Abstract
Histone deacetylation plays an important role in epigenetic control of gene expression. HD2 is a plant-specific histone deacetylase that is able to mediate transcriptional repression in many biological processes. To investigate the epigenetic and transcriptional mechanisms of longan fruit senescence, one histone deacetylase 2-like gene, DlHD2, and two ethylene-responsive factor-like genes, DlERF1 and DlERF2, were cloned and characterized from longan fruit. Expression of these genes was examined during fruit senescence under different storage conditions. The accumulation of DlHD2 reached a peak at 2 d and 30 d in the fruit stored at 25 °C (room temperature) and 4 °C (low temperature), respectively, or 6 h after the fruit was transferred from 4 °C to 25 °C, when fruit senescence was initiated. However, the DlERF1 transcript accumulated mostly at the later stage of fruit senescence, reaching a peak at 5 d and 35 d in the fruit stored at 25 °C and 4 °C, respectively, or 36 h after the fruit was transferred from low temperature to room temperature. Moreover, application of nitric oxide (NO) delayed fruit senescence, enhanced the expression of DlHD2, but suppressed the expression of DlERF1 and DlERF2. These results indicated a possible interaction between DlHD2 and DlERFs in regulating longan fruit senescence, and the direct interaction between DlHD2 and DlERF1 was confirmed by yeast two-hybrid and bimolecular fluorescence complementation (BiFC) assays. Taken together, the results suggested that DlHD2 may act with DlERF1 to regulate gene expression involved in longan fruit senescence.
Keywords: ERFs, fruit senescence, HD2, histone deacetylase, longan
Introduction
The regulation of gene expression at the transcription level underlies many biological aspects including growth and development, metabolic and physiological balances, and responses to the environmental stimuli (Vlachonasios et al., 2003). Gene expression depends on not only DNA sequences such as promoters and cis-acting regulatory elements, but also epigenetic factors such as histone modifications and chromatin remodelling. In eukaryotes, post-translational modification of histones plays an important role in gene regulation (Turner, 2000; Lagaće et al., 2003). A number of post-translational modifications of histone have been observed, including acetylation, methylation, phosphorylation, ubiquination, and ADP-ribosylation (Strahl and Allis, 2000). All histone modifications are removable, which may therefore provide a versatile way for regulating gene expression during plant development and response to environmental stimuli. Acetylation of the histones catalysed by histone acetyltransferases (HATs) is generally associated with increased transcriptional activation, whereas deacetylation of histones by histone deacetylases (HDACs) is correlated with transcriptional repression (Berger, 2002; Sridha and Wu, 2006).
Three types of HDACs, namely reduced potassium dependency 3 (RPD3), histone deacetylase 1 (HDA1), and silent information regulation 2 or sirtuin 2 (SIR2), have been found in plants, yeast, and animals (Pandey et al., 2002; Yang and Seto, 2003). In addition, plants contain a unique type of HDACs called histone deacetylase2 (HD2), which is unrelated to the RPD3, HDA1, and SIR2 HDAC types (Sridha and Wu, 2006). The first HD2 was isolated as an acidic nucleolar phosphoprotein from maize embryo (Lusser et al., 1997), suggesting a possible role for HD2 in the expression of rRNAs. Later, four HD2 proteins, AtHD2A, AtHD2B, AtHD2C and AtHD2D, were identified in Arabidopsis (Wu et al., 2000b, 2003). AtHD2A and AtHD2B were shown to be involved in establishing leaf polarity in Arabidopsis (Ueno et al., 2007) while the Arabidopsis AtHD2C gene played a role in abscisic acid (ABA) response and abiotic stress (Sridha and Wu, 2006). ScHD2a, an orthologue of AtHD2A in Solanum chacoense (a wild species related to potato), was strongly induced in ovules after fertilization (Lagaće et al., 2003). In addition, HvHDAC2-1 and HvHDAC2-2 from barley were found to respond to plant stress-related hormones, such as jasmonic acid (JA), ABA, and salicylic acid (SA) (Demetriou et al., 2009). Collectively, these findings suggest that HD2-type HDACs play an important role in plant development and stress response. Furthermore, HD2 may modulate gene expression in a complex containing RPD3-like HDACs such as AtHDA6 and AtHD1, although it is yet to be determined whether any HD2 members interact with RPD3-like proteins (Chen and Tian, 2007). Recently, DNA methyltransferase 2 (AtDNMT2) interacting with AtHD2s has been identified in Arabidopsis (Song et al., 2010). It has been demonstrated that chromatin structure including the structure imposed by the nucleosome implies that transcription factors (TFs) work together with large multisubunit complexes that remodel nucleosomes to facilitate DNA accessibility and to enable transcription (Depège-Fargeix et al., 2011). Unfortunately, it remains largely unknown whether HD2 can interact with TFs to regulate gene expression in plants.
Ethylene response factors (ERFs) constitute one of the largest TF gene families in plants, with 122 members in Arabidopsis and 139 members in rice, and contain a conserved DNA-binding domain (AP2/ERF domain) (Nakano et al., 2006). The ERF proteins have diverse biological functions in plant growth and development, such as leaf epidermal cell density, flower development, and embryo development (Elliott et al., 1996; Boutilier et al., 2002), as well as hormonal signalling mediated by ethylene (Yin et al., 2010), cytokinin (Rashotte et al., 2006), brassinosteroid (Alonso et al., 2003; Hu et al., 2004), and ABA (Zhu et al., 2010). ERFs are also involved in biotic and abiotic stress responses via direct interaction with GC-rich cis-elements [e.g. the GCC-box and the dehydration-responsive element (DRE)] in the promoter of their target genes (Jofuku et al., 1994; Okamuro et al., 1997; Hao et al., 1998; Liu et al., 1998; Gilmour et al., 2000; Aharoni et al., 2004). So far, the isolation and characterization of ERF genes related to fruit development and ripening have been reported only in a few fruit, such as tomato (Tournier et al., 2003; Sharma et al., 2010), apple (Wang et al., 2007), plum (El-Sharkawy et al., 2009), and kiwifruit (Yin et al., 2010). In tomato, it was found that overexpression of LeERF1 induced ethylene triple response on etiolated seedlings, whereas antisense lines exhibited longer shelf life (Li et al., 2007). LeERF2 was shown to act as a positive regulator in the feedback loop of ethylene induction (Wu et al., 2002; Tournier et al., 2003). Moreover, LeERF2 regulated ethylene production in tomato and tobacco by modulating ethylene biosynthesis genes through interaction with the NtACS3 and LeACO3 promoters (Zhang et al., 2009). However, little is known about the involvement of ERF in fruit senescence.
Longan is a non-climacteric subtropical fruit with high value (Jiang et al., 2002). The edible portion of longan fruit is a fleshy and translucent white aril. However, the fruit senesce rapidly after harvest, with rapid appearance of pericarp browning and aril breakdown, resulting in reduced market value (Jiang et al., 2002). Aril breakdown involves loss of turgidity and translucency, and, thus, the fruit become bland in taste. The disorder starts near the pericarp and appears to be more prevalent at the distal end (Jiang et al., 2002). Accordingly, it is important to understand longan fruit senescence at the molecular level and then to optimize post-harvest handling to extend storage life or maintain quality of the fruit. Increasing evidence has shown that nitric oxide (NO) as a free radical gas may have anti-senescence and anti-ripening properties (Leshem and Haramaty, 1996) and can extend the post-harvest life of fresh horticultural products such as strawberry (Wills et al., 2000), carnations (Bowyer et al., 2003), pear (Sozzi et al., 2003), and longan (Duan et al., 2007). In a previous study, it was shown that expansin (EXP), xyloglucan endotransglucosylase (XET), and endo-β-1,4-glucanase (EGase) related to cellular wall metabolism are involved in longan fruit senescence (Zhong et al., 2008; Xiao et al., 2009). However, little attention has been paid to the epigenetic and transcriptional mechanisms of longan fruit senescence. In this study, one HD2 gene and two ERF genes were isolated from longan fruit. The expression patterns of these genes in relation to senescence of longan fruit stored under various conditions, and their responses to NO treatment, were investigated. Moreover, the direct interaction between HD2 and ERF1 was detected by yeast two-hybrid assay and the bimolecular fluorescence complementation (BiFC) assay, suggesting that DlHD2 may mediate longan fruit senescence by interacting with DlERF1.
Materials and methods
Plant materials
Longan (Longan chinensis Sonn. cv. Shixia) fruit at physiological maturity (∼90 d after anthesis), exhibiting yellow-brown colour in peel and optimal eating quality of the aril (Jiang et al., 2002), were harvested, then transported to the laboratory within 2 h, and finally selected for freedom from visual defects and for uniformity of weight, shape, and maturity.
