Abstract
Glutaredoxins (GRXs) belong to the antioxidant and signalling network involved in the cellular response to oxidative stress in bacterial and eukaryotic cells. In spite of the high number of GRX genes in plant genomes, the biological functions and physiological roles of most of them remain unknown. Here the functional characterization of the Arabidopsis GRXS13 gene (At1g03850), that codes for two CC-type GRX isoforms, is reported. The transcript variant coding for the GRXS13.2 isoform is predominantly expressed under basal conditions and is the isoform that is induced by photooxidative stress. Transgenic lines where the GRXS13 gene has been knocked down show increased basal levels of superoxide radicals and reduced plant growth. These lines also display reduced tolerance to methyl viologen (MeV) and high light (HL) treatments, both conditions of photooxidative stress characterized by increased production of superoxide ions. Consistently, lines overexpressing the GRXS13.2 variant show reduced MeV- and HL-induced damage. Alterations in GRXS13 expression also affect superoxide levels and the ascorbate/dehydroascorbate ratio after HL-induced stress. These results indicate that GRXS13 gene expression is critical for limiting basal and photooxidative stress-induced reactive oxygen species (ROS) production. Together, these results place GRXS13.2 as a member of the ROS-scavenging/antioxidant network that shows a particularly low functional redundancy in the Arabidopsis GRX family.
Keywords: Arabidopsis, glutaredoxin, GRXS13, high light stress, methyl viologen, oxidative stress response, photooxidative stress
Introduction
A common feature of plant responses to environmental stress is an oxidative burst of reactive oxygen species (ROS) in different cellular compartments, mainly chloroplasts, peroxisomes, and the apoplastic space (Gechev et al., 2006; Holuigue et al., 2007; Miller et al., 2007). This oxidative burst, together with the metabolic reprogramming that occurs in response to stress, alters the cellular redox equilibrium producing oxidative stress (Miller et al., 2007; Foyer and Noctor, 2009). The augmented ROS levels are sensed and controlled by a battery of ROS-scavenging systems, antioxidant buffers such as glutathione (GSH) and ascorbate (ASC), and oxidoreductases such as glutaredoxins (GRXs), thioredoxins, peroxiredoxins (PRXs), and peroxidases (Dietz et al., 2006; Gechev et al., 2006; Rouhier et al., 2008b; Foyer and Noctor, 2011).
GRXs have been shown to play antioxidant roles in the cellular response to oxidative stress in bacteria, yeast, and mammals (Meyer et al., 2009). In contrast, a role for GRXs in plant responses to oxidative stress has not been well established yet. GRXs are small disulphide oxidoreductases that catalyse the reduction of disulphide bridges and protein–GSH adducts (S-glutathionylated proteins) using the reducing power of GSH, which is constantly regenerated by an NADPH-dependent GSH reductase system (Rouhier et al., 2008b). S-Glutathionylation can protect proteins’ thiol groups from irreversible inactivation, and it is also a mechanism for redox modification of proteins that alters their biological activity (Meyer et al., 2009). GRXs efficiently catalyse S-deglutathionylation and also seem to catalyse S-glutathionylation of proteins (Gallogly and Mieyal, 2007; Gallogly et al., 2008), thus becoming key enzymes in controlling this protein modification mechanism (Michelet et al., 2006; Rouhier et al., 2008b). Through their disulphide oxidoreductase activity, GRXs play a role in redox signalling and indirectly in ROS scavenging, by reducing and recovering enzymes such as PRXs that detoxify H2O2 (Noguera-Mazon et al., 2006; Rouhier et al., 2008a). Furthermore, it has also been reported that GRXs are able to reduce H2O2 and dehydroascorbate (DHA) directly, contributing to the ROS-scavenging cellular capacity (Zaffagnini et al., 2008; Foyer and Noctor, 2011).
In plants, the family of GRXs is more extensive than in other organisms: 31 members in Arabidopsis, compared with four in Escherichia coli, 6–7 in Saccharomyces cerevisiae, and three in humans (Rouhier et al., 2006). Plant GRXs are named GRXS or GRXC, depending on whether there is an S or C residue at the fourth position of the active site (CxxS or CxxC), and are classified as CC-, CPYC-, and CGFS-type according to the conserved residues at this site (Supplementary Fig. S1 available at JXB online) (Rouhier et al., 2004, 2006). CC-type GRXs (also called ROXY) are exclusive of higher plants and represent the most numerous group (21 members in Arabidopsis; Supplementary Fig. S1) (Rouhier et al., 2008b). The other two types, CPYC (six members) and CGFS (four members), are homologous to the classic dithiolic and monothiolic types of GRXs characterized in prokaryotes and other eukaryotes (Supplementary Fig. S1) (Meyer et al., 2009).
In spite of the high number of GRX genes in plant genomes, little is currently known concerning the functional diversity of this protein family in plants. In Arabidopsis only eight out of the 31 predicted GRXs have been functionally characterized by genetic approaches (indicated by asterisks in Supplementary Fig. S1): three monothiol CGFS-type (GRXS14, GRXS15, and GRXS17), two dithiol CPYC-type (GRXC1 and GRXC2), and only three of the most numerous CC-type (GRXC7/ROXY1, GRXC8/ROXY2 and GRXC9). Concerning the CGFS- and CPYC-type, evidence from genetic studies indicates that GRXS14 and GRXS15, located in chloroplasts and mitochondria, respectively, are involved in protection against protein oxidative damage produced by H2O2 treatment (Cheng et al., 2006; Bandyopadhyay et al., 2008; Cheng, 2008). GRXS17 seems to play a critical role in redox homeostasis and auxin sensing to mediate temperature-dependent post-embryonic growth (Cheng et al., 2011). Also the redundant proteins GRXC1 and GRXC2, localized in the cytoplasm and nucleus, have a vital function at early stages after pollination (Riondet et al., 2011). Interestingly, biochemical evidence shows that in Arabidopsis all monothiol GRXs (GRXS16, GRXS15, GRXS14, and GRXS17) and at least half of the dithiol GRXs (GRXC5, GRXC2, and GRXC1) are able to bind Fe–S clusters and can be implicated in the biogenesis and transfer of these clusters to acceptor proteins (Bandyopadhyay et al., 2008; Rouhier et al., 2010; Couturier et al., 2011; Riondet et al., 2011). Then, Arabidopsis CPYC and CGFS GRXs, like their homologues characterized in prokaryotes and other eukaryotes, are not only involved in redox reactions associated with their thiol reductase activity, but also could act as scaffold and carrier proteins implicated in the Fe–S cluster assembly machinery (Rouhier et al., 2010).
Concerning the CC-type GRXs, only three of the 21 members of this class have been characterized; two of them have a nucleocytoplasmic localization (GRXC7/ROXY1 and GRXC8/ROXY2) and are redundantly involved in flower development (Xing et al., 2005; Xing and Zachgo, 2008; Li et al., 2009); the third GRX (GRXC9) has a nuclear localization and is involved in plant defence responses against pathogens (Ndamukong et al., 2007). Although the function of the remaining 18 CC-type GRXs from Arabidopsis is not known, at least seven of them have been shown to rescue the floral phenotype of the roxy1/roxy2 mutant (Li et al., 2009).
Here the functional characterization of Arabidopsis GRXS13, a gene coding for two CC-type GRX isoforms, is reported. Interestingly, it is shown that knocking down the GRXS13 gene produces a phenotype of deficiency in superoxide radical detoxification. This deficiency is observed under basal conditions and also under photooxidative stress caused by methyl viologen (MeV) treatment and by high light (HL) irradiation. Consistently, overexpression of GRXS13.2, the gene variant that is predominantly expressed in basal conditions and induced by photooxidative stress, enhances protection against stress-induced damage associated with increases in the ASC/DHA ratio. Together, these results indicate that GRXS13 gene expression is critical to limit basal and photooxidative stress-induced ROS production, being crucial for protection against oxidative cellular damage in Arabidopsis. It can also be concluded that GRXS13.2 shows a particularly low functional redundancy in the Arabidopsis GRX family (Meyer et al., 2009).
Materials and methods
Plant materials and growth conditions
Wild-type (WT) and transgenic Arabidopsis thaliana plants were from the Columbia (Col-0) ecotype. Seeds were surface-sterilized, germinated, and grown on one-half Murashige and Skoog (MS) medium with 1% sucrose and 0.8% agar (or 0.27% phytagel agar, Sigma-Aldrich) in a growth chamber (16 h light, 100 μmol m−2 s−1, 22±2 °C).
