-x-GQE-x(3)-[K/R] motif) were taken from NCBI (http://www.ncbi.nlm.nih.gov/) and SEED (http://www.theseed.org/) databases. Plant genomes and expresed sequence tags (ESTs) were searched for COG0354 sequences using BLAST algorithms at the NCBI and Joint Genome Institute (http://genome.jgi-psf.org/) databases. Sequences were aligned using Multalin (http://multalin.toulouse.inra.fr/multalin/) or ClustalW; phylogenetic analyses were carried out with MEGA version 4.0 (Tamura et al., 2007). Organellar targeting was predicted using TargetP (http://www.cbs.dtu.dk/services/TargetP/), WoLF PSORT (http://wolfpsort.org/), and Predotar (http://urgi.versailles.inra.fr/predotar/predotar.html). Ambiguous (mitochondrial/plastidial) targeting was evaluated using the COSMOSS Ambiguous Targeting Predictor (http://www.cosmoss.org/bm/ATP).
Plant materials and growth conditions
Arabidopsis thaliana (ecotype Col-0) seeds were sterilized, imbibed at 4 °C for 5 d, and germinated on 0.5× MS salts containing 0.5% (w/v) sucrose. Seedlings were transferred after 14 d either into potting soil and supplemented biweekly with Peters Professional 20:20:20 fertilizer, or (for protein expression analysis) to hydroponic culture (Gibeaut et al., 1997). Photosynthetic photon flux density was 150 μmol quanta m−2 s−1 with a 16 h photoperiod (22 °C day, 18 °C night). Seeds of pea (Pisum sativum cv. Laxton's Improved Progress 9) were imbibed overnight in running tap water, sown in moist vermiculite, and grown at 16–18 °C under fluorescent light (75 μmol quanta m−2 s−1) with a 12 h photoperiod for 11 d.
In vitro organellar import assays
For dual import assays (Rudhe et al., 2002), cDNAs encoding full-length At4g12130 and At1g60990 were PCR amplified using primers At4g12130-pGEM-fwd and -rev, and At1g60990-pGEM-fwd and -rev, respectively (Supplementary Table S1 available at JXB online); forward primers included a Kozak sequence. Amplicons were cloned into pGEM-4Z (Promega). Coupled in vitro transcription–translation, isolation of pea chloroplasts and mitochondria, dual import assays, thermolysin treatment, and repurification of organelles were performed as described (Pribat et al., 2010). Soybean alternative oxidase (Rudhe et al., 2002) and pine phenylalanine hydroxylase (Pribat et al., 2010) served as positive controls for mitochondrial and chloroplast targeting, respectively.
Transient expression of GFP fusion proteins
The N-terminal 126 residues of At4g12130 or 122 residues At1g60990 were fused, via a tetraglycine linker, upstream and in-frame with green fluorescent protein (GFP) as follows. The first 378 bp of At4g12130 or 366 bp of At1g60990 were amplified from cDNAs (ABRC stock no. U17502 and Riken stock no. RAFL21-91-J21, respectively) using primers At4g12130-GFP-fwd and -rev, and At1g60990-GFP-fwd and -rev, respectively (Supplementary Table S1 at JXB online). Amplicons were digested with appropriate restriction enzymes and ligated into pTH2 (Niwa, 2003). Protoplasts were prepared from suspension culture cells of Arabidopsis or tobacco (Nicotiana tabacum) cv. BY-2, transformed, and stained with MitoTracker Orange (Molecular Probes) as described (Ravanel et al., 2001). Controls for GFP targeting and epifluorescence microscopy were as described (Pinon et al., 2005). BY-2 protoplasts lack chlorophyll, which enables co-localization by epifluorescence microscopy of signals from GFP and the MitoTracker mitochondrial marker. Mitochondrial localization was therefore tested in BY-2 cells, and chloroplastic localization in Arabidopsis cells.
Protein expression analyses
Roots and stems were harvested from 6-week-old hydroponically cultured plants; other organs (except seedlings) were from soil-grown plants. Tissues were ground in liquid N2. For immunoblot analysis, 100 mg tissue samples were extracted in 0.1 M TRIS-HCl (pH 7.5), 0.2 M KCl, 3 mM MgCl2, 1 mM EDTA, 1 mM EGTA, 0.05% (v/v) Nonidet P-40, 0.1% (w/v) polyvinylpyrrolidone, 2% (w/v) polyvinylpolypyrrolidone, 1 mM dithiothreitol, 1 mM benzamidine, 1 mM phenylmethylsulphonyl fluoride, 5 mM ϵ-aminocaproic acid, and centrifuged to clear. Protein was estimated by dye binding (Bradford, 1976) with bovine serum albumin as standard. Denaturing gel electrophoresis and immunoblotting were as described (Turner et al., 2005); antisera were diluted 1:1000. Antisera were raised in rabbits (Cocalico Biologicals Inc.) against hexahistidine-tagged, denatured At4g12130 or At1g60990 prepared as described (Waller et al., 2010).