Treatments
The selected longan fruit were divided randomly into four groups and then placed into unsealed plastic bags (0.04 mm in thickness). Each bag contained 50 individual fruit and 14 bags were used as one group. For NO treatment, the fruit from Groups 1 and 2 were exposed to air (control) or 40 μl l−1 NO for 4 h in six closed chambers (5.0 l) at 25 °C. After these treatments, the fruit were placed into unsealed plastic bags and then stored at 25 °C (room temperature) for 5 d until the control fruit were completely senescent. Fruit were sampled daily. The fruit from Group 3 were stored at 4 °C (low temperature) for 45 d and then sampled at 5 d intervals, while the fruit from Group 4 were stored at 4 °C for 40 d and then transferred to 25 °C for further storage, and the fruit were sampled at 0, 6, 12, 24, 36, and 48 h, respectively. Each sampling contained 60 individual fruit withdrawn randomly from six bags. The whole aril of 20 individual fruit were mixed thoroughly as one replicate, frozen in liquid nitrogen. and stored at –80 °C until use. All assessments were conducted in three biological replicates.
Evaluation of fruit senescence
The aril breakdown index was used to indicate the longan fruit senescence. The index was measured by determining the area ratio of the aril which had broken down to the whole aril using 30 individual fruit per sample, according to the methods of Zhong et al. (2008) and Xiao et al. (2009).
RNA extraction and isolation of longan DlHD2, DlERF1, and DlERF2 full-length cDNAs
Total RNA from longan fruit was extracted using the hot borate method of Wan and Wilkins (1994). Frozen aril tissues (10 g) were ground to a fine powder in a mortar using a pestle in the presence of liquid nitrogen. Following the RNA extraction, potentially contaminating DNA was eliminated by the treatment with DNase I digestion using the RNase-free kit (Promega, Madison, WI, USA). The DNA-free total RNA was then used as template for reverse transcription-PCR (RT-PCR). The first-strand cDNA of the product was subjected to PCR amplification. To isolate HD2 cDNA from longan fruit, two synthetic degenerate oligonucleotide primers were designed with regard to the N-terminal (MEFWGVE) and the internal peptide sequences (HVATPHP) of the HD2 protein (Aravind and Koonin, 1998). Degenerate primers for the ERF were designed based on the report of Tournier et al. (2003). The isolated fragments were cloned into the pMD20-T vector (TaKaRa, Dalian Division), sequenced, and finally compared with the database sequence using the BLAST program (http://www.ncbi.nlm.nih.gov/BLAST).
Next, 3′- or 5′-rapid amplification of cDNA ends (RACE)-PCR was performed using cDNA end amplification kits (Takara, Dalian Division) according to the manufacturer's protocol. In order to amplify 3′ and 5′ end fragments, the specific primers were designed based on the nucleotide sequences of the cDNA fragments already cloned by RT-PCR. The 3′- and 5′-RACE-PCR products were cloned and sequenced as described above. The primer sequences are provided in Supplementary Table S1 available at JXB online.
Bioinformatics analysis
Identification of nucleotide sequences from RT-PCR clones was established using the NCBI Blast program (http://www.ncbi.nlm.nih.gov/BLAST). Alignments were carried out on Clustalx 1.83 and GeneDoc software, and a phylogenetic tree was constructed using the Neighbor–Joining method in the MEGA 4 programme visualized by TreeView software. The theoretical isoelectric points (pIs) and mass values for mature peptides were calculated using the PeptideMass program (http://us.expasy.org/tools/peptide-mass.html).
Transcriptional activation analysis in yeast cells
The open reading frames (ORFs) of DlHD2, DlERF1, and DlERF2 were amplified by PCR with gene-specific primers (listed in Supplementary Table S2 at JXB online) and subcloned into pGBKT7 (Clontech, USA) (Supplementary Fig. S1). According to the protocol of the manufacturer, pGBKT7-DlERF1, pGBKT7-DlERF2, pGBKT7-DlHD2, the positive control pGBKT7-53+pGADT7-T, and the negative control pGBKT7 plasmids were each used to transform the AH109 yeast strain. The transformed strains were streaked onto SD/–Trp or SD/–Trp–His–Ade plates. The transactivation activity of each protein was evaluated according to their growth status and the β-galactosidase activity.
Subcellular localization analysis
The coding region sequences of DlERF1, DlERF2, and DlHD2 without the stop codon were amplified by PCR (the primers are listed in Supplementary Table S3 at JXB online) from the full-length clone of pMD-DlERF1, pMD-DlERF2, and pMD-DlHD2, and subcloned into the pUC-GFP vector, in-frame with the green fluorescent protein (GFP) sequence, resulting in the 35S::gene-GFP vectors under the control of the Cauliflower mosaic virus (CaMV) 35S promoter (Supplementary Fig. S2). The fusion constructs and the control GFP vector were introduced into onion epidermal cells by particle bombardment using a Bio-Rad (Hercules, CA, USA) Biolistic Particle Delivery System PDS-1000. GFP fluorescence was observed with a laser scan confocal microscope. All transient expression assays were repeated at least three times.
Northern blot analysis
Total RNA (10 μg) was separated on a 1.2% agarose–formadehyde gel and capillary blotted onto a positively charged nylon membrane (Biodyne® B, 0.45 μm, PALL Co., Sarasota, FL, USA). The RNA was fixed to the membrane by baking for 2 h at 80 °C and then cross-linked to the membranes using an ultraviolet cross-linker (Amersham Biosciences, Piscataway, NJ, USA). The membranes were pre-hybridized for >3 h in SDS buffer solution containing 50% (v/v) deionized formamide, 5× SSC, 7% SDS, 2% blocking reagent (Roche Diagnostics, Mannheim, Germany), 50 mM sodium phosphate (pH 7.0), and 0.1% N-lauroylsarcosine (w/v). Hybridization was performed overnight in the same buffer solution containing the gene-specific digoxigenin (DIG)-labelled probe at 45 °C. The probe was prepared with a DIG probe synthesis kit (Roche Applied Science, Mannheim, Germany) according to the manufacturer's instruction. The probes were synthesized from the 3'-untranslated regions of the genes and each membrane was only hybridized once with each probe to ensure that the hybridization was specific and reliable. Following hybridization, membranes were washed twice for 10 min with 2× SSC containing 0.1% SDS at 25 °C, followed by washing twice for 30 min in 0.1× SSC containing 0.1% SDS at 62 °C. The signals were detected with chemiluminescence using CDP-Star™ (Roche Diagnostics) as described by the manufacturer. The most stable reference gene, iron superoxide dismutase (FeSOD) (Lin and Lai, 2010), was used as an internal control. Membranes were scanned with a densitometer (Bio-Rad Fluor-S Multimager) and then the hybridization signals were quantified by using the Bio-Rad Quantity One software. The relative expression abundance of each gene was expressed as the ratio of target gene intensity to FeSOD intensity. The specific primers used for synthesis of the DIG-labelled probes are available in Supplementary Table S4 at JXB online.
Yeast two-hybrid assays
Yeast two-hybrid assays were performed using the Matchmaker GAL4-based two hybrid system 3 (Clontech, USA). The ORFs of DlHD2, DlERF1, and DlERF2 were amplified by PCR and subcloned into pGBKT7 and pGADT7 vectors as bait and prey, respectively (Supplementary Fig. S3 at JXB online). The primers used for PCR cloning of the cDNAs are listed in Supplementary Table S5. Construct pairs pGBKT7-53+pGADT7-T (positive control), pGBKT7-Lam+pGADT7-T (negative control), pGBKT7-DlHD2+pGADT7-DlERF1, pGBKT7-HD2+pGADT7-DlERF2, and pGBKT7-DlERF2+pGADT7-DlHD2 were transformed into the yeast strain AH109 by the lithium acetate method. Transformed yeast cells are first grown on a minimal medium/–Leu–Trp according to the manufacturer's instructions (Clontech), and then plated onto a minimal medium/–Leu–Trp–His–Ade containing 5 mM 3-amino-1,2,4-triazole (3-AT) at 30 °C to test the possible interactions between DlHD2 and DlERFs.
BiFC assay in Arabidopsis mesophyll protoplasts
The vectors used in the BiFC assay (pUC-pSPYNE or pUC-pSPYCE) were obtained from the laboratory of Harter and Kudla (Walter et al., 2004). For generation of the BiFC vectors (Supplementary Fig. S4 at JXB online), the full-length coding sequence of DlERF1 was fused with the N-terminal fragment of yellow fluorescent protein (YFP) in the pUC-pSPYNE vector to form the DlERF1-pSPYNE construct, while the full-length coding sequence of DlHD2 was cloned into pUC-pSPYCE as a fusion with the C-terminal fragment of YFP to produce the DlHD2-pSPYCE construct. Both empty vectors (pUC-pSPYNE/pUC-pSPYCE) and expression of DlERF1 alone (DlERF1-pSPYNE/pUC-pSPYCE) were used as negative controls, while the bZIP63-pSPYNE and bZIP63-pSPYCE vectors were used as a positive control (Walter et al., 2004). The resulting constructs were used for transient assays by polyethylene glycol (PEG) transfection of Arabidopsis protoplasts isolated from 4-week-old wild-type (Columbia) plants according to previously reported procedures (Yoo et al., 2007). mCherry-VirD2NLS was included in each transfection to serve as a control for successful transfection as well as for nuclear localization (Lee et al., 2008). Transfected cells were imaged using a TCS SP5 Confocal Spectral Microscope Imaging System (Leica), with an argon blue laser at 488 nm, a beamsplitter for excitation at 500 nm, and a spectral detector set between 515 nm and 540 nm. The primers used in BiFC assay are also listed in Supplementary Table S5 at JXB online.