Plasmid constructs and plant transformation
Arabidopsis lines overexpressing (OE) and silencing (Sil) the GRXS13 gene were obtained by Agrobacterium-mediated transformation using the Gateway™ (Invitrogen) cloning system. To overexpress the GRXS13 gene (At1g03850.2 variant), a 453 bp DNA fragment corresponding to the coding sequence was amplified by PCR using cDNA from salicylic acid-treated Arabidopsis seedlings as a template, and specific primers containing Gateway recombination sites (indicated in bold) (F, CACCATGCAAAAAGCAATTCG; and R, AAGCCATAAAGCCCCAGCTTGTC). The amplified fragment was subcloned into Gateway donor vector (pENTR/SD/D-TOPO), verified by sequencing, and transferred to the destination vector pBADC-myc for the 35S::GRXS13.2-Myc construct. To silence the GRXS13 gene (both variants), a DNA fragment of 219 bp from the first exon was amplified with specific primers containing Gateway recombination sites (indicated in bold) (F, CACCATGCAAAAAGCAATTCG; and R, GCCTCTCCTTGCAAACACCACC). This fragment was cloned into the pENTR/SD/D-TOPO vector and transferred, in both the sense and antisense directions, into the pK7GWIWG2 (II) vector to overexpress a GRXS13-RNAi harpin (35S::GRXS13-RNAi). Binary constructs were verified by sequencing and transferred to Agrobacterium (GV3101 strain), which was used to transform Arabidopsis plants by the floral dip method (Clough and Bent, 1998). Seeds of the T1 generation were selected on one-half MS medium with 50 mg l−1 kanamycin (35S::GRXS13-RNAi lines) or 50 mg l−1 sodium glufosinate (BASTA) (35S::GRXS13.2-Myc lines). Transgene presence was verified by PCR and all lines used were homozygous.
Stress-inducing treatments and gene expression analysis
The basal expression of GRXS13.1 (At1g03850.1) and GRXS13.2 (At1g03850.2) was evaluated in different tissues. For this, samples were obtained from 15-day-old seedlings grown in MS medium, from mature seeds, or from different tissues of 5-week-old plants grown in soil (leaves, stems, roots, and flowers). To evaluate the expression of GRXS13.1, GRXS13.2, GRXC9 (At1g28480), and GST-1 (At1g02930) genes after MeV and HL stress treatments, 15-day-old seedlings grown in a medium with 0.27% phytagel agar and collected at 09:00 h (2 h post-light) were used. For MeV treatments, seedlings were transferred to a 50 μM MeV solution in MS liquid medium (one-half Murashige and Skoog, 1% sucrose) and incubated under standard conditions except for the use of continuous light (100 μmol m−2 s−1). Control samples were incubated in MS medium. A stock solution of 10 mM MeV (Sigma) was freshly prepared in water. For HL stress treatments, seedlings were transferred to MS liquid medium and incubated under HL (1000 μmol m−2 s−1) applied by means of two 500 W halogen lamps (Halux, CE-82 R75). The temperature was controlled at 24±2 °C by a circulating water bath. Control samples were exposed to low light (100 μmol m−2 s−1). After treatment, tissue samples were frozen in liquid nitrogen and stored at –80 °C.
Total RNA was obtained from frozen samples using the TRIzol® Reagent (Invitrogen) according to the manufacturer’s instructions. The TRIzol-based method described by Meng and Feldman (2010) was used to obtain RNA from seeds. cDNA was synthesized from each sample (2 μg of total RNA) with an ImProm II Kit (Promega). qPCR was performed with Sensimix™ Plus SYBR® (Quantace) in a STRATAGENE® MX300P™ thermocycler. YLS8 (At5g08290) was used to normalize GRXS13.1 and GRXS13.2 expression in different tissues (Czechowski et al., 2005). Clathrin (At4g24550) was used to normalize the stress-induced expression of GRXS13.1, GRXS13.2, and GRXC9 genes, while UBQ10 (At4g05320) was used to normalize GST-1 expression. Primer sequences designed for each gene are shown in Supplementary Table S1 at JXB online.
Immunoblot to detect GRXS13-Myc protein
To evaluate the level of expression of the GRXS13.2-Myc protein in Arabidopsis lines expressing the 35S::GRXS13.2-Myc construct, a commercial monoclonal anti c-myc antibody (Invitrogen Co., CA, USA) was used. Total protein extracts were obtained from WT and transgenic lines, by homogenizing 0.5 g of 15-day-old Arabidopsis seedlings in extraction buffer [50 mM TRIS pH 8.0, 0.5 M NaCl, 0.5 mM EDTA, 1 mM dithiothreitol (DTT), 1 mM benzamidine, 0.2 μg ml−1 leupeptin, 1 mM phenylmethylsulphonyl fluoride, and 10% glycerol]. After centrifugation at 22 000 g for 30 min, supernatants were transferred to clean tubes and stored at –80 °C. Protein concentration was determined by the Bradford method, using the BioRad Protein Assay (Bio-Rad Laboratories, Inc., CA, USA).
For immunoblot analysis, protein extracts (30 μg of protein per lane) were separated on 15% SDS–polyacrylamide gels (PAGE) and transferred to Immobilon™ PDVF membranes (Millipore Co., Bedford, MA, USA) by semi-dry electroblotting. The membrane was blocked for 1 h in TRIS-buffered saline (TBS-T: 20 mM TRIS pH 7.5, 150 mM NaCl, and 0.1% Tween-20) with 5% non-fat dried milk at room temperature. To detect the myc-tagged GRXS13.2 protein, the membrane was incubated with the anti-myc antibody (1:200 dilution). After washing in TBS (four times), the membrane was incubated with the anti-mouse horseradish peroxidase-conjugated secondary antibody (1:2500 dilution) (Goat anti-mouse, KPL, USA). After washing in TBS-T (15 min four times), the immunocomplexes were visualized using a chemiluminescence kit (Western Lightning Plus Chemiluminescence Reagent, PerkinElmer) according to the manufacturer’s instructions.
In vivo quantification of superoxide radical levels
To quantify in vivo levels of superoxide radicals in seedlings of WT and transgenic lines, the dihydroethidine (DHE)-based assay developed by Georgiou et al. (2005) was used, with the following modifications. A 100 mg aliquot of 15-day-old seedlings, either control or treated with HL for 12 h, was immersed in 2 ml of 0.1 mM DHE fresh solution for 40 min in the dark. Plant tissue was washed with distilled water, frozen in liquid nitrogen, and homogenized in a mortar with 3 ml of 40 mM TRIS-HCl pH 7.5 buffer. The homogenate was centrifuged for 15 min at 20 600 g. In the clear extract, 2-hydroxy ethidium (2OH-E) the product of DHE oxidation by the superoxide radical, was measured with a spectrofluorometer (Luminescence Spectrometer LS50B, PerkinElmer), using an excitation wavelength of 480 nm and an emission wavelength of 590 nm. Superoxide anion levels were expressed as nanomoles of 2OH-E formed, using the extinction coefficient of 2OH-E ξ=6.4 mM−1 cm−1.
Tolerance to MeV treatments
To evaluate Arabidopsis lines for tolerance to MeV treatment, seeds were surface sterilized, spread on solid MS medium (one-half MS medium, 1% sucrose, and 0.27% phytagel agar) supplemented with 0.25 μM MeV, stratified at 4 °C for 48 h in the dark, and then germinated and grown for 15 d under controlled conditions. Seedlings were scored for survival rates (percentage of green seedlings with respect to the total germinated seedlings) after the treatment.
To evaluate tolerance to MeV in adult plants, a 2 μl drop of a 15 μM MeV solution was placed on the surface of leaves from 5-week-old plants. Plants were then maintained under constant light (100 μmol m−2 s−1) and standard conditions in a growth chamber. After 2 d, necrotic lesions were scored and classified according to a three-point scale as follows: type I, round necrotic area smaller than the treated area; type II, necrosis occupying the treated area or reaching the edges of the treated leaf; and type III, necrosis progressing from the treated area towards the apical part of the leaf. Each point represents the mean value from four independent biological replicas. Each replica considers 4–5 leaves from at least 20 different plants.
Oxidative membrane damage after HL-induced stress
To evaluate the response of Arabidopsis lines to HL stress treatments, the oxidative membrane damage was estimated by the extent of ion leakage. For each sample, two 15-day-old seedlings were floated abaxial side up onto a 12-well plate with 2 ml of MilliQ water, and incubated under HL (1000 μmol m−2 s−1) for different time periods. The conductivity of the bathing solution was then measured at 22 °C with a conductimeter. Seedlings were then returned to the bathing solution, introduced into sealed tubes, and sterilized by autoclaving. After cooling to 22 °C, the conductivity of the bathing solution was measured again and this value was referred to as 100%. For each sample, ion leakage was expressed as percentage leakage referred to its corresponding 100%.
Determination of ASC and DHA levels
Control (100 μmol m−2 s−1, 12 h) and HL-treated (1000 μmol m−2 s−1, 12 h) seedlings (15 d old) were frozen in liquid nitrogen and the levels of ASC and DHA were determined as described (Gillespie and Ainsworth, 2007).