Folate dependence tests
The ΔygfZ and ΔfolP::kanr ΔygfZ E. coli strains (K12 MG1655 background) were as described (Waller et al., 2010). Both harboured pACYC-RP (Stratagene), which carries genes for rare arginine and proline tRNAs. Each of these strains was transformed with pBAD24 containing the truncated At4g12130 or At1g60990 cDNAs described previously (Waller et al., 2010). Cells for tRNA analysis were cultured at 37 °C in Antibiotic Medium 3 (Difco) plus 300 μM thymidine. Antibiotic concentrations (μg ml−1) were: ampicillin 25; chloramphenicol 10; and kanamycin 25. L-Arabinose 0.02% (w/v) was added to induce gene expression. Bulk nucleic acids were isolated from stationary phase cells and enriched for tRNA (Bailly et al., 2008) before Nucleobond AXR 400 column chromatography purification (Macherey-Nagel, Germany). The purified tRNA was then hydrolysed and analysed by liquid chromatography–tandem mass spectrometry (LC-MS/MS) as described (Phillips et al., 2008).
Knockout mutant of At4g12130 and complementation
A T-DNA mutant for At4g12130 (Arabidopsis Biological Resource Center stock no. SAIL_646_F03) was identified in the GABI-Kat collection (Rosso et al., 2003). Heterozygous and wild-type segregants were identified by PCR using At4g12130 gene-specific primers up- and downstream from the insertion site, At4g12130-GS-LP and -RP, and a primer in the left border of the insert, SAIL-LB2 (Supplementary Table S1 at JXB online). The insertion site was confirmed by amplicon sequencing. In the progeny of a backcross of a heterozygote to the wild type, the BASTA resistance marker was shown to co-segregate with the At4g12130 mutation, indicating the absence of inserts at other loci. BASTA resistance was scored on plates containing 0.5× MS salts, 0.5% (w/v) sucrose, and 10 μg ml−1 BASTA. For complementation, a full At4g12130 genomic clone with 2000 bp of upstream promoter sequence and flanking NotI sites was amplified using primers At4g12130-COM-fwd and -rev (Supplementary Table S1), cut with NotI, and cloned into NotI-digested pART27 (Gleave, 1992). The sequence-verified construct was introduced into Arabidopsis by pMP90::Agrobacterium tumefaciens-mediated transformation of floral tissue (Clough and Bent, 1998). Transgenic lines were selected on kanamycin (50 μg ml−1).
Results
Molecular characterization and phylogenetic analysis
As noted above, the Arabidopsis genome encodes two COG0354 proteins, At4g12130 and At1g60990, either of which can functionally replace E. coli COG0354 (Waller et al., 2010). Both Arabidopsis proteins have N-terminal extensions relative to their bacterial homologues (Fig. 1A); these extensions are predicted to target At4g12130 to mitochondria and At1g60990 to plastids. Both Arabidopsis proteins have orthologues in many land plants, as judged by TBLASTN searches of genome and EST databases, so that Arabidopsis is a representative model to study COG0354 in plants.
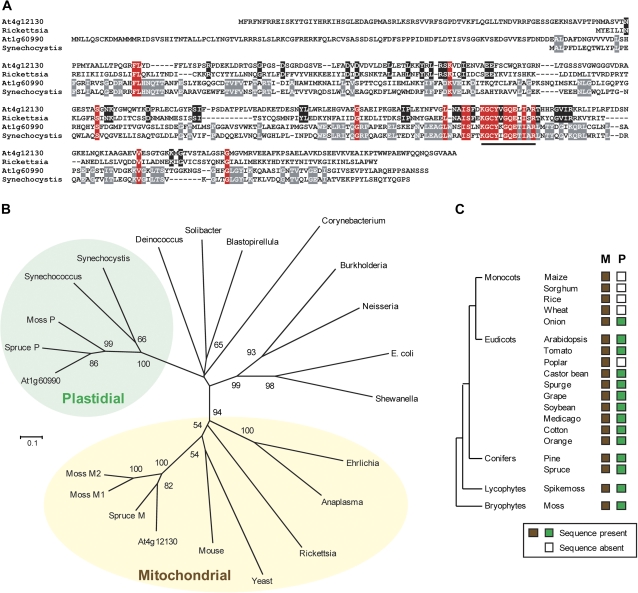
Fig. 1.