Results
Isolation and sequence analysis of full-length cDNA of DlHD2, DlERF1, and DlERF2
One HD2-like and two ERF-like full-length cDNAs were isolated from longan aril and designated as DlHD2, DlERF1, and DlERF2, respectively. DlHD2 contained an ORF of 931 bp. It encoded a putative protein of 306 amino acids, with a predicted mol. wt of 32.8 kDa and a calculated pI of 4.86. Significant homology was identified between DlHD2 and histone deacetylase HD2 from other organisms, based on their deduced protein sequences. The deduced DlHD2 protein contained all the structural features of the plant-specific HD2-type proteins (Aravind and Koonin, 1998), such as an N-terminal domain with the invariable pentapeptide (MEFWG), a central region containing an extended acidic domain, required for repression, followed by a central acidic region rich in glutamic and/or aspartic acid, and a single C2H2-type zinc finger domain in the C-terminus, which may enable high affinity DNA binding or mediate protein–protein interactions (Dangl et al., 2001; Zhou et al., 2004) (Supplementary Fig. S5 at JXB online). A BLAST search of GenBank revealed that DlHD2 shared 55% identity with RcHD2a (XP_002527449) from bean and 53% identity with NtHD2a (ACZ54945) from tobacco at the protein level. Phylogenetic analysis further showed that DlHD2 was closely related to HD2a from Ricinus communis (Supplementary Fig. S6).
DlERF1 had an ORF of 813 bp, encoding a predicted polypeptide of 270 amino acids, with a predicted mol. wt of 29.8 kDa and a calculated pI of 5.62, while DlERF2 exhibited an ORF of 966 bp, encoding 321 amino acids with a predicted mol. wt of 35.3 kDa and a calculated pI of 4.96. The deduced amino acid sequences of the two DlERFs comprised a conserved DNA-binding ERF/AP2 domain, which is a typical characteristic of the plant ERF gene families (Supplementary Fig. S7 at JXB online). In addition, both DlERF1 and DlERF2 contained two key amino acid residues, the 14th alanine (A14) and the 19th aspartate (D19), which were reported to contribute to a functional GCC-box binding activity in many ERFs (Sakuma et al., 2002). Phylogenetic analysis also indicated that the ERF family proteins were classified into four groups, Classs I, II, III, or IV (Supplementary Fig. S8). The first three groups were previously identified according to their functions and structures (Fujimoto et al., 2000). Class IV, a new member of the ERF subfamily containing a conserved MCGGAIL signature sequence in the N-terminus, was later added to the classification (Tournier et al., 2003). As shown in Supplementary Fig. S8, DlERF1 belonged to the Class III ERFs (Fujimoto et al., 2000; Nakano et al., 2006). The ERF genes in this group had a CMIX-2 motif and, thus, were subclassified into three types based on the different constituents of their putative mitogen-activated protein (MAP) kinase phosphorylation sites, the CMIX-5 motif, and the CMIX-6 motif (Nakano et al., 2006). DlERF1 had a higher similarity to NtERF4 and SlERF4 than to other types of ERFs, in which both the CMIX-5 motif and the CMIX-6 motif were found at the C-terminus (Supplementary Fig. S9). However, DlERF2 belonged to Class IV ERFs (Tournier et al., 2003; Nakano et al., 2006), characterized by a conserved N-terminal signature and the MCGGAII/L motif (CMVII-1 motif) (Supplementary Fig. S10). Overall, these results suggested that DlERF1 and DlERF2 might exhibit diverse functions.
DlERF1 showed transcriptional activation activity in yeast
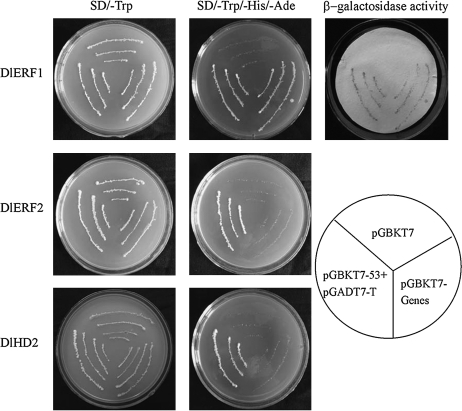
To investigate the transcriptional activities of DlHD2, DlERF1, and DlERF2, a transient expression assay using a GAL4-responsive reporter system was performed. For the assay of transcriptional activity, the entire ORF of DlHD2, DlERF1, or DlERF2 was cloned into the same pGBKT7 vector (Supplementary Fig. S1 at JXB online). The yeast strain AH109 harbouring two reporter genes, lacZ and His3, was transformed with the fusion plasmids pGBKT7-DlHD2, pGBKT7-DlERF1, and pGBKT7-DlERF2, the positive control pGBKT7-53+pGADT7-T, and the negative control pGBKT7, respectively. As shown in Fig. 1, the transformed yeast cells harbouring pGBKT7-DlERF1 and pGBKT7-53+pGADT7-T (the positive control) grew well in the SD medium lacking tryptophan, histidine and adenine, and showed β-galactosidase activity, whereas the cells containing pGBKT7-DlHD2, pGBKT7-DlERF2, or pGBKT7 (the negative control) did not grow or showed no β-galactosidase activity. These data suggested that DlERF1 functioned as a transcriptional activator in yeast.
Fig. 1.
Transcriptional activation analysis of DlERFs and DlHD2 in yeast. DlERF1, DlERF2, and DlHD2 were each fused with the GAL4 DNA-binding domain and expressed in yeast strain AH109. The vectors pGBKT7 or pGBKT7-53+pGADT7-T were expressed in yeast as a negative or a positive control, respectively. Yeast clones transformed with the different vectors were grown on SD plates without tryptophan or tryptophan, histidine and adenine for 3 d at 30 °C. Transcription activation was monitored by the detection of yeast growth and β-galactosidase assay.
Subcellular localization of DlHD2 and DlERFs
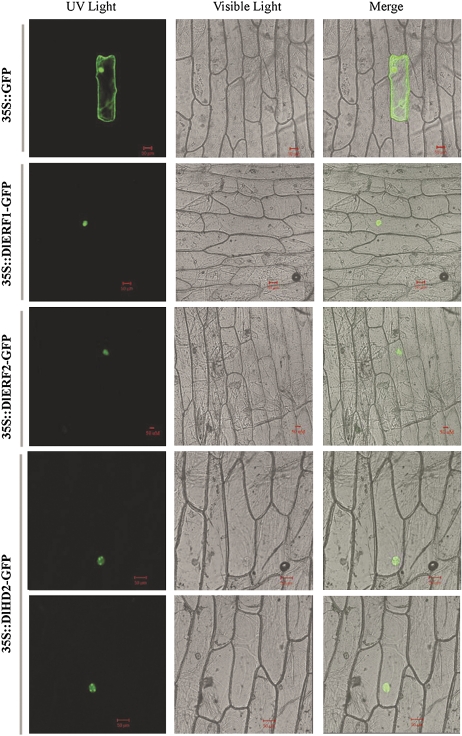
To validate the subcellular localization of DlHD2 and DlERF proteins, the coding regions of the DlHD2, DlERF1, and DlERF2 were fused in-frame with the GFP gene (Supplementary Fig. S2 at JXB online), and then the resulting constructs were bombarded into onion epidermal cells. As shown in Fig. 2, the fluorescence of DlERF1–GFP and DlERF2–GFP was localized exclusively to the nucleus, while the fluorescence of GFP alone was observed in the entire cells. Interestingly, the fluorescence of DlHD2–GFP was probably localized exclusively to the three nucleoli in the nucleus. In addition, a similar subcellular localization of DlHD2 was also observed in Arabidopsis mesophyll protoplasts (Supplementary Fig. S11), indicating that DlHD2 may also act directly on rRNA gene chromatin (Lusser et al., 1997).
Fig. 2.
Subcellular localization of DlHD2 and DlERFs in onion epidermal cells. Cells were bombarded with constructs carrying GFP, DlERF1–GFP, DlERF2–GFP, or DlHD2–GFP as described in the Materials and methods. GFP, DlERF1–GFP, DlERF2–GFP, and DlHD2–GFP fusion proteins were transiently expressed under control of the CaMV 35S promoter in onion epidermal cells and observed with a laser scanning confocal microscope. Images were taken in the dark field for green fluorescence, while the outline of the cell and the combination were photographed in a bright field. The length of the bar is indicated in the photographs.