Results
GRXS13.2 is the variant that contributes most to the transcriptional activity of the GRXS13 gene
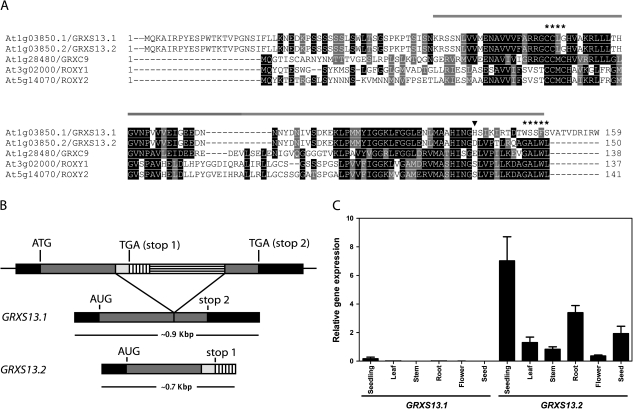
GRXS13 (At1g03850) codes for two 85.5% identical protein isoforms: GRXS13.1 and GRXS13.2 that belong to the group of CC-type GRXs from Arabidopsis (Supplementary Fig. S1 at JXB online) (Rouhier et al., 2004). Alignment of amino acid sequences for the GRXS13 variants and the other previously characterized CC-type GRXs from Arabidopsis (GRXC9, ROXY1, and ROXY2) is shown in Fig. 1A. The closest CC-type GRX that clusters together with the GRXS13 isoforms is GRXC9 (encoded by At1g28480), that shows 42% and 47% identity with GRXS13.1 and GRXS13.2, respectively (Fig. 1A; Supplementary Fig. S1). GRXS13 isoforms are coded by two transcript variants (Fig. 1B) and share their amino acid sequences from the N-terminus up to position 136 (Fig. 1A, arrowhead). This region contains most of the characteristic GRX motifs (amino acids 53–149; http://www.uniprot.org/uniprot/Q84TF4) including the predicted CCLG active site (Fig. 1A, B). These isoforms differ in their C-terminal end; while GRXS13.1 contains a unique 23 amino acid sequence that has no homology with other GRXs, GRXS13.2 has a 14 amino acid sequence that contains the conserved GAL/IWL motif (Fig. 1A, B). This motif was proven to be essential for ROXY1 protein function and is conserved among the CC-type GRXs—including GRXS13.2—that complement ROXY1 function in the roxy1-2 Arabidopsis mutant (Li et al., 2009) (Fig. 1A). The sequence conservation and functional complementation suggest that GRXS13.2 is the predominant isoform. Supporting this hypothesis, analyses of the full-length (FL) cDNAs and expressed sequence tags (ESTs) found for the GRXS13 gene in the Arabidopsis database (TAIR, www.arabidopsis.org) indicate that 87% of the transcripts correspond to the GRXS13.2 variant, while the rest correspond to the GRXS13.1 variant (Supplementary Table S2).
Fig. 1.
The GRXS13 gene codes for two CC-type GRX isoforms, GRXS13.2 being the predominantly expressed isoform in Arabidopsis seedlings. (A) Multiple sequence alignment of amino acid sequences for GRXS13 isoforms and the three previously characterized CC-type GRXs from Arabidopsis (GRXC9, ROXY1, and ROXY2). The glutaredoxin domain (amino acids 53–149; http://www.uniprot.org/uniprot/Q84TF4) is indicated by the line drawn above the sequences; the CCLG active site and the GALWL motif are indicated by asterisks (*); GRXS13.1 and GRXS13.2 are identical up to residue 136 (arrowhead). Black boxes are used to indicate amino acid identity, while grey boxes indicate amino acid similarity. (B) Transcript processing of GRXS13 gene variants. The 23 amino acid C-terminus of GRXS13.1 is encoded by the second exon, while the 14 amino acid C-terminus of GRXS13.2 is encoded by part of the gene intron. (C) Expression pattern of GRXS13 gene variants in Arabidopsis seedlings (15 d old), in leaves, stems, roots, and flowers from adult plants (5 weeks old), and in seeds. GRXS13.1 and GRXS13.2 transcript levels were quantified by qRT-PCR and normalized by clathrin transcript levels. Data are presented as normalized transcript levels and correspond to mean values ±SD from three biological replicas.
To evaluate whether GRXS13.2 is the predominant variant expressed in Arabidopsis tissues, the basal normalized levels of transcripts for both variants were compared using qRT-PCR. The results confirmed that GRXS13.2 is the variant predominantly expressed under basal conditions in all tissues. The levels of GRXS13.2 transcript in these tissues were 2–3 orders of magnitude higher than those of GRXS13.1 (Fig. 1C). The tissues displaying the highest accumulation of GRXS13.2 transcript were seedlings, roots, and seeds (Fig. 1C).
As shown below, GRXS13.2 was responsive to stress treatments while the GRXS13.1 variant was not (Fig. 2). Taking together the experimental results, the relative abundance in FL cDNA and ESTs databases (Supplementary Table S2), and the ability of GRXS13.2 to complement ROXY1 function, it can be argued that GRXS13.2 is the variant that predominantly contributes to the transcriptional activity of the GRXS13 gene.
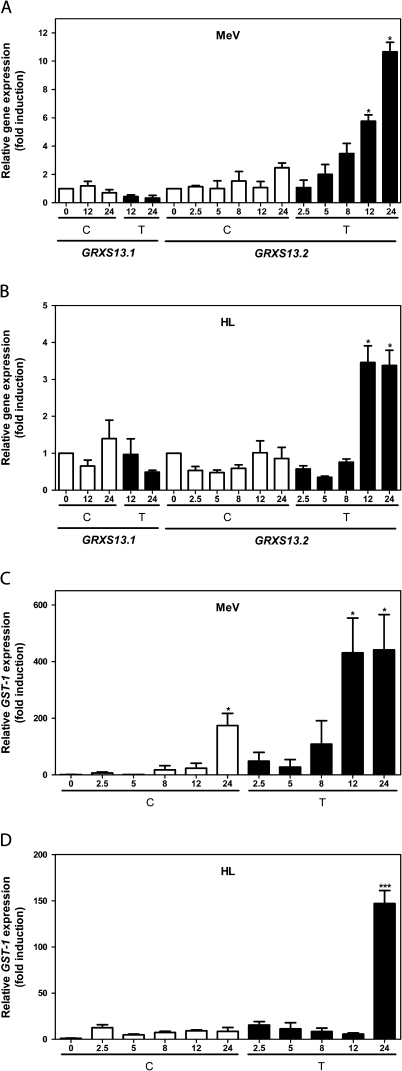
Fig. 2.
The GRXS13.2 gene variant is induced by methyl viologen (MeV) and high light (HL) stress treatments. Transcript levels for GRXS13.1 and GRXS13.2 (A, B) and GST-1 (C, D) were measured using qRT-PCR in 15-day-old Arabidopsis seedlings at different times (in hours) after treatments (black bars) with 50 μM MeV (A, C) or with HL (1000 μmol m−2 s−1) (B, D). Control treatments were performed in MS medium (white bars). Transcript levels of GRXS13.1 and GRXS13.2 variants were normalized by comparison with a reference gene, and the extent of gene induction was calculated with regard to the basal level (time 0) for the corresponding variant. Clathrin was used to normalize GRXS13.1 and GRXS13.2 expression, while UBQ10 was used to normalize GST-1 expression. Data corresponds to mean values ±SD from three biological replicas. Statistical analysis was performed using the paired Mann–Whitney test with the GraphPad Prism 5 program. Values showing statistically significant differences relative to the corresponding WT samples are indicated by asterisks (*P < 0.05 and ***P < 0.001).
The GRXS13.2 gene variant is induced by MeV and HL treatments
To determine whether GRXS13 gene variants are responsive to oxidative stress, Arabidopsis seedlings were treated with 50 μM MeV (1,1'-dimethyl-4,4'-bipyridylium chloride) in the presence of light (100 μmol m−2 s−1), or with HL (1000 μmol m−2 s−1). Both treatments are known to produce photooxidative stress characterized by increased production of superoxide at the photosystem I (PSI) complex. Transcript levels of GRXS13.1 and GRXS13.2 variants were normalized by comparison with a reference gene, and the extent of gene induction was calculated with regard to the basal level (time 0) for the corresponding variant. As shown in Fig. 2A and B, these treatments produced an increase in GRXS13.2 mRNA levels, which reached statistical significance after 12 h of treatment. In contrast, the GRXS13.1 variant was not responsive to these stress treatments (Fig. 2A, B). GST1, a marker gene for oxidative stress (Rentel and Knight, 2004), was used as a positive control to verify that treatments were effective (Fig. 2C, D). These results indicate that the GRXS13.2 variant is responsive to photooxidative stress and support evidence that indicates that this is the main transcriptional active variant of GRXS13.