Primary structure, phylogeny, and distribution of plant COG0354 proteins. (A) Amino acid sequence alignment of At4g12130, At1g60990, and a close bacterial homologue of each (Rickettsia prowazekii and Synechocystis sp. PCC 6803, respectively). Residues conserved in all four sequences are in red; those conserved only in At4g12130 and Rickettsia are in black, and those conserved only in At1g60990 and Synechocystis are in grey. Dashes are gaps to maximize alignment. The COG0354 signature motif (KGC-[YF]-x-GQE-x(3)-[KR]) is red underlined; note that this motif is slightly longer than that defined by Teplyakov et al. (2004). (B) Neighbor–joining phylogenetic tree of COG0354 proteins from diverse plants and bacteria; yeast Iba57p and mouse C1orf69 are also included. Bootstrap values (1000 replicates) are next to branches; values <50% are not shown. Branch length indicates the number of inferred amino acid changes per position. At4g12130, other plant proteins predicted to be mitochondrial (M), and yeast and mouse sequences fall into the same clade (‘Mitochondrial’) as Rickettsia and other Rickettsiales (Anaplasma, Erlichia). At1g60990 and other plant proteins predicted to be plastidial (P) fall into the same clade (‘Plastidial’) as cyanobacteria. (C) Occurrence of orthologues of At4g12130 (M) and At1g60990 (P) in genomes and/or EST collections of land plants. Full species names: maize, Zea mays; sorghum, Sorghum bicolor; rice, Oryza sativa; wheat, Triticum aestivum; onion, Allium cepa; tomato, Solanum lycopersicum; poplar, Populus trichocarpa; castor bean, Ricinus communis; spurge, Euphorbia esula; grape, Vitis vinifera; soybean, Glycine max; medicago, Medicago truncatula; cotton, Gossypium hirsutum; orange, Citrus sinensis; pine, Pinus taeda; spruce, Picea glauca; spikemoss, Selaginella moellendorfii; moss, Physcomitrella patens.
Phylogenetic analysis of COG0354 proteins from plants, yeast, mouse, and diverse bacteria showed that At4g12130 and its plant orthologues fall into the same clade as Rickettsia and other α-proteobacteria, which are the closest living relatives of the mitochondrial progenitor (Williams et al., 2007). This clade also contained the yeast and mouse COG0354 proteins, which have both been shown to be mitochondrial (Gelling et al., 2008; Pagliarini et al., 2008) (Fig. 1B). Conversely, At1g60990 and its counterparts in other plants are in the same clade as proteins from cyanobacteria, the group that gave rise to plastids (Gould et al., 2008) (Fig. 1B). These clades are referred to here as ‘mitochondrial’ and ‘plastidial’, respectively (Fig. 1B). Alignment of the sequences of the Arabidopsis proteins with their closest bacterial homologues (Fig. 1A) underscores the marked divergence between mitochondrial- and plastidial-type proteins. While there is substantial overall sequence identity within each type, there is little identity between the types outside the COG0354 signature motif KGC-[YF]-x-GQE-x(3)-R.
Although many plant genomes encode both COG0354 proteins, some seem to lack the plastidial type (Fig. 1C). Those without this type include poplar, and all grasses for which genome sequences and/or deep EST collections exist. EST evidence indicates that a plastidial-type protein occurs in other monocots, for example onion (Fig. 1C).
Subcellular localization of Arabidopsis COG0354 proteins
The subcellular location of the two Arabidopsis COG0354 proteins was investigated first using dual import assays, in which radiolabelled full-length translation products are incubated with a mixture of purified chloroplasts and mitochondria (Rudhe et al., 2002). In reactions with At4g12130, mitochondria contained a labelled product that was smaller than the full-length precursor and resistant to attack by thermolysin, as expected for a translocated protein, but no thermolysin-resistant protein was present in chloroplasts (Fig. 2A). Conversely, in reactions with At1g60990, chloroplasts contained a smaller, thermolysin-resistant product but mitochondria did not (Fig. 2A). To complement this approach, subcellular localization was analysed in tobacco BY-2 protoplasts and Arabidopsis protoplasts using fusions between the N-terminus of each COG0354 protein and GFP. Expression of the At4g12130::GFP fusion in BY-2 cells resulted in a punctate pattern of fluorescence matching that of the MitoTracker mitochondrial marker (Fig. 2B). In contrast, the At1g60990::GFP fusion expressed in Arabidopsis cells gave larger punctate fluorescent areas that coincided with chlorophyll autofluorescence (Fig. 2B). Thus both in vitro and in vivo approaches indicate that, as predicted, the At4g12130 protein is targeted to mitochondria and the At1g60990 protein is targeted to chloroplasts. High throughput proteomic data also support a chloroplastic location for At1g60990 (Olinares et al., 2010).
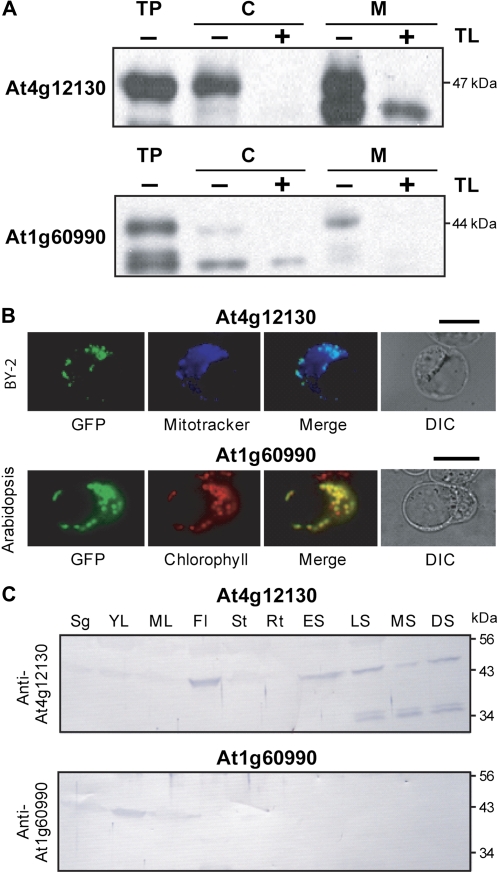
Fig. 2.