Expression patterns of DlHD2, DlERF, and DlERF2 during senescence of longan fruit stored under different conditions
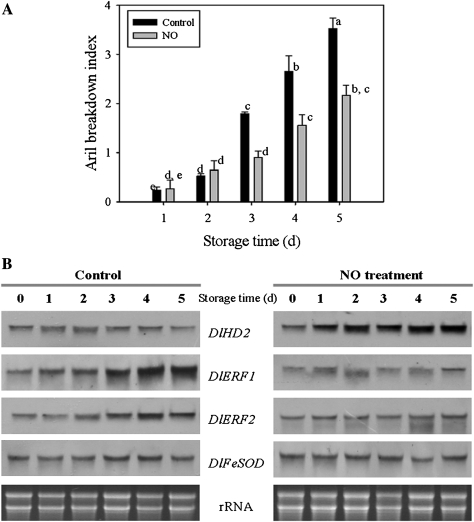
To understand the possible role of the DlHD2, DlERF1, and DlERF2 during longan fruit senescence, the expression patterns of DlHD2, DlERF1, and DlERF2 in fruit stored under different conditions, namely room temperature (25 °C), low temperature (4 °C), the transfer from low temperature to room temperature, and application of NO, were analysed. Longan fruit began to show senescence symptoms after 2 d of storage at 25 °C, as reflected by the aril breakdown index, which increased rapidly during storage, particularly after 3 d of storage (Fig. 3A). Accumulation of DlHD2 mRNA increased in the first 2 d, but decreased afterwards as the aril broke down. The mRNAs of both DlERF1 and DlERF2 showed a gradual increase acccompanying the aril breakdown, especially at days 4 and 5 (Fig. 3B; Supplementary Fig. S12). As expected, NO pre-treatment delayed longan fruit senescence, as the aril breakdown index of the NO-treated fruit was significantly lower than that of the control fruit (Fig. 3A). Correspondingly, application of NO obviously enhanced the accumulation of DlHD2 mRNA but suppressed that of DlERF1 and DlERF2 mRNAs during fruit senescence (Fig. 3B; Supplementary Fig. S12 at JXB online).
Fig. 3.
Changes in the aril breakdown index (A), and differential expression patterns of DlHD2, DlERF1 and DlERF2 (B) in aril tissues of control and NO-treated longan fruit stored at room temperature (25 °C) for 5 d. In A, each value represents the means of three replicates, and vertical bars indicate the SE. Different letters indicate a statistical difference at the 5% level among data groups according to the Duncan's multiple range test. In B, total RNA (10 μg per lane) was used for RNA gel blot analysis and hybridization with DIG-labelled probes. Hybridization with DIG-labelled DlFeFOD probes and ethidium bromide-stained rRNA are shown as the internal loading controls.
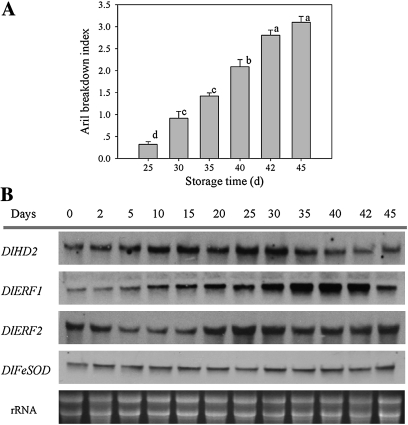
Storage at low temperature obviously delayed the appearance of fruit senescence symptoms. Aril breakdown symptoms were observed only after 25 d of storage at 4 °C and became significant after 30 d of storage (Fig. 4A). The DlHD2 transcript continuously increased within the first 30 d, with higher accumulations at days 25 and 30, then decreased gradually afterwards (days 35–45) (Fig. 4B; Supplementary Fig. S13A at JXB online). With a rapid increase in aril breakdown index, the DlERF1 transcript was first increased, especially at days 35–42 of storage, then decreased at day 45 (Fig. 4B; Supplementary Fig. S13B). However, the DlERF2 transcript decreased during the first 2–15 d, then increased and finally reached a plateau during 20–45 d of storage (Fig. 4B; Supplementary Fig. S13C).
Fig. 4.
Changes in the aril breakdown index (A), and DlHD2, DlERF1 and DlERF2 mRNAs (B) in aril tissues of longan fruit stored at low temperature (4 °C) for 45 d. In A, each value represents the means of three replicates, and vertical bars indicate the SE. Different letters indicate a statistical difference at the 5% level among data groups according to the Duncan's multiple range test. In B, total RNA (10 μg per lane) was used for RNA gel blot analysis and hybridization with DIG-labelled probes. Hybridization with DIG-labelled DlFeFOD probes and ethidium bromide-stained rRNA are shown as the internal loading controls.
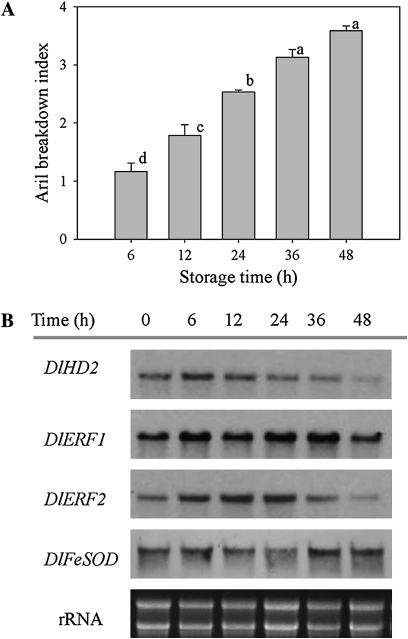
The aril breakdown index increased markedly when fruit were removed from 4 °C to 25 °C. The index increased progressively, and then reached a maximum level at 48 h (Fig. 5A). DlHD2 transcripts decreased progressively during the whole 48 h of storage, while DlERF1 and DlERF2 transcripts increased, reached their peaks at 36 h and 24 h, respectively, and finally decreased (Fig. 5B; Supplementary Fig. S14).
Fig. 5.
Changes in the aril breakdown index (A), and DlHD2, DlERF1 and DlERF2 mRNAs (B) in aril tissues of longan fruit stored for 40 days at 4 °C and then transferred to 25 °C, and stored for 48 h at this temperature. In A, each value represents the means of three replicates, and vertical bars indicate the SE. Different letters indicate a statistical difference at the 5% level among data groups according to the Duncan's multiple range test. In B, total RNA (10 μg per lane) was used for RNA gel blot analysis and hybridization with DIG-labelled probes. Hybridization with DIG-labelled DlFeFOD probes and ethidium bromide-stained rRNA are shown as the internal loading controls.
Interaction between DlHD2 and DlERF1
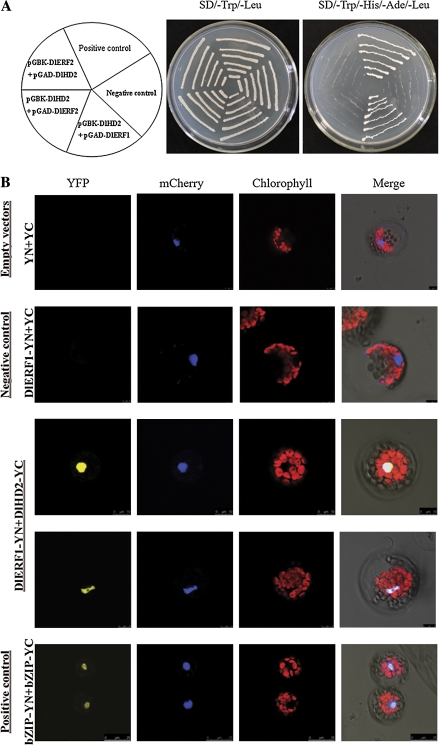
Collectively, based on the expression characteristics of DlHD2, DlERF1, and DlERF2 in which the DlHD2 transcript obviously decreased during fruit senescence (aril breakdown) while the DlERF1 and DlERF2 transcripts increased, it is interesting to analyse the possible interaction between DlHD2 and DlERFs. DlHD2, DlERF1, and DlERF2 coding sequences were subcloned into pGADT7 and pGBKT7 vectors for yeast two-hybrid assay (Supplementary Fig. S3 at JXB online). Since DlERF1 had a transactivation activity in yeast when fused with the BD domain (Fig. 1), DlERF1 was only fused with the AD domain in the yeast two-hybrid analysis. Similar to pGBKT7-53+pGADT7-T (positive control), yeast cells co-transformed with pGBKT7-DlHD2+pGADT7-DlERF1 could grow on selective medium lacking Trp, Leu, His, and Ade in the presence of 5 mM 3-AT (Fig. 6A). In contrast, yeast cells harbouring pGBKT7-Lam+pGADT7-T (negative control), pGBKT7-DlHD2+pGADT7-DlERF2, and pGBKT7-DlERF2+pGADT7-DlHD2 could not grow on the selective medium under the same condition (Fig. 6A). These results indicated that DlHD2 may physically interact with DlERF1.
Fig. 6.