GRXS13 gene knock down reduces plant growth and increases basal production of superoxide radicals
To carry out a gain–loss functional analysis for the GRXS13.2 gene variant, homozygous Arabidopsis lines that either knock down the GRXS13 gene (Sil lines expressing the 35S::GRXS13-RNAi construct) or overexpress the GRXS13.2 variant (OE lines expressing 35S::GRXS13.2-Myc construct) were generated.
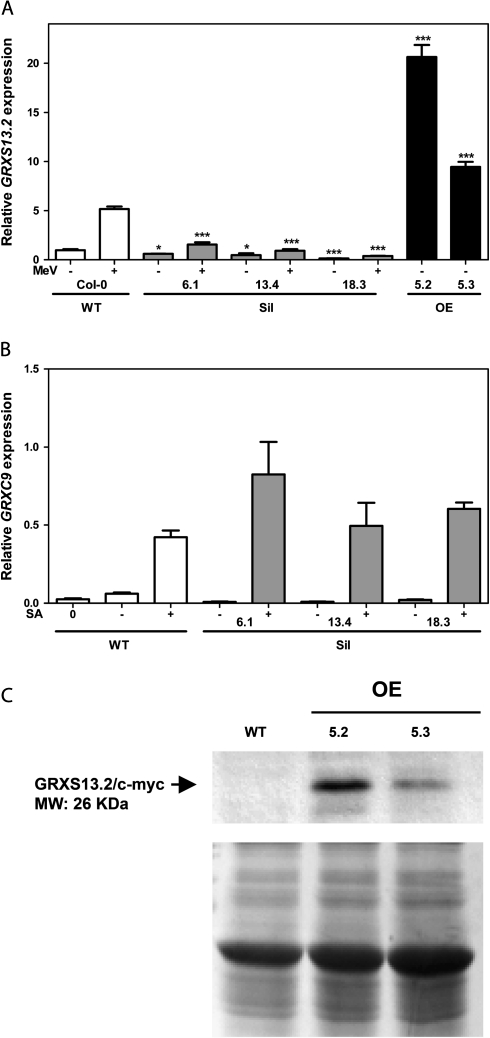
The efficacy of the knocking down of GRXS13.2 gene expression was assessed in the three characterized Sil lines (named 6.1, 13.4, and 18.3) by measuring GRXS13.2 transcript levels in response to MeV. As shown in Fig. 3A, Sil lines showed reduced levels of GRXS13.2 transcript. Basal and MeV-induced levels of this transcript ranged between 13–61% and 7–30%, respectively, of those detected in WT plants (Fig. 3A). To confirm the specificity of the silencing, the activity of GRXC9, the nearest GRX gene that shares 46% sequence identity with the GRXS13 gene in the fragment used for the RNAi (RNA interference) harpin was measured. Because the GRXC9 gene has been described as a salicylic acid (SA)-responsive gene (Ndamukong et al., 2007; Blanco et al., 2009), its activation by SA was studied in the GRXS13-silenced lines. As shown in Fig. 3B, this gene was not silenced in any of the three Sil lines, confirming the specificity of the knocking down procedure.
Fig. 3.
Characterization of Arabidopsis transgenic lines silencing (Sil) and overexpressing (OE) the GRXS13 gene. Homozygous 35S::GRXS13-RNAi lines (Sil lines 6.1, 13.4, and 18.3) and 35S::GRXS13.2-Myc lines (OE lines 5.2 and 5.3) were obtained using the Gateway® cloning technology and Agrobacterium-mediated transformation. Transcript levels of GRXS13.2 (A) and GRXC9 (B) were determined by qRT-PCR in WT (white bars), Sil lines (grey bars), and OE lines (black bars), treated or not with methyl viologen (+/– MeV, 50 μM) for 12 h (A) or salicylic acid (+/– SA, 0.5 mM) for 2.5 h (B). Expression data are indicated as relative levels with respect to the basal level in WT plants and represent mean values ±SD from three biological replicas. Data were analysed by two-way ANOVA with Bonferroni post-test. Values showing statistically significant differences relative to the corresponding WT samples are indicated by asterisks (*P < 0.05 and ***P < 0.001). (C) Immunoblot for GRXS13.2-Myc protein detection in OE lines. A 30 μg aliquot of total protein extracts from WT (Col-0) and two transgenic OE lines (5.2 and 5.3) was loaded on an SDS–polyacrylamide gel, transferred to a membrane, and the GRXS13.2-Myc protein was detected with a commercial monoclonal anti-c-myc antibody (Invitrogen) (upper panel, arrowhead). Coomassie Brilliant Blue staining (lower panel) indicates that equivalent amounts of proteins were loaded.
Two 35S::GRXS13.2-Myc lines that overexpress a C-terminally myc-tagged GRXS13.2 protein (OE lines 5.2 and 5.3) were also characterized. These OE lines show high basal levels of GRXS13.2 transcripts: 4- and 1.8-fold higher compared with those found in MeV-treated WT plants (Fig. 3A). Increased transcript levels correlated with overexpression of the Myc-tagged GRXS13.2 protein, as detected by immunoblot with anti-Myc antibodies (Fig. 3C).
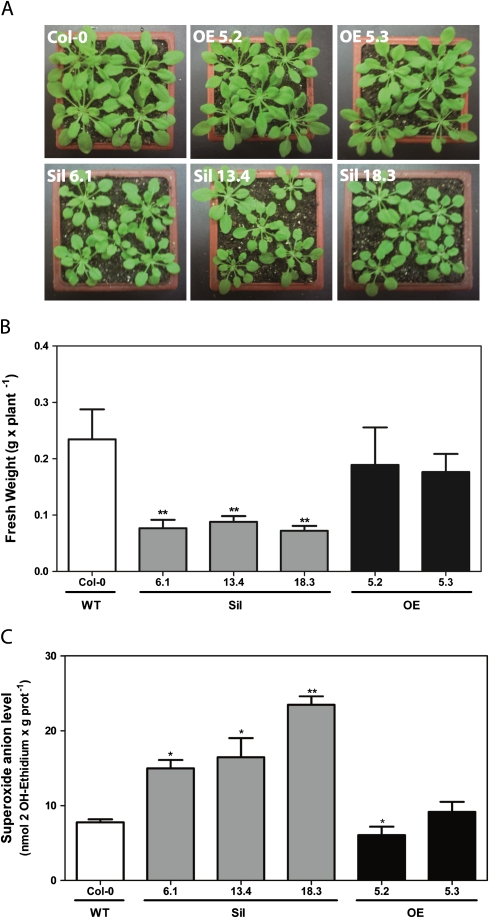
Using the Sil and OE lines, the aim was to determine whether alterations in GRXS13 gene expression produced any effect on plant growth. Six weeks after germination in soil under short-day conditions, the plants obtained from Sil lines showed reduced growth compared with the WT, as determined by fresh weight quantification (Fig. 4A, B). This phenotype is not evidenced under long-day conditions. In contrast, OE plants did not show any significant difference in growth compared with the WT, under either short-day or long-day conditions (Fig. 4A, B).
Fig. 4.
Effect of knocking down and overexpressing the GRXS13 gene on plant growth and superoxide radical basal levels. (A and B) GRXS13 gene knock down reduced plant growth. Plant biomass (determined by fresh weight) was measured in 6-week-old plants from WT (Col-0, white bar), Sil lines (grey bars), and OE lines (black bars) grown under short-day conditions (8 h light). Data represent mean values ±SD from five biological replicas. (C) GRXS13 gene knock down increased basal levels of superoxide ion. In vivo levels of superoxide radical were determined in 15-day-old seedlings from WT (white bar), Sil lines, (grey bars), and OE lines (black bars), through the oxidized fluorescent probe 2-hydroxy ethidium (2OH-E). Data represent mean values ±SD from four biological replicas. Data were analysed by the paired Mann–Whitney test using the GraphPad Prism 5 program. Values showing statistically significant differences relative to the corresponding WT samples are indicated by asterisks (*P <0.05 and **P < 0.01).