Organellar targeting and expression of Arabidopsis COG0354 proteins. (A) Protein import into isolated pea chloroplasts and mitochondria. Full-length At4g12130 and At1g60990 sequences were translated in vitro in the presence of [3H]leucine. The translation products were incubated for 20 min (At4g12130) or 5 min (At1g60990) in the light with mixed chloroplasts (C) and mitochondria (M), which were then separated on an 8% (v/v) Percoll gradient, without (–) or with (+) prior thermolysin (TL) treatment to remove adsorbed proteins. Proteins were separated by SDS–PAGE and visualized by fluorography. Samples were loaded on the basis of equal chlorophyll or mitochondrial protein content next to aliquots of the respective translation products (TP). The molecular masses of the full-length proteins are indicated. (B) Top panels: transient expression in tobacco BY-2 protoplasts of GFP fused to the predicted targeting sequence of At4g12130. Bottom panels: transient expression in Arabidopsis protoplasts of GFP fused to the predicted targeting sequence of At1g60990. GFP (green pseudo-colour), MitoTracker (blue pseudo-colour), and chlorophyll (red pseudo-colour) fluorescence were observed by epifluorescence microscopy. Cells were observed by differential interference contrast (DIC) microscopy. Scale bars=20 μM. (C) Immunoblot analysis of At4g12130 and At1g60990 expression in selected Arabidopsis organs. Lanes contained 15 μg of protein; blots were probed with antiserum to At4g12130 or At1g60990, or with pre-immune serum. Pre-immune serum gave no signals. Samples were from 14-day-old whole seedlings (Sg), roots from hydroponic culture (Rt), rosette leaves from plants at 16 d (YL) and 30 d (ML) old, stems (St), flowers (Fl), siliques at early (ES), mid (MS), and late (LS, yellowing) stages of development, and dry seeds (DS).
Expression patterns of At4g12130 and At1g60990
At4g12130 and At1g60990 protein levels were analysed by immunoblotting using rabbit antisera (Fig. 2C). At4g12130 protein was most abundant in flowers, siliques, and dry seeds, with cleavage products detectable in the latter samples. At1g60990 protein was most abundant in young leaves. These results agree broadly with transcriptome data, which show that At4g12130 mRNA is most strongly expressed in flowers, pollen, and seeds, whereas At1g60990 mRNA is expressed mainly in green tissues (Steinhauser et al., 2004; Xiang et al., 2011).
Evidence for folate dependence
The folate requirement of COG0354 can be established using a heterologous E. coli system in which the Fe/S enzyme MiaB serves as an in vivo reporter of COG0354 activity (Waller et al., 2010). MiaB mediates the conversion of N6-isopentenyladenosine (i6A) in tRNA to 2-methylthio-N6-isopentenyladenosine (ms2i6A); its in vivo activity can thus be gauged from the ms2i6A/i6A ratio in tRNA. As E. coli MiaB activity depends strongly on the action of COG0354, the ms2i6A/i6A ratio is a sensitive measure of COG0354 function (Fig. 3A). This ratio is >10 in wild-type E. coli cells, but can drop to <1 when the gene encoding COG0354 (ygfZ) is deleted (Ote et al., 2006; Waller et al., 2010).
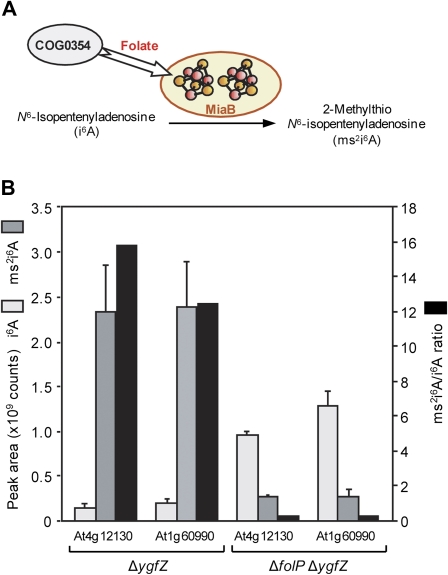
Fig. 3.