Physical interaction between DlHD2 and DlERF1 detected in yeast two-hybrid assays and in the BiFC system. (A) Interactions between DlHD2 and DlERF1 in the yeast two-hybrid assay. AH109 yeast strains were transformed with plasmids pGBKT7-53+pGADT7-T (positive control), pGBKT7-Lam+pGADT7-T (negative control), pGBKT7-DlHD2+pGADT7-DlERF1, pGBKT7-HD2+pGADT7-DlERF2, or pGBKT7-DlERF2+pGADT7-DlHD2. The ability of yeast cells to grow on synthetic medium lacking tryptophan, leucine, histidine, and adenine, and containing 5 mM 3-amino-1,2,4-triazole (3-AT) was scored as a positive interaction. (B) BiFC visualization of the DlHD2 and DlERF1 interaction in transiently co-expressed Arabidopsis mesophyll protoplasts. DlHD2 protein was fused with the C-terminus of YFP and DlERF1 protein was fused with the N-terminus of YFP. The mCherry-VirD2NLS was included in each transfection to serve as a control for successful transfection as well as for nuclear localization. Empty vectors (pUC-pSPYNE/pUC-pSPYCE) and expression of DlERF1 alone (DlERF1-pSPYNE/pUC-pSPYCE) were used as negative controls, while the Arabidopsis nuclear protein bZIP63 served as the positive control. The length of the bar is indicated in the photographs.
The interaction between DlHD2 and DlERF1 was further tested in plant cells using BiFC. DlERF1 tagged with pSPYNE (split YFP N-terminal fragment expression, Supplementary Fig. S4 at JXB online) and DlHD2 tagged with pSPYCE (split YFP C-terminal fragment expression, Supplementary Fig. S4) were transiently co-expressed in Arabidopsis leaf mesophyll protoplasts by PEG transfection (Walter et al., 2004; Yoo et al., 2007). As shown in Fig. 6B, cells transfected with DlERF1-pSPYNE and DlHD2-pSPYCE, and the positive control exhibited YFP fluorescence. Similar results were also observed when DlHD2-pSPYNE was co-transformed with DlERF1-pSPYCE (Supplementary Fig. S16). In contrast, the combined expression of unfused pSPYNE and pSPYCE, and the expression of DlERF1 alone did not induce any YFP signals. The BiFC results not only demonstrated the in vivo interaction between the two proteins tested but also showed the specific localization of the interacting proteins in the nucleus, which is consistent with the subcellular localization of DlHD2 and DlERF1 in the nuclear compartment (Fig. 2). These results imply that the protein complex of DlHD2 and DlERF1 may function in the nucleus.
Discussion
Characterization of DlHD2 and DlERFs
Plants differ from other eukaryotes in that they possess a new HDAC family, the HD2 family (Lusser et al., 1997; Wu et al., 2003). In this study, a HD2-type HDAC gene, designated as DlHD2, was cloned and characterized from longan aril. Alignment of DlHD2 with HD2 proteins from other organisms showed that it contained all the features of HD2 proteins (Supplementary Fig. S5 at JXB online), including an invariable pentapeptide (MEFWG), an acidic domain, and a single C2H2-type zinc finger domain (Lusser et al., 1997). NCBI Blast revealed that DlHD2 exhibited the highest homology with RcHD2a in castor bean, with 55% amino acid identity. Similar to ZmHD2 (Lusser et al., 1997) and AtHD2A (Earley et al., 2006), DlHD2 was also localized exclusively in the cell nucleoli (Fig. 2, Supplementary Fig. S11), indicating that DlHD2 may be involved in the regulation of rRNA genes (Lusser et al., 1997).
ERF proteins were first identified as TFs, which possess GCC-box binding activity (Ohme-Takagi and Shinshi, 1995; Büttner and Singh, 1997). Although sequence identity can be as low as 13% among the different ERFs, all ERFs exhibited a highly conserved AP2/ERF DNA-binding domain of 57–66 amino acids (Tournier et al., 2003; Cao et al., 2006). Based on the sequence features of the AP2/ERF domain, the ERF family has been categorized into four classes (Fujimoto et al., 2000; Tournier et al., 2003). In this study, two DlERF genes from longan fruit were isolated and characterized. These DlERF genes were found to fall into two different classes of the previously characterized ERF proteins (Fujimoto et al., 2000; Tournier et al., 2003). DlERF1 contains a putative MAP kinase phosphorylation site in the C-terminal region (Supplementary Fig. S9 at JXB online) and therefore belongs to Class III ERFs, which include AtERF5 and AtERF6 from Arabidopsis, and SlERF4 from tomato. AtERF5, AtERF6, and SlERF4 have been shown to function as transcription activators (Fujimoto et al., 2000; Tournier et al., 2003). Similarly, transcriptional activation and subcellular localization analysis also indicated that DlERF1 could function as a transcriptional activator (Figs 1, 2). DlERF2 was categorized as a ‘Class IV’ ERF based on the location of the putative nuclear localization signal (NLS) site and the presence of the conserved N-terminal CMVII-1 motif (MCGGAII/L) (Supplementary Fig. S10) (Tournier et al., 2003). Although the function of the motif is still unknown, it is unlikely to be required for nuclear localization or for binding to the GCC-box (Tournier et al., 2003). Further studies are needed to unravel the function of the ‘Class IV’ ERFs (El-Sharkawy et al., 2009).
Possible roles of DlHD2 and DlERFs in longan fruit senescence
Plant development is an intricate process involving a series of highly organized mechanisms, including both genetic and epigenetic regulation. There is extensive evidence to show that plant HDACs act as global transcriptional regulators to play crucial roles in a range of plant developmental processes (Hollender and Liu, 2008; Chung et al., 2009). It has been reported that the down-regulation of AtHD1 by antisense inhibition or T-DNA insertion in Arabidopsis can induce various developmental defects, including early senescence, serrated leaves, aerial rosettes, defects in floral organ identity, and late flowering (Wu et al., 2000a; Tian and Chen, 2001; Tian et al., 2005). In rice, overexpression of the rice HD1 homologue OsHDAC1-3 in transgenic rice resulted in an increased growth rate and altered architecture (Jang et al., 2003). However, whether epigenetic regulation is involved in fruit senescence remains unknown. In this study, the DlHD2 transcript was highly expressed before fruit senescence but decreased dramatically during senescence (Figs 3–5; Supplementary Figs S12–S14 at JXB online), indicating that DlHD2 might be involved in the regulation of the early stage of longan fruit senescence. Histone modifications including acetylation, methylation, phosporylation, ubiquitination, sumoylation, and ADP-ribosylation are important aspects of epigenetic changes, among which acetylation and deacetylation are well characterized (Xu et al., 2005; Chung et al., 2009; Yaish et al., 2011; Yu et al., 2011). In particular, the lysines residues (K) including K9, K14, K18, K23, K27, and K56 in histone H3 and K5, K8, K12, and K16 in H4, as well as other lysines in histones H2A and H2B, are specifically acetylated or deacetylated by various HATs or HDACs within a given histone (Peng et al., 2008). For example, OsHDAC1 epigenetically repressed the expression of OsNAC6 by deacetylating K9, K14, and K18 in histone H3, and K5, K12, and K16 in histone H4 during seedling root growth in rice (Chung et al., 2009). Recently, functional interplay between Arabidopsis HDAC and demethylase through HDA6 and FLD interaction in flowering control by increased levels of histone H3 acetylation and H3K4 trimethylation in FLC, MAF4, and MAF5 has been reported (Yu et al., 2011). These studies indicate that histone acetylation regulates chromatin structure and gene expression throughout the plant life cycle. In the present work, western blot analysis of histone H3 acetylation showed that levels of histone H3 acetylation in fruit stored at 25 °C increased progressively accompanying the aril breakdown, especially at days 3–5 (Supplementary Fig. S15), which was in parallel with the patterns of change of DlERF1 and DlERF2 (Fig. 3B), whereas it was the opposite of DlHD2 expression (Fig. 3B). In addition, levels of histone H3 acetylation were found to be lower in NO-treated fruit than in control fruit (Supplementary Fig. S15). These results indicate that DlHD2 may be involved in the epigenetic regulation of longan fruit senescence by histone acetylation. Histone H3 acetylation has been regarded as a positive marker of histone modification associated with gene activation (Chen et al., 2010). However, whether DlHD2 might regulate the expression of DlERF1 or DlERF2 by histone acetylation needs to be further elucidated.