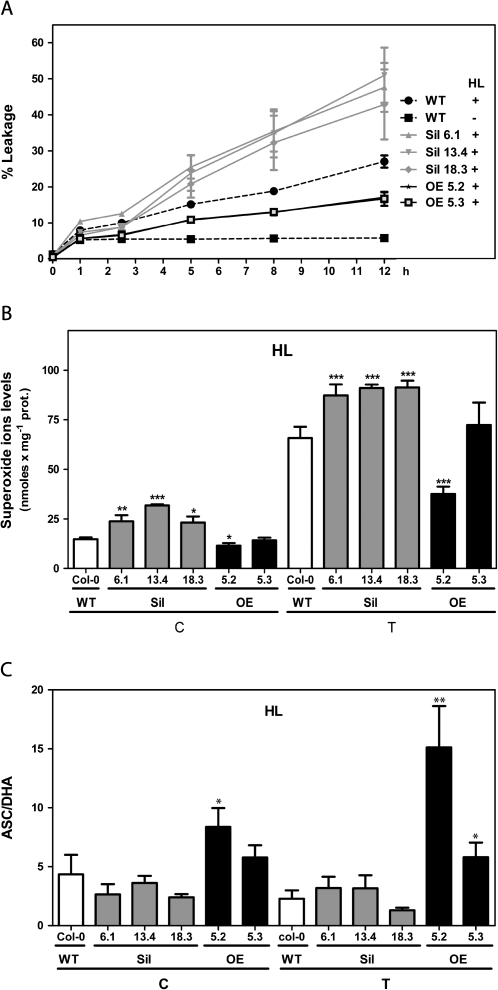
To evaluate whether GRXS13 plays a role in the basal capacity of plants to detoxify ROS, in vivo levels of superoxide radicals were determined in WT, Sil, and OE lines using the fluorescent probe DHE (Georgiou et al., 2005). Interestingly, the three Sil lines showed higher levels of superoxide radicals compared with the WT (Figs 4C, 6C). In contrast, the OE line 5.2 had a significantly lower basal level of superoxide compared with the WT (Figs 4C, 6C). This line displayed the highest GRXS13.2 expression (Fig. 3A).
Fig. 6.
Effect of knocking down and overexpressing GRXS13.2 on oxidative damage, levels of superoxide radicals, and ASC/DHA after HL stress. Arabidopsis seedlings (15 d old) from WT, Sil, and OE lines were treated under HL (T, 1000 μmol m−2 s−1) or under control conditions (C, 100 μmol m−2 s−1) for the indicated periods of time. (A) A time course for the oxidative damage of membranes, evidenced by ion leakage measurements, is shown for WT (dotted black lines), Sil lines (grey lines), and OE lines (black lines). Control treatment for WT plants only is shown (black squares); Sil and OE lines behaved similarly to the WT. Lines for both OE lines are superimposed. Data represent mean values ±SD from four biological replicas. (B and C) The superoxide radical levels (B) and the ASC/DHA ratios (C) were determined in WT (white bar), Sil lines (grey bars), and OE lines (black bars) under control conditions (C) and after 12 h of treatment with HL stress (T). Data represent mean values ±SD from four biological replicas. Statistical analysis was performed using paired Mann–Whitney test with the GraphPad Prism 5 program. Values that show statistically significant differences compared with the corresponding WT samples are indicated by asterisks (*P < 0.05, **P < 0.01, and ***P < 0.001).
The fact that knocking down GRXS13 inhibits plant growth and is associated with increased levels of superoxide radicals suggests that GRXS13 expression is required for normal superoxide detoxification under basal conditions.
The GRXS13 gene plays a role in protection against photooxidative stress
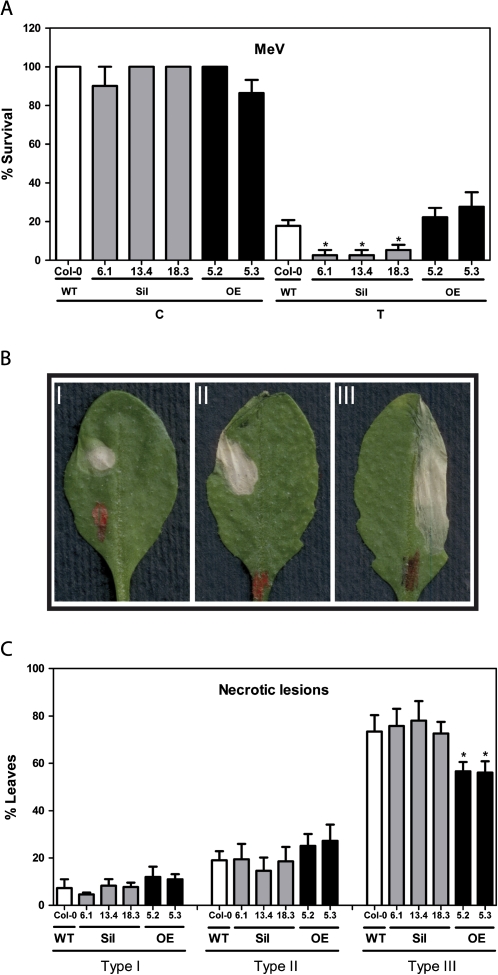
Given the fact that GRXS13.2 is a photooxidative stress-responsive gene required for basal superoxide detoxification, it was hypothesized that its induction is essential for Arabidopsis plants’ tolerance to this type of stress. To assess the role of GRXS13 in the tolerance of plants to MeV treatment, the effect of silencing and overexpressing the gene on seedling survival in a medium supplemented with 0.25 μM MeV was studied. For this purpose, seeds were germinated in this medium and 15 d later the survival rate (percentage of green seedlings with respect to the total germinated seedlings) was calculated. As shown in Fig. 5A (white bars), WT plants showed 82% reduction in survival rate. Interestingly, Sil lines showed a higher reduction in survival (95–97%) after MeV exposure (Fig. 5A, grey bars). OE lines did not show significant differences in survival compared with the WT (Fig. 5A, black bars). The development of tolerance to MeV was also assessed in WT, Sil, and OE lines, by evaluating the development of necrotic lesions in leaves whose surface was exposed to 2 μl of a 15 μM MeV solution. After 2 d of exposure to MeV, lesions were scored according to the extension of the necrotic area. They were classified as type I, II, and III (Fig. 5B, see the Materials and methods) (de Leon et al., 2002). No differences in the severity of lesions between WT and Sil plants were detected. In contrast, OE plants had significantly fewer type III lesions compared with the WT (Fig. 5C). In sum, these results indicate that GRXS13 plays a role in tolerance to photooxidative stress produced by MeV.
Fig. 5.
Effect of knocking down and overexpressing GRXS13.2 on the tolerance to methyl viologen (MeV) treatment. (A) Silencing of the GRXS13 gene reduced the ability of Arabidopsis plants to survive in the presence of MeV. Seeds from WT (white bars), Sil lines (grey bars), and OE lines (black bars) were germinated on MS medium alone (C) or supplemented with 0.25 μM MeV (T) under controlled conditions (16 h light, 100 μmol m−2 s−1, 22±2 °C). At day 4, seedlings were scored for survival rates (percentage of green seedlings with respect to the total germinated seedlings); these scores were compared with the corresponding values of untreated WT seedlings (100%). Data represent mean values ±SD from six biological replicas. (B and C) Overexpressing GRXS13.2 reduces necrotic symptoms after MeV treatment. Analysis of necrotic symptoms that developed in leaves of 5-week-old plants of WT (white bars), Sil lines (grey bars), and OE lines (black bars) after 2 d of MeV treatment (a 2 μl drop of a 15 μM MeV solution). Representative examples of symptoms which developed after MeV treatments are shown in (B). According to the extension of the leaf necrotic area, lesions were scored in a three-point scale I, II, and III, as described in the Materials and methods. (C) The percentage of leaves showing each of the symptoms at day 2 after MeV treatment was scored. Statistical analyses were performed using two-way ANOVA with Bonferroni post-test with the GraphPad Prism 5 program. Values that show statistically significant differences relative to the corresponding WT samples are indicated by an asterisk (P < 0.05).
To assess whether GRXS13 plays a role in protecting plants against HL stress, the oxidative membrane damage in Arabidopsis seedlings from WT, Sil, and OE lines treated with HL (1000 μmol m−2 s−1) was assessed. In WT plants, HL treatment produced a significant increase in ion leakage reaching up to 27% of basal levels after 12 h of treatment (dotted lines in Fig. 6A). Compared with the WT, Sil lines showed a higher increase in ion leakage that reached between 43% and 51% the basal level after 12 h of HL treatment (grey lines in Fig. 6A). In contrast, OE lines showed significantly less ion leakage than the WT (17–18%) after 12 h of treatment (black lines in Fig. 6A). As a control treatment plants were exposed to 100 μmol m−2 s−1 light intensity; this treatment produced a small increase in ion leakage (6–9%) after 12 h of treatment in all lines (Fig. 6A).
Considering the effect of GRXS13.2 expression on the basal levels of superoxide radical (Fig. 4C), the levels of this radical were then measured in WT, Sil, and OE lines subjected to HL stress conditions. As shown in Fig. 6B, after 12 h HL treatment the three Sil lines showed higher levels of superoxide than WT plants. On the other hand, only one of the overexpressor lines (OE line 5.2) displayed reduced levels of superoxide compared with WT plants (Fig. 6B). It was then investigated whether changes in ROS levels correlated with alterations in the levels and redox state of ASC. The ASC/DHA ratio did not differ between WT and Sil plants. In contrast, in OE lines, the ASC/DHA ratio was significantly higher than in WT plants (Fig. 6C). The total levels of ASC+DHA were not significantly different between transgenic lines and WT plants (data not shown).