Evidence that At4g12130 and At1g60990 require folate for activity in vivo. (A) The reaction catalysed by the tRNA modification enzyme MiaB, and the relationship between MiaB, the COG0354 protein, and folate. MiaB has two [4Fe–4S] clusters (Hernández et al., 2007), which are depicted schematically. (B) LC-MS/MS quantification of i6A and ms2i6A, and the ms2i6A/i6A ratio, in tRNA of the ΔygfZ or ΔfolP ΔygfZ strains expressing At4g12130 or At1g60990, grown in Antibiotic Medium 3 plus 300 μM thymidine, 0.02% L-arabinose, and appropriate antibiotics. Data are means and SE for three independent samples.
To test folate dependence of the Arabidopsis COG0354 proteins, each was expressed from a plasmid in an E.coli ygfZ deletant strain (ΔygfZ). As expected from the observation that At4g12130 and At1g60990 both complement the growth phenotypes of this strain (Waller et al., 2010), they both gave high ms2i6A/i6A ratios in the wild-type range (Fig. 3B). However, when the folP gene (encoding the folate synthesis enzyme dihydropteroate synthase) was also deleted, the ratios were <1 (Fig. 3B). Since ΔfolP strains completely lack folates (Waller et al., 2010), this result indicates that the action of both Arabidopsis COG0354 proteins requires folate, as has been shown for animal and protist COG0354 proteins (Waller et al., 2010).
An insertional mutant of At4g12130 and its complementation
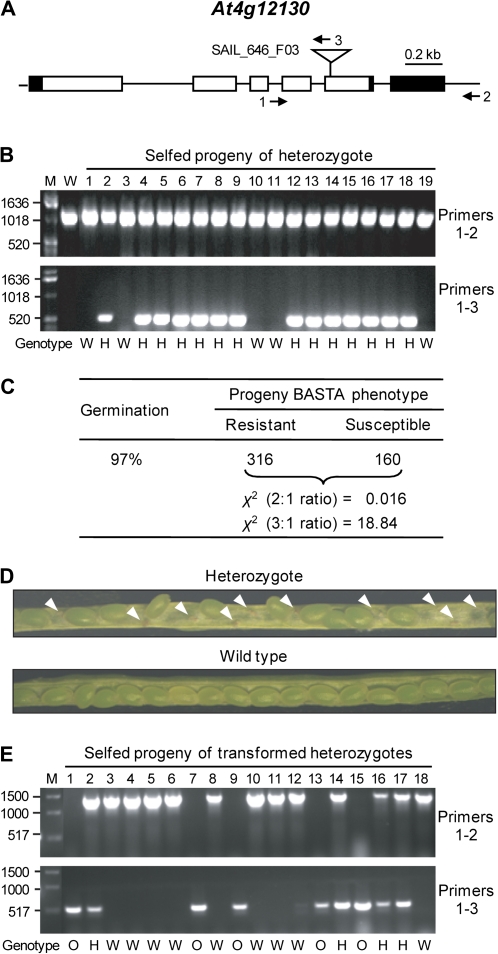
A T-DNA mutant carrying a single insertion in exon 5 of At4g12130 was identified in the GABI-Kat collection (Fig. 4A); the insertion interrupts the At4g12130 open reading frame 69 residues upstream of the C-terminus. The selfed progeny of heterozygotes contained only wild-type and heterozygous individuals (Fig. 4B), and segregated for the BASTA resistance marker in a 2:1 (resistant:susceptible) ratio (Fig. 4C). These observations both point to embryo lethality. Consistent with embryo lethality, immature siliques of heterozygous plants contained aborted seeds at about one-quarter of the positions; such defective seeds were absent from wild-type siliques (Fig. 4D). To confirm that the lethal phenotype is due to ablation of At4g12130, heterozygous mutants were transformed with a genomic clone of At4g12130 that included 2000 bp of upstream promoter sequence. The selfed progeny of transformants included individuals homozygous for the mutant At4g12130 locus, as determined by PCR (Fig. 4E). Of 68 progeny screened, eight were complemented homozygotes, 38 were wild type, and 22 were heterozygotes. The complemented plants were normal in appearance. These complementation data validate the conclusion that At4g12130 is essential for embryo development.
Fig. 4.
Evidence that At4g12130 is essential for embryo development. (A) Scheme showing the position of the T-DNA insertion in the At4g12130 gene. Introns are represented by lines and exons by boxes; 5′- and 3′-untranslated regions are in black. The positions of primers used for genomic PCR are shown by numbered arrows. Key to primers: 1, At4g12130-GS-fwd; 2, At4g12130-GS-rev; 3, TDNA-LB. (B) PCR analysis of genomic DNA of 19 representative selfed progeny of heterozygotes, showing the absence of mutant homozygotes; a wild-type control (W) is included. Lane M shows molecular size markers (bp). The progeny genotypes [i.e. wild type (W) or heterozygous (H)] are indicated beneath the gel images. (C) Segregation of BASTA resistance in the selfed progeny of heterozygous plants. The hypothesis of a 2:1 ratio (resistant:susceptible) is accepted by the χ2 test; a 3:1 ratio is strongly rejected (P=0.001). (D) Dissection of representative siliques from self-pollinated heterozygous and wild-type plants. Arrowheads mark aborted seeds. (E) PCR analysis of genomic DNA of 18 representative selfed progeny of heterozygotes transformed with a genomic clone of At4g12130 that included the native promoter. Note the presence of mutant homozygotes (O) as well as heterozygous (H) and wild-type (W) individuals.