The ERF family of TFs, with a highly conserved signature element including an ERF domain responsible for the DNA binding (e.g. the GCC-box and DRE) activity, is essential for plant development and plant responses to different environmental stress factors (Okamuro et al., 1997; Sakuma et al., 2002; Cao et al., 2006; Nakano et al., 2006; Sharma et al., 2010). ERF genes associated with fruit ripening and senescence have been reported. For example, transcripts of tomato LeERF2 (Tournier et al., 2003) and apple MdERF1 (Wang et al., 2007) were accumulated during fruit ripening, and the expression of plum PsERF2a and PsERF2b (El-Sharkawy et al., 2009) was enhanced in flowers after fertilization, while kiwifruit AdERF4 and AdERF6 were markedly stimulated at the later fruit senescence stage (Yin et al., 2010). Similarly, the DlERF1 and DlERF2 transcripts were markedly accumulated at the beginning of longan fruit senescence, and then reached higher levels at the later stage of senescence (Figs 3–5; Supplementary Figs S12–S14 at JXB online). In addition, NO is a simple diatomic free radical gas and can act as a signalling molecule (Guo and Crawford, 2005). Leshem and Pinchasov (2000) suggested that there may be an antagonistic effect of NO against ethylene during fruit maturation and senescence. As shown in Fig. 3, NO pre-treatment could effectively delay fruit senescence and suppress the expression of DlERF1 and DlERF2. These results indicated that DlERF genes were positively involved in regulating longan fruit senescence. ERFs can regulate the target gene expression in the ethylene signal transduction pathway or might relate to cellular wall metabolism by binding to the GCC-box in the promoter region. These target genes in turn may regulate the firmness, aroma, taste, colour, and shelf life of harvested fruit (Nath et al., 2006). Interestingly, AdERF9 significantly suppressed the activity of the AdXET5 promoter, which does not have a GCC-box or DRE (Yin et al., 2010). In tomato, Pti4, an ERF-like TF, could also regulate defence-related gene expression via either GCC- or non-GCC-boxes (Chakravarthy et al., 2003). In previous studies, it was demonstrated that cellular wall-modifying genes such as DlEXP, DlXET, and DlEGase genes were associated with longan aril breakdown (Zhong et al., 2008; Xiao et al., 2009). Further research is required to determine whether DlERF genes are involved in fruit senescence via regulating DlEXP, DlXET, or DlEGase genes
Interaction of DlHD2 with DlERF1 in longan fruit senescence
Both histone acetylation and deacetylation are promoter dependent, locus specific, and reversible, which may therefore provide a versatile way to regulate gene expression during plant development or the plant response to environmental stimuli (Tian et al., 2005; Yu et al., 2011). It is generally accepted that HATs can be recruited through transcriptional activators whereas the HDACs interact with transcriptional repressors. For instance, Arabidopsis HAT protein could interact with the transcriptional activator CBF1 to modulate cold-regulated gene expression (Stockinger et al., 2001). On the other hand, the Class I RPD3 family of HDACs, HDA19, may act in a protein complex with AtERF7, a transcription repressor (Song et al., 2005), to regulate abiotic stress response genes. It was found that AtERF7 interacted with the Arabidopsis homologue of a human global co-repressor of transcription, AtSin3 (Silverstein and Ekwall, 2005), which, in turn, may interact with HDA19, indicating that AtERF7, AtSin3, and HDA19 can form a transcriptional repressor complex to regulate the ABA- and drought-related response in Arabidopsis (Song et al., 2005). HDACs seem to interact specifically with transcriptional repressors, but they also interact with certain transcriptional activators. For example, Arabidopsis HDA19 interacted with the transcriptional activators WRKY38 and WRKY62 (Kim et al., 2008). Overexpression of HDA19 specifically prevented activation of transcription by WRKY38 and WRKY62 and enhanced plant resistance to pathogens (Kim et al., 2008). Interestingly, it was also found that longan DlHD2 played a role in regulating longan fruit senescence opposite to those of DlERF1 and DlERF2 in the present study. Moreover, DlHD2 could interact with the transcriptional activator DlERF1 (Fig. 6; Supplementary Fig. S16 at JXB online), which suggested that they might act in the same protein complex to regulate gene expression involved in fruit senescence. Further experiments such as pull-down and co-immunoprecipitation assays will be needed to confirm the interaction of DlHD2 with DlERF1. More importantly, further studies are required to assess the biological significance of the interaction between DlHD2 and DlERF1.
In summary, the longan fruit DlHD2 and DlERF genes were isolated and characterized. Moreover, DlHD2 and DlERF genes were differentially expressed during longan fruit senescence. Protein–protein interaction analysis indicated that DlHD2 physically interacted with DlERF1, suggesting that they may act together to regulate gene expression involved in fruit senescence. To the best of our knowledge, this is the first report on the interaction between a HD2-type HDAC and an ERF TF. These results suggested the involvement of the epigenetic regulation of gene expression during longan fruit senescence.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Schematic maps of the constructs used in the transcriptional activation analysis in yeast cells.
Figure S2. Schematic maps of the constructs used in the subcellular localization analysis.
Figure S3. Schematic maps of bait and prey constructs used in the yeast two-hybrid assay.
Figure S4. Schematic maps of the constructs for the BiFC assay.
Figure S5. Amino acid sequence alignment of the DlHD2 protein with other plant HD2 proteins.
Figure S6. Phylogenetic tree of the deduced amino acid sequences of DlHD2 and other plant HD2s.
Figure S7. Amino acid alignment of the AP2/ERF domain of DlERFs and other ERF proteins.
Figure S8. Phylogenetic analysis of DlERFs with other AP2/ERF proteins.
Figure S9. Amino acid sequence alignment of Group III ERFs.
Figure S10. Amino acid sequence alignment of Group IV ERFs.
Figure S11. Subcellular localization of DlHD2 in Arabidopsis mesophyll protoplasts.
Figure S12. Relative quantification of DlHD2 (A), DlERF1 (B), and DlERF2 (C) in aril tissues of control and NO-treated longan fruit stored at room temperature.
Figure S13. Relative quantification of DlHD2 (A), DlERF1 (B), and DlERF2 (C) in aril tissues of longan fruit stored at low temperature.
Figure S14. Relative quantification of DlHD2 (A), DlERF1 (B), and DlERF2 (C) in aril tissues of longan fruit stored for 40 d at 4 °C and then transferred to 25 °C.
Figure S15. Western blot analysis of histone H3 acetylation levels in aril tissues of control and NO-treated longan fruit stored at room temperature (25 °C) for 5 d.
Figure S16. BiFC visualization of the DlHD2 and DlERF1 interaction in transiently co-expressed Arabidopsis mesophyll protoplasts.
(see Table S1. Primer sequences used for cloning DlHD2, DlERF1, and DlERF2.
(see Table S2. Primer sequences used for subcloning into pGBK-T7.
(see Table S3. Primer sequences used for fusing GFP.
(see Table S4. Primer sequences used for synthesis of DIG-labelled probes for northern blotting.
(see Table S5. Primer sequences used for yeast two-hybrid and BiFC assays.
Acknowledgments
The authors thank Professor Jörg Kudla (Institut für Biologie und Biotechnologie der Pflanzen, Universität Münster) and Professor Da-peng Zhang (School of Life Science, Tsinghua University) for the gift of BiFC vectors. We are grateful to Dr Jun-xian He (Biology Programme, The Chinese University of Hong Kong) for his helpful discussions and critical English language editing. This work was supported in part by the Guangdong Modern Agricultural Industry Technology System (LNSG2010-12) and the Guangdong Science Foundation (06200670).