Taken together, these results indicate that expression of the GRXS13.2 gene variant is required to limit ROS accumulation and therefore to protect tissues from damage induced by photooxidative stress treatments.
Discussion
Plants are continuously exposed to conditions of environmental stress that increase ROS production and may produce cellular oxidative damage. Thus, it is not surprising that compared with bacterial, yeast, and animal systems, plants are equipped with a more diverse and complex battery of ROS scavenger and antioxidant enzymatic systems (Lillig et al., 2008; Rouhier et al., 2008b; Foyer and Noctor, 2011). In this context, the large number of GRX genes in plants, and the few examples of ascribed physiological functions for most of them, particularly for the most numerous CC-type, suggests a considerable functional redundancy. Interestingly, here is reported that GRXS13, a gene belonging to the plant-exclusive CC-type GRX gene family in Arabidopsis, is a key piece of the stress-inducible antioxidant system that protects plants against oxidative damage. GRXS13 shows less functional redundancy than other GRX genes characterized as having this function in other organisms (Meyer et al., 2009). In fact, knocking down GRXS13 is enough to produce a deficient phenotype under basal and photooxidative stress conditions, while overexpressing the predominant gene variant (GRXS13.2) improves stress tolerance by enhancing the plant’s antioxidant capacity.
ROS production and detoxification under photooxidative stress
The photooxidative stress models used in this work lead to increased production of ROS. HL stress treatment mainly increases production of superoxide radicals by photoreduction of molecular oxygen at PSI, but also produces singlet oxygen at PSII and increases H2O2 levels in the peroxisomes by increasing rates of photorespiratory metabolism (Asada, 1999; Krieger-Liszkay et al., 2008). On the other hand, treatment of plants with MeV under light enhances superoxide production at the thylakoids due to autooxidation of MeV radicals, which are generated by electrons coming from PSI (Asada, 2000; Krieger-Liszkay et al., 2011).
Rapid scavenging of ROS produced in the thylakoids, prior to their diffusion from the generation site, is needed to protect target molecules in different cellular compartments. For this purpose and as a first line of defence, plants are equipped with constitutive and inducible chloroplastic isoforms of superoxide dismutase to detoxify superoxide, and enzymes from the ASC- and GSH-dependent systems to detoxify H2O2. These enzymes include ascorbate peroxidases (APXs), glutathione S-transferases (GSTs), GRX, PRXs, and enzymes in charge of regenerating reduced GSH and ASC (Asada, 2006; Dixon et al., 2009; Maruta et al., 2010; Foyer and Noctor, 2011). Interestingly, genes coding for key components of the cytosolic antioxidant/ROS scavenging system—such as APX2, APX1, and CAT1—are also rapidly induced under photooxidative stress, which suggests that they provide a second line of cellular defence (Karpinski et al., 1997). The importance of these chloroplastic and cytosolic ROS detoxification systems working under basal and stressed conditions has been made clear by the growth deficiency and reduced tolerance to photooxidative stress phenotypes reported for mutants and knock down lines for these enzymes (Danna et al., 2003; Rizhsky et al., 2003; Davletova et al., 2005; Tarantino et al., 2005; Myouga et al., 2008; Maruta et al., 2010).
In this context, the functional analysis of the Arabidopsis GRXS13 gene reported here indicates that GRXS13.2 is an important component of the cellular ROS-scavenging/antioxidant system, which is essential for normal growth under short-day conditions and for tolerance to photooxidative stress conditions.
The basal phenotype of GRXS13 knock down lines—detected at the seedling stage where the GRXS13.2 expression level is higher (Fig. 1C)—is characterized by increased superoxide levels, revealing that GRXS13.2 is required to detoxify basal ROS production (Fig. 4C). This deficiency leads to reduced plant growth under short-day conditions, a phenotype that is not detected under long-day conditions (Fig. 4A, B). Interestingly, the same type of phenotype of growth deficiency under short-day but not long-day conditions was detected in a mutant for a rice NADPH thioredoxin reductase (NTRC), an enzyme that uses NADPH to reduce 2-Cys peroxiredoxin, which in turn reduces H2O2 (Perez-Ruiz et al., 2006). This mutant, like the GRXS13 Sil lines characterized here, is deficient in protection against oxidative stress (Perez-Ruiz et al., 2006). The growth phenotype of these lines indicates that the redox function of GRXS13.2, as well as of NTRC, is more important at night than during the day. This could be explained because at night there are no light-driven electrons through ferredoxin for regeneration of ASC and thioredoxins. Instead, ROS detoxifying systems such as GRXs and NTRC—that use the reducing power of NADPH produced by the Calvin cycle—could be active and therefore relatively more important during darkness (Perez-Ruiz et al., 2006).
The results reported here support a role for GRXS13.2 as an important component, not only for basal but also for the stress-inducible ROS-scavenging/antioxidant system. It is shown that the GRXS13.2 gene variant is induced by photooxidative stress produced by treatment with MeV and HL radiation (Fig. 2A, B). Supporting a role for its induction, GRXS13 knock down lines showed increased susceptibility to these stress conditions (Figs 5, 6A). Consistently, overexpression of GRXS13.2 increases tolerance to HL- and MeV-induced damage (Figs 5C, 6A). In the case of MeV tolerance assays, the effect of overexpression was only detected in assays of oxidative damage in mature leaves (Fig. 5B, C) but not in germination assays (Fig. 5A). There is no explanation as yet as to why OE affects the tolerance to oxidative stress in mature leaves while this effect is not detected in germinating seedlings. It could be speculated that, considering that the basal abundance of GRXS13.2 transcript is 5.4 times higher in seedlings that in mature leaves (Fig. 1C), the overexpression must produce a less dramatic change in levels in seedlings than in leaves.
Thus, it can be concluded that the expression of the GRXS13.2 gene variant, induced by stresses that promote superoxide production in the chloroplast, is important to limit cellular ROS accumulation and is therefore essential to protect tissues from photooxidative-induced damage.
Role of GRXs in the antioxidant response to oxidative stress
There is evidence that plant GRXs, like GRXs from other organisms, show direct and indirect antioxidant activities. Indeed, biochemical studies performed with purified recombinant GRXs from rice and poplar indicate that these proteins are able to reduce peroxides and DHA directly (Lee et al., 2002; Rouhier et al., 2002a). Furthermore, an indirect antioxidant role for plant GRXs is supported by evidence indicating that they are able to regenerate type II PRXs, and a plastidial methionine sulphoxide reductase (MSRB1) (Asada et al., 1975; Rouhier et al., 2001; Rouhier et al., 2002b; Brehelin et al., 2003; Noguera-Mazon et al., 2006; Gama et al., 2007).
In Arabidopsis, functional genetic studies allowed a role in the oxidative stress response to be assigned for two genes coding for monothiol CGFS-type GRXs, GRXS14 and GRXS15 (Cheng et al., 2006; Cheng, 2008). Although biochemical evidence with the recombinant GRXS14 and GRXS15 proteins, as well as other CGFS- and CPYC-type GRXs indicates their involvement in the assembly and transfer of Fe–S clusters to proteins (Bandyopadhyay et al., 2008; Rouhier et al., 2008b), information about catalytic activity that could explain the role that these two GRXs play in the oxidative stress response triggered by environmental stress is still lacking.
On the other hand, only three of the 21 CC-type GRXs have been characterized in Arabidopsis: the GRXC7/ROXY1 and GRXC8/ROXY2 redundant proteins involved in flower development (Xing et al., 2005; Xing and Zachgo, 2008; Li et al., 2009); and the GRXC9 protein involved in plant defence responses against pathogens (Ndamukong et al., 2007). Although the physiological role of ROXY1/2 and GRXC9 proteins seems to be different, they share their ability to bind bZIP transcription factors from the TGA family (Ndamukong et al., 2007; Murmu et al., 2010). Whether this binding promotes redox modification of these factors is still unknown. Surprisingly, GRXS13.2 is one of the CC-type GRXs that complement ROXY1 function in the roxy1-2 Arabidopsis mutant (Li et al., 2009).
In this context, the functional characterization of the GRXS13 gene reported here indicates that GRXS13.2, the predominant isoform coded by this gene, is part of the cellular ROS-scavenging/antioxidant system that is functional under basal and stressed conditions. In contrast, the physiological role of the GRXS13.1 isoform is not known, and evidence reported here supports the idea that is not involved in the cellular ROS-scavenging/antioxidant system. In fact, this protein lacks the GAL/IWL motif conserved in most CC-type GRXs (Fig. 1A). The basal level of its transcript in all tissues assayed is 2–3 orders of magnitude lower than that of the GRX13.2 transcript (Fig. 1C), and this basal level is not altered by photooxidative stress (Fig. 2A, B).