Discussion
Bioinformatic analysis and targeting studies showed that plants, as a group, have two COG0354 proteins with distinct subcellular locations and origins. One is mitochondrial, is related to the mitochondrial COG0354 proteins of other eukaryotes, and is most probably derived from the α-proteobacterial progenitor of mitochondria. The other COG0354 protein is unique to plants, is located in plastids, and probably originated from cyanobacteria. This dual distribution is consistent with the presence of independent Fe/S cluster assembly machinery in mitochondria and plastids, and underscores how these types of machinery mirror the endosymbiotic history of the plant cell (Balk and Pilon, 2011). Also, the presence of COG0354 proteins in both organelles in which Fe/S clusters are made is consistent with the evidence that COG0354 family proteins interact physically with other Fe/S assembly components (Gelling et al., 2008; Hu et al., 2009), namely that they form part of the assembly complex.
It is at first sight surprising that certain plant taxa—grasses and poplar—have no plastidial-type COG0354, presumably as a result of relatively recent losses in independent angiosperm lineages. However, inasmuch as either plant protein can complement the E. coli COG0354 deletant (Waller et al., 2010), it seems likely that they could replace each other in plants, in which case dual targeting of the mitochondrial-type protein to mitochondria and plastids would provide a simple explanation. There are many examples of such dual targeting in plants (Mackenzie et al., 2005; Millar et al., 2006). In this connection it is noteworthy that the COSMOSS targeting algorithm (Mitschke et al., 2009) predicts that the maize, rice, and poplar mitochondrial-type COG0354 proteins are dual targeted and that their Arabidopsis, grape, and castor bean counterparts are not (scores for the former trio being 0.70–0.76 and the latter 0.47–0.51). As the genomes of grasses and poplar encode other components of the plastidial SUF Fe/S assembly machinery, the lack of a plastidial-type COG0354 in these plants is not attributable to the absence of the whole cluster assembly system.
The expression pattern of the Arabidopsis plastidial-type COG0354 protein (At1g60990) is consistent with a chloroplast location, being highest in young leaves. In contrast, the mitochondrial-type protein (At4g12130) is expressed principally in seeds and flowers. Within flowers, microarray data (Steinhauser et al., 2004) and proteome analysis (Sheoran et al., 2006) point to pollen as a main site of expression. The high expression of At4g12130 mRNA and its translation product in pollen and seeds—tissues with low water contents—shows that both message and protein are stable to drying and so are available upon hydration to support repair processes (Bewley, 1997). Since desiccation entails oxidative stress (Smirnoff, 1993), expression in drying and dry tissues is consistent with the evidence from E. coli that COG0354 helps maintain Fe/S protein function during oxidative stress (Waller et al., 2010). High-level At4g12130 expression in seeds may also contribute to the embryo lethality observed when this gene is ablated, although such lethality is characteristic of most components of the Fe/S cluster assembly machinery (Balk and Pilon, 2011).
The tests of recombinant plant COG0354 proteins in the surrogate E. coli system established that both of them require folate to function, as do COG0354 proteins from other kingdoms (Waller et al., 2010). This conclusion rests on the failure of either plant protein to restore activity to the MiaB Fe/S ‘reporter’ protein in a strain rendered completely folate deficient by deletion of the biosynthetic gene folP. FolP mediates the coupling of the pterin and p-aminobenzoate moities of folate (de Crécy-Lagard et al., 2007), and deletion of folP leaves pterin synthesis unaffected but greatly reduces MiaB activity (Waller et al., 2010). The results thus make it very improbable that pterins can substitute for folates, and place the folate requirement for MiaB activity upstream of MiaB along with the requirement for the COG0354 protein itself (Fig. 3A). While the data for the folP deletant do not identify the type of folate that plant COG0354 proteins require, results with E. coli COG0354 implicate tetrahydrofolate as the active form (Waller et al., 2010). The ability of the plant proteins to substitute for the E. coli protein implies that they, too, require tetrahydrofolate. As both mitochondria and chloroplasts contain tetrahydrofolates (Hanson and Gregory, 2011) the necessary folate cofactor would be available to both COG0354 proteins in planta.
In summary, the present data establish COG0354 proteins as components of both the mitochondrial and the plastidial Fe/S assembly machinery, and show that these types of machinery have a hitherto unsuspected folate requirement. The data also show that the mitochondrial COG0354 protein is required for embryo development. Finally, the loss of the plastidial-type protein from certain lineages raise interesting questions about the evolutionary forces that drove the loss of this protein and about its function in the species that retain it.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Oligonucleotide primers used in this study.