Glossary
Abbreviations
- 3-AT
3-amino-1,2,4-triazole
- ACO
1-aminocyclopropane-1-carboxylic acid oxidase
- ACS
1-aminocyclopropane-1-carboxylic acid synthase
- BiFC
bimolecular fluorescence complementation
- DRE
dehydration-responsive element
- EGase
endo-β-1,4-glucanase
- ERF
ethylene-responsive factor
- EXP
expansin
- GFP
green fluorescent protein
- HAT
histone acetyltransferase
- HD2
histone deacetylase 2
- HDAC
histone deacetylase
- NO
nitric oxide
- PEG
polyethylene glycol
- RPD3
reduced potassium dependency 3
- SIR2
silent information regulator 2
- TF
transcription factor
- XET
xyloglucan endotransglucosylase
- YFP
yellow fluorescent protein
References
- Aharoni A, Dixit S, Jetter R, Thoenes E, van Arkel G, Pereira A. The SHINE clade of AP2 domain transcription factors activates wax biosynthesis, alters cuticle properties, and confers drought tolerance when overexpressed in Arabidopsis. The Plant Cell. 2004;16:2463–2480. doi: 10.1105/tpc.104.022897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Solano R, Wisman E, Ferrar S, Ausubel FM, Ecker JR. Five components of the ethylene response pathway identified in a screen for weak ethylene-insensitive mutants in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2003;100:2992–2997. doi: 10.1073/pnas.0438070100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Second family of histone deacetylases. Science. 1998;280:1167. [Google Scholar]
- Berger SL. Histone modification in transcriptional regulation. Current Opinion in Genetics and Development. 2002;12:142–148. doi: 10.1016/s0959-437x(02)00279-4. [DOI] [PubMed] [Google Scholar]
- Boutilier K, Ovringa R, Sharma VK, et al. Ectopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. The Plant Cell. 2002;14:1737–1749. doi: 10.1105/tpc.001941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowyer MC, Wills RBH, Badiyan D, Ku VVV. Extending the postharvest life of carnations with nitric oxide—comparison of fumigation and in vivo delivery. Postharvest Biology and Technology. 2003;30:281–286. [Google Scholar]
- Büttner M, Singh KB. Arabidopsis thaliana ethylene responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proceedings of the National Academy of Sciences, USA. 1997;94:5961–5966. doi: 10.1073/pnas.94.11.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Song F, Goodman RM, Zheng Z. Molecular characterization of four rice genes encoding ethylene-responsive transcriptional factors and their expressions in response to biotic and abiotic stress. Journal of Plant Physiology. 2006;163:1167–1178. doi: 10.1016/j.jplph.2005.11.004. [DOI] [PubMed] [Google Scholar]
- Chakravarthy S, Tuori RP, D'Ascenzo MD, Fobert PR, Despres C, Martin GB. The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. The Plant Cell. 2003;15:3033–3050. doi: 10.1105/tpc.017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LT, Luo M, Wang YY, Wu KQ. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. Journal of Experimental Botany. 2010;61:3345–3353. doi: 10.1093/jxb/erq154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Tian L. Roles of dynamic and reversible histone acetylation in plant development and polyploidy. Biochimica et Biophysica Acta. 2007;1769:295–307. doi: 10.1016/j.bbaexp.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung PJ, Kim YS, Jeong JS, Park SH, Nahm BH, Kim JK. The histone deacetylase OsHDAC1 epigenetically regulates the OsNAC6 gene that controls seedling root growth in rice. The Plant Journal. 2009;59:764–776. doi: 10.1111/j.1365-313X.2009.03908.x. [DOI] [PubMed] [Google Scholar]
- Dangl M, Brosch G, Haas H, Loidl P, Lusser A. Comparative analysis of HD2 type histone deacetylases in higher plants. Planta. 2001;213:280–285. doi: 10.1007/s004250000506. [DOI] [PubMed] [Google Scholar]
- Demetriou K, Kapazoglou A, Tondelli A, Francia E, Stanca MA, Bladenopoulos K, Tsaftaris AS. Epigenetic chromatin modifiers in barley: I. Cloning, mapping and expression analysis of the plant specific HD2 family of histone deacetylases from barley, during seed development and after hormonal treatment. Physiologia Plantarum. 2009;136:358–368. doi: 10.1111/j.1399-3054.2009.01236.x. [DOI] [PubMed] [Google Scholar]
- Depège-Fargeix N, Javelle M, Chambrier P, Frangne N, Gerentes D, Perez P, Rogowsky PM, Vernoud V. Functional characterization of the HD-ZIP IV transcription factor OCL1 from maize. Journal of Experimental Botany. 2011;62:293–305. doi: 10.1093/jxb/erq267. [DOI] [PubMed] [Google Scholar]
- Duan XW, Su XG, You YL, Qu HX, Li YB, Jiang YM. Effect of nitric oxide on pericarp browning of harvested longan fruit in relation to phenolic metabolism. Food Chemistry. 2007;104:571–574. [Google Scholar]
- Earley K, Lawrence RJ, Pontes O, Reuther R, Enciso AJ, Silva M, Neves N, Gross M, Viegas W, Pikaard CS. Erasure of histone acetylation by Arabidopsis HDA6 mediates large-scale gene silencing in nucleolar dominance. Genes and Development. 2006;20:1283–1293. doi: 10.1101/gad.1417706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Betzner AS, Huttner E, Oakes MP, Tucker WQ, Gerentes D, Perez P, Smyth DR. AINTEGUMENTA, an APETALA2-like gene of Arabidopsis with pleiotropic roles in ovule development and floral organ growth. The Plant Cell. 1996;8:155–168. doi: 10.1105/tpc.8.2.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sharkawy I, Sherif S, Mila I, Bouzayen M, Jayasankar S. Molecular characterization of seven genes encoding ethylene-responsive transcriptional factors during plum fruit development and ripening. Journal of Experimental Botany. 2009;60:907–922. doi: 10.1093/jxb/ern354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Talagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. The Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour SJ, Sebolt AM, Salazar MP, Everard JD, Thomashow MF. Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiology. 2000;124:1854–1865. doi: 10.1104/pp.124.4.1854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo FQ, Crawford NM. Arabidopsis nitric oxide synthase1 is targeted to mitochondria and protects against oxidative damage and dark-induced senescence. The Plant Cell. 2005;17:3436–3450. doi: 10.1105/tpc.105.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao D, Ohme-Takagi M, Sarai A. Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plants. Journal of Biological Chemistry. 1998;273:26857–26861. doi: 10.1074/jbc.273.41.26857. [DOI] [PubMed] [Google Scholar]
- Hollender C, Liu Z. Histone deacetylase genes in Arabidopsis development. Journal of Integrative Plant Biology. 2008;50:875–885. doi: 10.1111/j.1744-7909.2008.00704.x. [DOI] [PubMed] [Google Scholar]
- Hu YX, Wang YX, Liu XF, Li JY. Arabidopsis RAVI is down-regulated by brassinosteroid and may act as a negative regulator during plant development. Cell Research. 2004;14:8–15. doi: 10.1038/sj.cr.7290197. [DOI] [PubMed] [Google Scholar]
- Jang IC, Pahk YM, Song SI, Kwon HJ, Nahm BH, Kim JK. Structure and expression of the rice class-I type histone deacetylase genes OsHDAC1-3: OsHDAC1 overexpression in transgenic plants leads to increased growth rate and altered architecture. The Plant Journal. 2003;33:531–541. doi: 10.1046/j.1365-313x.2003.01650.x. [DOI] [PubMed] [Google Scholar]
- Jiang YM, Zhang ZQ, Joyce DC, Ketsa S. Postharvest biology and handling of longan fruit (Dimocarpus longan Lour.) Postharvest Biololgy and Technology. 2002;26:241–252. [Google Scholar]
- Jofuku KD, Boer BGW, Van Montagu V, Okamuro JK. Control of Arabidopsis flower and seed development by the homeotic gene. APETALA2. The Plant Cell. 1994;6:1211–1225. doi: 10.1105/tpc.6.9.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KC, Lai Z, Fan B, Chen Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with Histone Deacetylase 19 in basal defense. The Plant Cell. 2008;20:2357–2371. doi: 10.1105/tpc.107.055566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagaće M, Chantha SC, Major G, Matton DP Fertilization induces strong accumulation of a histone deacetylase (HD2) and of other chromatin-remodeling proteins in restricted areas of the ovules. Plant Molecular Biology. 2003;53:759–769. doi: 10.1023/B:PLAN.0000023665.36676.89. [DOI] [PubMed] [Google Scholar]
- Lee LY, Fang MJ, Kuang LY, Gelvin SB. Vectors for multi-color bimolecular fluorescence complementation to investigate protein–protein interactions in living plant cells. Plant Methods. 2008;4:24. doi: 10.1186/1746-4811-4-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leshem YY, Haramaty E. The characterization and contrasting effects of the nitric oxide free radical in vegetative stress and senescence of Pisum sativum Linn. foliage. Journal of Plant Physiology. 1996;148:258–263. [Google Scholar]
- Leshem YY, Pinchasov Y. Non-invasive photoacoustic spectroscopic determination of relative endogenous nitric oxide and ethylene content stoichiometry during the ripening of strawberries Fragaria anannasa (Duch.) and avocados. Persea Americana (Mill.).Journal of Experimental Botany. 2000;51:1471–1473. [PubMed] [Google Scholar]
- Li YC, Zhu BZ, Xu WT, Zhu HL, Chen AJ, Xie YH, Shao Y, Luo YB. LeERF1 positively modulated ethylene triple response on etiolated seedling, plant development and fruit ripening and softening in tomato. Plant Cell Reports. 2007;26:1999–2008. doi: 10.1007/s00299-007-0394-8. [DOI] [PubMed] [Google Scholar]
- Lin YL, Lai ZX. Reference gene selection for qPCR analysis during somatic embryogenesis in longan tree. Plant Science. 2010;178:359–365. [Google Scholar]