Although the precise biochemical role that the GRXS13.2 protein plays in this system is still unknown, based on the present results it is possible to speculate about its possible function. It was shown that knocking down GRXS13 increases the levels of superoxide accumulated under basal and HL stress conditions (Figs 4C, 6B), without altering the levels of ASC/DHA (Fig. 6C). It has been reported that increased levels of ROS, particularly those produced by HL stress, have a low impact on the redox status of ASC (Maruta et al., 2010; Foyer and Noctor, 2011). Interestingly, overexpressing the GRXS13.2 variant decreases superoxide levels (Fig. 6B, line 5.2), which correlates with increases in the ASC/DHA ratio (Fig. 6C). Therefore, the effect of GRXS13.2 expression on superoxide levels could be indirectly due to the contribution of GRXs to the H2O2 scavenger capacity. It can be speculated that a deficiency in this capacity in GRXS13 knock down lines could explain superoxide radical accumulation, due to the inhibitory effect of H2O2 on superoxide dismutase activities, as previously reported (Asada et al., 1975). On the other hand, the effect of overexpressing GRXS13.2 on ASC/DHA levels indicates that the GRXS13.2 isoform regulates the cellular dehydroascorbate reductase (DHAR) activity, either by itself harbouring DHAR activity or by modulating the DHAR activity of other proteins.
Concerning possible substrates/interactors of GRXS13.2, information contained in the CCSB (Center for Cancer Systems Biology) interactome database (http://interactome.dfci.harvard.edu/A_thaliana/index.php?page=networklist) showed direct interaction between GRXS13 and CDPK4 (calcium-dependent protein kinase 4), which was previously described in a search for CDPK4 interactors in a yeast two-hybrid cDNA library (Uno et al., 2009). The possible effect of this interaction on GRXS13.2 function is still unknown.
Future characterization of the enzymatic activity and target substrates/interactors of GRXS13.2 will shed light on the mechanism by which this protein is involved in superoxide detoxification and stress tolerance.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Phylogenetic relationships and classification of the 31 GRXs from Arabidopsis.
Table S1. Primers used to quantify transcripts from the indicated genes by qRT-PCR.
Table S2. Evidence of expression for GRXS13 gene variants in Arabidopsis databases.
Acknowledgments
This work was supported by the National Commission for Science and Technology CONICYT [FONDECYT grants nos 1060494 and 1100656] and the Millennium Science Initiative [Nucleus for Plant Functional Genomics, grants nos P06-009-F and P10-062-F]. DL and EO were supported by PhD fellowships, and MS by a postdoctoral fellowship from CONICYT [grant no. PSD 74].
References
- Asada K. The water–water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annual Review of Plant Physiology and Plant Molecular Biology. 1999;50:601–639. doi: 10.1146/annurev.arplant.50.1.601. [DOI] [PubMed] [Google Scholar]
- Asada K. The water–water cycle as alternative photon and electron sinks. Philosophical Transactions of the Royal Society B: Biological Sciences. 2000;355:1419–1430. doi: 10.1098/rstb.2000.0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K. Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiology. 2006;141:391–396. doi: 10.1104/pp.106.082040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asada K, Takahashi Y, Maeda Y, Enmanji K. Superoxide dismutases from a blue-green alga, Plectonema boryanum. Journal of Biological Chemistry. 1975;250:2801–2807. [PubMed] [Google Scholar]
- Bandyopadhyay S, Gama F, Molina-Navarro MM, Gualberto JM, Claxton R, Naik SG, Huynh BH, Herrero E, Jacquot JP, Rouhier N. Chloroplast monothiol glutaredoxins as scaffold proteins for the assembly and delivery of [2Fe–2S] clusters. EMBO Journal. 2008;27:1122–1133. doi: 10.1038/emboj.2008.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco F, Salinas P, Cecchini NM, Jordana X, Van Hummelen P, Alvarez ME, Holuigue L. Early genomic responses to salicylic acid in Arabidopsis. Plant Molecular Biology. 2009;70:79–102. doi: 10.1007/s11103-009-9458-1. [DOI] [PubMed] [Google Scholar]
- Brehelin C, Meyer EH, de Souris JP, Bonnard G, Meyer Y. Resemblance and dissemblance of Arabidopsis type II peroxiredoxins: similar sequences for divergent gene expression, protein localization, and activity. Plant Physiology. 2003;132:2045–2057. doi: 10.1104/pp.103.022533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng NH. AtGRX4, an Arabidopsis chloroplastic monothiol glutaredoxin, is able to suppress yeast grx5 mutant phenotypes and respond to oxidative stress. FEBS Letters. 2008;582:848–854. doi: 10.1016/j.febslet.2008.02.006. [DOI] [PubMed] [Google Scholar]
- Cheng NH, Liu JZ, Brock A, Nelsono RS, Hirschi KD. AtGRXcp, an Arabidopsis chloroplastic glutaredoxin, is critical for protection against protein oxidative damage. Journal of Biological Chemistry. 2006;281:26280–26288. doi: 10.1074/jbc.M601354200. [DOI] [PubMed] [Google Scholar]
- Cheng NH, Liu JZ, Liu X, et al. Arabidopsis monothiol glutaredoxin, AtGRXS17, is critical for temperature-dependent postembryonic growth and development via modulating auxin response. Journal of Biological Chemistry. 2011;286:20398–20406. doi: 10.1074/jbc.M110.201707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Couturier J, Stroher E, Albetel AN, et al. Arabidopsis chloroplastic glutaredoxin C5 as a model to explore molecular determinants for iron–sulfur cluster binding into glutaredoxins. Journal of Biological Chemistry. 2011;286:27515–27527. doi: 10.1074/jbc.M111.228726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danna CH, Bartoli CG, Sacco F, Ingala LR, Santa-Maria GE, Guiamet JJ, Ugalde RA. Thylakoid-bound ascorbate peroxidase mutant exhibits impaired electron transport and photosynthetic activity. Plant Physiology. 2003;132:2116–2125. doi: 10.1104/pp.103.021717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davletova S, Rizhsky L, Liang HJ, Zhong SQ, Oliver DJ, Coutu J, Shulaev V, Schlauch K, Mittler R. Cytosolic ascorbate peroxidase 1 is a central component of the reactive oxygen gene network of Arabidopsis. The Plant Cell. 2005;17:268–281. doi: 10.1105/tpc.104.026971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Leon IP, Sanz A, Hamberg M, Castresana C. Involvement of the Arabidopsis alpha-DOX1 fatty acid dioxygenase in protection against oxidative stress and cell death. The Plant Journal. 2002;29:61–72. doi: 10.1046/j.1365-313x.2002.01195.x. [DOI] [PubMed] [Google Scholar]
- Dietz KJ, Jacob S, Oelze ML, Laxa M, Tognetti V, de Miranda SMN, Baier M, Finkemeier I. The function of peroxiredoxins in plant organelle redox metabolism. Journal of Experimental Botany. 2006;57:1697–1709. doi: 10.1093/jxb/erj160. [DOI] [PubMed] [Google Scholar]
- Dixon DP, Hawkins T, Hussey PJ, Edwards R. Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. Journal of Experimental Botany. 2009;60:1207–1218. doi: 10.1093/jxb/ern365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxidants and Redox Signaling. 2009;11:861–905. doi: 10.1089/ars.2008.2177. [DOI] [PubMed] [Google Scholar]
- Foyer CH, Noctor G. Ascorbate and glutathione: the heart of the redox hub. Plant Physiology. 2011;155:2–18. doi: 10.1104/pp.110.167569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Current Opinion in Pharmacology. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Gallogly MM, Starke DW, Leonberg AK, Ospina SME, Mieyal JJ. Kinetic and mechanistic characterization and versatile catalytic properties of mammalian glutaredoxin 2: implications for intracellular roles. Biochemistry. 2008;47:11144–11157. doi: 10.1021/bi800966v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gama F, Keech O, Eymery F, Finkemeier I, Gelhaye E, Gardestrom P, Dietz KJ, Rey P, Jacquot JP, Rouhier N. The mitochondrial type II peroxiredoxin from poplar. Physiologia Plantarum. 2007;129:196–206. [Google Scholar]