Acknowledgments
This work was supported in part by National Science Foundation grant no. MCB-0839926 and by an endowment from the C. V. Griffin Sr Foundation. We thank J. Whelan for the soybean alternative oxidase clone, H. J. Klee and K. M. Folta for advice, and O. Frelin for assistance with transgenic plants.
References
- Bailly M, Blaise M, Roy H, Deniziak M, Lorber B, Birck C, Becker HD, Kern D. tRNA-dependent asparagine formation in prokaryotes: characterization, isolation and structural and functional analysis of a ribonucleoprotein particle generating Asn-tRNAAsn. Methods. 2008;44:146–163. doi: 10.1016/j.ymeth.2007.11.012. [DOI] [PubMed] [Google Scholar]
- Balk J, Pilon M. Ancient and essential: the assembly of iron–sulfur clusters in plants. Trends in Plant Science. 2011;16:218–226. doi: 10.1016/j.tplants.2010.12.006. [DOI] [PubMed] [Google Scholar]
- Beinert H. Iron–sulfur proteins: ancient structures, still full of surprises. Journal of Biological and Inorganic Chemistry. 2000;5:2–15. doi: 10.1007/s007750050002. [DOI] [PubMed] [Google Scholar]
- Bewley JD. Seed germination and dormancy. The Plant Cell. 1997;9:1055–1066. doi: 10.1105/tpc.9.7.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- de Crécy-Lagard V, El Yacoubi B, de la Garza RD, Noiriel A, Hanson AD. Comparative genomics of bacterial and plant folate synthesis and salvage: predictions and validations. BMC Genomics. 2007;8:245. doi: 10.1186/1471-2164-8-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelling C, Dawes IW, Richhardt N, Lill R, Mühlenhoff U. Mitochondrial Iba57p is required for Fe/S cluster formation on aconitase and activation of radical SAM enzymes. Molecular and Cellular Biology. 2008;28:1851–1861. doi: 10.1128/MCB.01963-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibeaut DM, Hulett J, Cramer GR, Seemann JR. Maximal biomass of Arabidopsis thaliana using a simple, low-maintenance hydroponic method and favorable environmental conditions. Plant Physiology. 1997;115:317–319. doi: 10.1104/pp.115.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave AP. A versatile binary vector system with a T-DNA organizational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Molecular Biology. 1992;20:1203–1207. doi: 10.1007/BF00028910. [DOI] [PubMed] [Google Scholar]
- Gould SB, Waller RR, McFadden GI. Plastid evolution. Annual Review of Plant Biology. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- Hanson AD, Gregory JF 3rd. Folate biosynthesis, turnover, and transport in plants. Annual Review of Plant Biology. 2011;62:4.1–4.21. doi: 10.1146/annurev-arplant-042110-103819. [DOI] [PubMed] [Google Scholar]
- Hernández HL, Pierrel F, Elleingand E, García-Serres R, Huynh BH, Johnson MK, Fontecave M, Atta M. MiaB, a bifunctional radical- S-adenosylmethionine enzyme involved in the thiolation and methylation of tRNA, contains two essential [4Fe–4S] clusters. Biochemistry. 2007;46:5140–5147. doi: 10.1021/bi7000449. [DOI] [PubMed] [Google Scholar]
- Hu P, Janga SC, Babu M, et al. Global functional atlas of Escherichia coli encompassing previously uncharacterized proteins. PLoS Biology. 2009;7:e96. doi: 10.1371/journal.pbio.1000096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron–sulfur clusters. Annual Review of Biochemistry. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- Lill R. Function and biogenesis of iron–sulfur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- Lin CN, Syu WJ, Sun WS, Chen JW, Chen TH, Don MJ, Wang SH. A role of ygfZ in the Escherichia coli response to plumbagin challenge. Journal of Biomedical Science. 2010;17:84. doi: 10.1186/1423-0127-17-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackenzie SA. Plant organellar protein targeting: a traffic plan still under construction. Trends in Cell Biology. 2005;15:548–554. doi: 10.1016/j.tcb.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Millar AH, Whelan J, Small I. Recent surprises in protein targeting to mitochondria and plastids. Current Opinion in Plant Biology. 2006;9:610–615. doi: 10.1016/j.pbi.2006.09.002. [DOI] [PubMed] [Google Scholar]
- Mitschke J, Fuss J, Blum T, Höglund A, Reski R, Kohlbacher O, Rensing SA. Prediction of dual protein targeting to plant organelles. New Phytologist. 2009;183:224–235. doi: 10.1111/j.1469-8137.2009.02832.x. [DOI] [PubMed] [Google Scholar]
- Nilsson R, Schultz IJ, Pierce EL, et al. Discovery of genes essential for heme biosynthesis through large-scale gene expression analysis. Cell Metabolism. 2009;10:119–130. doi: 10.1016/j.cmet.2009.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa Y. A synthetic green fluorescent protein gene for plant biotechnology. Plant Biotechnology. 2003;20:1–11. [Google Scholar]