- Liu Q, Kasuga M, Sakuma Y, Abe H, Miura S, Yamaguchi-Shinozaki K, Shinozaki K. Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. The Plant Cell. 1998;10:1391–1406. doi: 10.1105/tpc.10.8.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusser A, Brosch G, Loidl A, Haas H, Loidl P. Identification of maize histone deacetylase HD2 as an acidic nucleolar phosphoprotein. Science. 1997;277:88–91. doi: 10.1126/science.277.5322.88. [DOI] [PubMed] [Google Scholar]
- Nakano T, Suzuki K, Fujimura T, Shinshi H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiology. 2006;140:411–432. doi: 10.1104/pp.105.073783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nath P, Trivedi PK, Sane VA, Sane AP. Role of ethylene in fruit ripening. In: Khan NA, editor. Ethylene action in plants. Berlin: Springer-Verlag; 2006. pp. 151–184. [Google Scholar]
- Ohem-Takagi M, Shinshi H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. The Plant Cell. 1995;7:173–182. doi: 10.1105/tpc.7.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 1997;94:7076–7081. doi: 10.1073/pnas.94.13.7076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey R, Muller A, Napoli CA, Selinger DA, Pikaard CS, Richards EJ, Bender J, Mount DW, Jorgensen RA. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Research. 2002;30:5036–5055. doi: 10.1093/nar/gkf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W, Togawa C, Zhang K, Kurdistani SK. Regulators of cellular levels of histone acetylation in. Saccharomyces cerevisiae. Genetics. 2008;179:277–289. doi: 10.1534/genetics.107.085068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashotte AM, Mason MM, Hutchison CE, Ferreria FJ, Schaller GE, Kieber JJ. A subset of Arabidopsis AP2 transcription factors mediate cytokinin responses in concert with a two-component pathway. Proceedings of the National Academy of Sciences, USA. 2006;103:11081–11085. doi: 10.1073/pnas.0602038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochemical Biophysical Research Communications. 2002;290:998–1009. doi: 10.1006/bbrc.2001.6299. [DOI] [PubMed] [Google Scholar]
- Sharma MK, Kumar R, Solanke AU, Sharma R, Tyagi AK, Sharma AK. Identification, phylogeny, and transcript profiling of ERF family genes during development and abiotic stress treatments in tomato. Molecular Genetics and Genomics. 2010;284:455–475. doi: 10.1007/s00438-010-0580-1. [DOI] [PubMed] [Google Scholar]
- Silverstein RA, Ekwall K. Sin3: a flexible regulator of global gene expression and genome stability. Current Genetics. 2005;47:1–17. doi: 10.1007/s00294-004-0541-5. [DOI] [PubMed] [Google Scholar]
- Song CP, Agarwal M, Ohta M, GuoY Halfter U, Wang P, Zhu JK. Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in ABA and drought stress responses. The Plant Cell. 2005;17:2384–2396. doi: 10.1105/tpc.105.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Wu K, Dhaubhadel S, An L, Tian L. Arabidopsis DNA methyltransferase AtDNMT2 associates with histone deacetylase AtHD2s activity. Biochemical and Biophysical Research Communications. 2010;396:187–192. doi: 10.1016/j.bbrc.2010.03.119. [DOI] [PubMed] [Google Scholar]
- Sozzi GO, Trinchero GF, Fraschina AA. Delayed ripening of ‘Bartlet’ pears treated with nitric oxide. Journal of Horticultural Science and Biotechnology. 2003;78:899–903. [Google Scholar]
- Sridha S, Wu K. Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. The Plant Journal. 2006;46:124–133. doi: 10.1111/j.1365-313X.2006.02678.x. [DOI] [PubMed] [Google Scholar]
- Stockinger EJ, Mao Y, Regier MK, Triezenberg SJ, Thomashow MF. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Research. 2001;29:1524–1533. doi: 10.1093/nar/29.7.1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- Tian L, Chen ZJ. Blocking histone deacetylation in Arabidopsis induces pleiotropic effects on plant gene regulation and development. Proceedings of the National Academy of Sciences, USA. 2001;98:200–205. doi: 10.1073/pnas.011347998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Fong MP, Wang JJ, Wei NE, Jiang H, Doerge RW, Chen ZJ. Reversible histone acetylation and deacetylation mediate genomewide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics. 2005;169:337–345. doi: 10.1534/genetics.104.033142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournier B, Sanchez-Ballesta MT, Jones B, Pesquet E, Regad F, Latché A, Pech JC, Bouzayen M. New members of the tomato ERF family show specific expression pattern and diverse DNA-binding capacity to the GCC box element. FEBS Letters. 2003;550:149–154. doi: 10.1016/s0014-5793(03)00757-9. [DOI] [PubMed] [Google Scholar]
- Turner BM. Histone acetylation and an epigenetic code. Bioessays. 2000;22:836–845. doi: 10.1002/1521-1878(200009)22:9<836::AID-BIES9>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Ueno Y, Ishikawa T, Watanabe K, Terakura S, Iwakawa H, Okada K, Machida C, Machida Y. Histone deacetylases and ASYMMETRIC LEAVES2 are involved in the establishment of polarity in leaves of Arabidopsis. The Plant Cell. 2007;19:445–457. doi: 10.1105/tpc.106.042325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachonasios KE, Thomashow MF, Triezenberg SJ. Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. The Plant Cell. 2003;15:626–638. doi: 10.1105/tpc.007922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schutze K, et al. Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. The Plant Journal. 2004;40:428–438. doi: 10.1111/j.1365-313X.2004.02219.x. [DOI] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA. A modified hot borate method significantly enhances the yield of high quality RNA from cotton (Gossypium hirsutum L.) Analytical Biochemistry. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- Wang A, Tan D, Takahashi A, Li TZ, Harada T. MdERFs, two ethylene-response factors involved in apple fruit ripening. Journal of Experimental Botany. 2007;58:3743–3748. doi: 10.1093/jxb/erm224. [DOI] [PubMed] [Google Scholar]
- Wills RBH, Ku VVV, Leshem YY. Fumigation with nitric oxide to extend the postharvest life of strawberries. Postharvest Biology and Technology. 2000;18:75–79. [Google Scholar]
- Wu K, Malik K, Tian L, Brown D, Miki B. Functional analysis of a RPD3 histone deacetylase homologue in. Arabidopsis thaliana. Plant Molecular Biology. 2000a;44:167–176. doi: 10.1023/a:1006498413543. [DOI] [PubMed] [Google Scholar]
- Wu K, Tian L, Malik K, Brown D, Miki B. Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. The Plant Journal. 2000b;22:19–27. doi: 10.1046/j.1365-313x.2000.00711.x. [DOI] [PubMed] [Google Scholar]
- Wu K, Tian L, Zhou C, Brown D, Miki B. Repression of gene expression by Arabidopsis HD2 histone deacetylases. The Plant Journal. 2003;34:241–247. doi: 10.1046/j.1365-313x.2003.01714.x. [DOI] [PubMed] [Google Scholar]
- Wu KQ, Tian LN, Hollingworth J, Brown DCW, Miki B. Functional analysis of tomato Pti4 in Arabidopsis. Plant Physiology. 2002;128:30–37. [PMC free article] [PubMed] [Google Scholar]
- Xiao R, Chen J, Chen J, Ou M, Jiang Y, Lin H, Lu W. Expression analysis of endo-1,4-β-glucanase genes during aril breakdown of harvested longan fruit. Journal of the Science of Food and Agriculture. 2009;89:1129–1136. [Google Scholar]
- Xu CR, Liu C, Wang YL, Li LC, Chen WQ, Xu ZH, Bai SN. Histone acetylation affects expression of cellular patterning genes in the Arabidopsis root epidermis. Proceedings of the National Academy of Sciences, USA. 2005;102:14469–14474. doi: 10.1073/pnas.0503143102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Seto E. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Current Opinion in Genetics and Development. 2003;13:143–153. doi: 10.1016/s0959-437x(03)00015-7. [DOI] [PubMed] [Google Scholar]
- Yaish MW, Colasanti J, Rothstein SJ. The role of epigenetic processes in controlling flowering time in plants exposed to stress. Journal of Experimental Botany. 2011;62:3727–3735. doi: 10.1093/jxb/err177. [DOI] [PubMed] [Google Scholar]
- Yin X, Allan AC, Chen K, Ferguson IB. Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiology. 2010;153:1280–1292. doi: 10.1104/pp.110.157081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nature Protocols. 2007;2:1565–1572. doi: 10.1038/nprot.2007.199. [DOI] [PubMed] [Google Scholar]
- Yu CW, Liu X, Luo M, Chen C, Lin X, Tian G, Lu Q, Cui Y, Wu K. HDA6 interacts with FLD and regulates flowering in. Arabidopsis. Plant Physiology. 2011;156:173–184. doi: 10.1104/pp.111.174417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang ZJ, Zhang HW, Quan RD, Wang XC, Huang RF. Transcriptional regulation of ethylene response factor LeERF2 in the expression of ethylene biosynthesis genes controls ethylene production in tomato and tobacco. Plant Physiology. 2009;150:365–377. doi: 10.1104/pp.109.135830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong YX, Chen JY, Feng HL, Kuang JF, Xiao R, Ou M, Xie H, Lu WJ. Transcriptional analysis of expansin and XET genes associated with aril breakdown in harvested longan fruit. Journal of the American Society for Horticultural Science. 2008;133:462–467. [Google Scholar]
- Zhou C, Labbe H, Sridha S, Wang L, Tian L, Latoszek-Green M, Yang Z, Brown D, Miki B, Wu K. Expression and function of HD2-type histone deacetylases in Arabidopsis development. The Plant Journal. 2004;38:715–724. doi: 10.1111/j.1365-313X.2004.02083.x. [DOI] [PubMed] [Google Scholar]
- Zhu Q, Zhang J, Gao X, Tong J, Xiao L, Li W, Zhang H. The Arabidopsis AP2/ERF transcription factor RAP2.6 participates in ABA, salt and osmotic stress responses. Gene. 2010;457:1–12. doi: 10.1016/j.gene.2010.02.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.