- Olinares PD, Ponnala L, van Wijk KJ. Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Molecular and Cellular Proteomics. 2010;9:1594–1615. doi: 10.1074/mcp.M000038-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ote T, Hashimoto M, Ikeuchi Y, Su'etsugu M, Suzuki T, Katayama T, Kato J. Involvement of the Escherichia coli folate-binding protein YgfZ in RNA modification and regulation of chromosomal replication initiation. Molecular Microbiology. 2006;59:265–275. doi: 10.1111/j.1365-2958.2005.04932.x. [DOI] [PubMed] [Google Scholar]
- Pagliarini DJ, Calvo SE, Chang B, et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips G, El Yacoubi B, Lyons B, Alvarez S, Iwata-Reuyl D, de Crécy-Lagard V. The biosynthesis of the 7-deazaguanosine modified tRNA nucleosides: a new role for GTP cyclohydrolase I. Journal of Bacteriology. 2008;190:7876–7884. doi: 10.1128/JB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon V, Ravanel S, Douce R, Alban C. Biotin synthesis in plants. The first committed step of the pathway is catalyzed by a cytosolic 7-keto-8-aminopelargonic acid synthase. Plant Physiology. 2005;139:1666–1676. doi: 10.1104/pp.105.070144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pribat A, Noiriel A, Morse AM, et al. Nonflowering plants possess a unique folate-dependent phenylalanine hydroxylase that is localized in chloroplasts. The Plant Cell. 2010;22:3410–3422. doi: 10.1105/tpc.110.078824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravanel S, Cherest H, Jabrin S, Grunwald D, Surdin-Kerjan Y, Douce R, Rébeillé F. Tetrahydrofolate biosynthesis in plants: molecular and functional characterization of dihydrofolate synthetase and three isoforms of folylpolyglutamate synthetase in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA. 2001;98:15360–15365. doi: 10.1073/pnas.261585098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso MG, Li Y, Strizhov N, Reiss B, Dekker K, Weisshaar B. An Arabidopsis thaliana T-DNA mutagenized population (GABI-Kat) for flanking sequence tag-based reverse genetics. Plant Molecular Biology. 2003;53:247–259. doi: 10.1023/B:PLAN.0000009297.37235.4a. [DOI] [PubMed] [Google Scholar]
- Rudhe C, Chew O, Whelan J, Glaser E. A novel in vitro system for simultaneous import of precursor proteins into mitochondria and chloroplasts. The Plant Journal. 2002;30:213–220. doi: 10.1046/j.1365-313x.2002.01280.x. [DOI] [PubMed] [Google Scholar]
- Sheftel A, Stehling O, Lill R. Iron–sulfur proteins in health and disease. Trends in Endocrinology and Metabolism. 2010;21:302–314. doi: 10.1016/j.tem.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Sheoran IS, Sproule KA, Olson DJ, Ross AR, Sawhney VK. Proteome profile and functional classification of proteins in Arabidopsis thaliana (Landsberg erecta) mature pollen. Sexual Plant Reproduction. 2006;19:185–196. [Google Scholar]
- Smirnoff N. The role of active oxygen in the response of plants to water deficit and desiccation. New Phytologist. 1993;125:27–58. doi: 10.1111/j.1469-8137.1993.tb03863.x. [DOI] [PubMed] [Google Scholar]
- Steinhauser D, Usadel B, Luedemann A, Thimm O, Kopka J. CSB.DB: a comprehensive systems-biology database. Bioinformatics. 2004;20:3647–3651. doi: 10.1093/bioinformatics/bth398. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Teplyakov A, Obmolova G, Sarikaya E, Pullalarevu S, Krajewski W, Galkin A, Howard AJ, Herzberg O, Gilliland GL. Crystal structure of the YgfZ protein from Escherichia coli suggests a folate-dependent regulatory role in one-carbon metabolism. Journal of Bacteriology. 2004;186:7134–7140. doi: 10.1128/JB.186.21.7134-7140.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner WL, Waller JC, Snedden WA. Identification, molecular cloning and functional characterization of a novel NADH kinase from Arabidopsis thaliana (thale cress) Biochemical Journal. 2005;385:217–223. doi: 10.1042/BJ20040292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waller JC, Alvarez S, Naponelli V, et al. A role for tetrahydrofolates in the metabolism of iron–sulfur clusters in all domains of life. Proceedings of the National Academy of Sciences, USA. 2010;107:10412–10417. doi: 10.1073/pnas.0911586107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams KP, Sobral BW, Dickerman AW. A robust species tree for the alphaproteobacteria. Journal of Bacteriology. 2007;189:4578–4586. doi: 10.1128/JB.00269-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang D, Venglat P, Tibiche C, et al. Genome-wide analysis reveals gene expression and metabolic network dynamics during embryo development in Arabidopsis. Plant Physiology. 2011;156:346–356. doi: 10.1104/pp.110.171702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Møller SG. Iron–sulfur cluster biogenesis systems and their crosstalk. Chembiochem. 2008;9:2355–2362. doi: 10.1002/cbic.200800384. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.