- Gechev TS, Van Breusegem F, Stone JM, Denev I, Laloi C. Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays. 2006;28:1091–1101. doi: 10.1002/bies.20493. [DOI] [PubMed] [Google Scholar]
- Georgiou CD, Papapostolou I, Patsoukis N, Tsegenidis T, Sideris T. An ultrasensitive fluorescent assay for the in vivo quantification of superoxide radical in organisms. Analytical Biochemistry. 2005;347:144–151. doi: 10.1016/j.ab.2005.09.013. [DOI] [PubMed] [Google Scholar]
- Gillespie KM, Ainsworth EA. Measurement of reduced, oxidized and total ascorbate content in plants. Nature Protocols. 2007;2:871–874. doi: 10.1038/nprot.2007.101. [DOI] [PubMed] [Google Scholar]
- Holuigue L, Salinas P, Blanco F, Garreton V. Salicylic acid and reactive oxygen species in the activation of stress defense genes. In: In Hayat S, Ahmad A., editors. Salicylic acid—a plant hormone. Dordrecht: Springer; 2007. pp. 197–246. [Google Scholar]
- Karpinski S, Escobar C, Karpinska B, Creissen G, Mullineaux PM. Photosynthetic electron transport regulates the expression of cytosolic ascorbate peroxidase genes in Arabidopsis during excess light stress. The Plant Cell. 1997;9:627–640. doi: 10.1105/tpc.9.4.627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger-Liszkay A, Fufezan C, Trebst A. Singlet oxygen production in photosystem II and related protection mechanism. Photosynthesis Research. 2008;98:551–564. doi: 10.1007/s11120-008-9349-3. [DOI] [PubMed] [Google Scholar]
- Krieger-Liszkay A, Kos PB, Hideg E. Superoxide anion radicals generated by methylviologen in photosystem I damage photosystem II. Physiologia Plantarum. 2011;142:17–25. doi: 10.1111/j.1399-3054.2010.01416.x. [DOI] [PubMed] [Google Scholar]
- Lee KO, Lee JR, Yoo JY, et al. GSH-dependent peroxidase activity of the rice (Oryza sativa) glutaredoxin, a thioltransferase. Biochemical and Biophysical Research Communications. 2002;296:1152–1156. doi: 10.1016/s0006-291x(02)02047-8. [DOI] [PubMed] [Google Scholar]
- Li S, Lauri A, Ziemann M, Busch A, Bhave M, Zachgo S. Nuclear activity of ROXY1, a glutaredoxin interacting with TGA factors, is required for petal development in Arabidopsis thaliana. The Plant Cell. 2009;21:429–441. doi: 10.1105/tpc.108.064477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillig CH, Berndt C, Holmgren A. Glutaredoxin systems. Biochimica et Biophysica Acta. 2008;1780:1304–1317. doi: 10.1016/j.bbagen.2008.06.003. [DOI] [PubMed] [Google Scholar]
- Maruta T, Tanouchi A, Tamoi M, Yabuta Y, Yoshimura K, Ishikawa T, Shigeoka S. Arabidopsis chloroplastic ascorbate peroxidase isoenzymes play a dual role in photoprotection and gene regulation under photooxidative stress. Plant and Cell Physiology. 2010;51:190–200. doi: 10.1093/pcp/pcp177. [DOI] [PubMed] [Google Scholar]
- Meng L, Feldman L. A rapid TRIzol-based two-step method for DNA-free RNA extraction from Arabidopsis siliques and dry seeds. Biotechnology Journal. 2010;5:183–186. doi: 10.1002/biot.200900211. [DOI] [PubMed] [Google Scholar]
- Meyer Y, Buchanan BB, Vignols F, Reichheld JP. Thioredoxins and glutaredoxins: unifying elements in redox biology. Annual Review of Genetics. 2009;43:335–367. doi: 10.1146/annurev-genet-102108-134201. [DOI] [PubMed] [Google Scholar]
- Michelet L, Zaffagnini M, Massot V, Keryer E, Vanacker H, Miginiac-Maslow M, Issakidis-Bourguet E, Lemaire SD. Thioredoxins, glutaredoxins, and glutathionylation: new crosstalks to explore. Photosynthesis Research. 2006;89:225–245. doi: 10.1007/s11120-006-9096-2. [DOI] [PubMed] [Google Scholar]
- Miller G, Suzuki N, Rizhsky L, Hegie A, Koussevitzky S, Mittler R. Double mutants deficient in cytosolic and thylakoid ascorbate peroxidase reveal a complex mode of interaction between reactive oxygen species, plant development, and response to abiotic stresses(1[W][OA]) Plant Physiology. 2007;144:1777–1785. doi: 10.1104/pp.107.101436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murmu J, Bush MJ, DeLong C, Li ST, Xu ML, Khan M, Malcolmson C, Fobert PR, Zachgo S. Arabidopsis basic leucine-zipper transcription factors TGA9 and TGA10 interact with floral glutaredoxins ROXY1 and ROXY2 and are redundantly required for anther development. Plant Physiology. 2010;154:1492–1504. doi: 10.1104/pp.110.159111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myouga F, Hosoda C, Umezawa T, Iizumi H, Kuromori T, Motohashi R, Shono Y, Nagata N, Ikeuchi M, Shinozaki K. A heterocomplex of iron superoxide dismutases defends chloroplast nucleoids against oxidative stress and is essential for chloroplast development in Arabidopsis. The Plant Cell. 2008;20:3148–3162. doi: 10.1105/tpc.108.061341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ndamukong I, Al Abdallat A, Thurow C, Fode B, Zander M, Weigel R, Gatz C. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. The Plant Journal. 2007;50:128–139. doi: 10.1111/j.1365-313X.2007.03039.x. [DOI] [PubMed] [Google Scholar]
- Noguera-Mazon V, Lemoine J, Walker O, Rouhier N, Salvador A, Jacquot JP, Lancelin JM, Krimm I. Glutathionylation induces the dissociation of 1-Cys D-peroxiredoxin non-covalent homodimer. Journal of Biological Chemistry. 2006;281:31736–31742. doi: 10.1074/jbc.M602188200. [DOI] [PubMed] [Google Scholar]
- Perez-Ruiz JM, Spinola MC, Kirchsteiger K, Moreno J, Sahrawy M. Rice NTRC is a high-efficiency redox system for chloroplast protection against oxidative damage. The Plant Cell. 2006;18:2356–2368. doi: 10.1105/tpc.106.041541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rentel MC, Knight MR. Oxidative stress-induced calcium signaling in Arabidopsis. Plant Physiology. 2004;135:1471–1479. doi: 10.1104/pp.104.042663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riondet C, Desouris JP, Montoya JG, Chartier Y, Meyer Y, Reichheld JP. 2011. A dicotyledon-specific glutaredoxin GRXC1 family with dimer-dependent redox regulation is functionally redundant with GRXC2. Plant, Cell and Environment. doi: 10.1111/j.1365-3040.2011.02355.x. (in press) [DOI] [PubMed] [Google Scholar]
- Rizhsky L, Liang HJ, Mittler R. The water–water cycle is essential for chloroplast protection in the absence of stress. Journal of Biological Chemistry. 2003;278:38921–38925. doi: 10.1074/jbc.M304987200. [DOI] [PubMed] [Google Scholar]
- Rouhier N, Couturier J, Jacquot JP. Genome-wide analysis of plant glutaredoxin systems. Journal of Experimental Botany. 2006;57:1685–1696. doi: 10.1093/jxb/erl001. [DOI] [PubMed] [Google Scholar]
- Rouhier N, Couturier J, Johnson MK, Jacquot JP. Glutaredoxins: roles in iron homeostasis. Trends in Biochemical Sciences. 2010;35:43–52. doi: 10.1016/j.tibs.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Jacquot JP. Exploring the active site of plant glutaredoxin by site-directed mutagenesis. FEBS Letters. 2002a;511:145–149. doi: 10.1016/s0014-5793(01)03302-6. [DOI] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Jacquot JP. Glutaredoxin-dependent peroxiredoxin from poplar. Protein–protein interaction and catalytic mechanism. Journal of Biological Chemistry. 2002b;277:13609–13614. doi: 10.1074/jbc.M111489200. [DOI] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Jacquot JP. Plant glutaredoxins: still mysterious reducing systems. Cellular and Molecular Life Sciences. 2004;61:1266–1277. doi: 10.1007/s00018-004-3410-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Gelhaye E, Sautiere PE, Brun A, Laurent P, Tagu D, Gerard J, de Fay E, Meyer Y, Jacquot JP. Isolation and characterization of a new peroxiredoxin from poplar sieve tubes that uses either glutaredoxin or thioredoxin as a proton donor. Plant Physiology. 2001;127:1299–1309. [PMC free article] [PubMed] [Google Scholar]
- Rouhier N, Koh CS, Gelhaye E, Corbier C, Favier F, Didiejean C, Jacquot JP. Redox based anti-oxidant systems in plants: biochemical and structural analyses. Biochimica et Biophysica Acta. 2008a;1780:1249–1260. doi: 10.1016/j.bbagen.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Rouhier N, Lemaire SD, Jacquot JP. The role of glutathione in photosynthetic organisms: emerging functions for glutaredoxins and glutathionylation. Annual Review of Plant Biology. 2008b;59:143–166. doi: 10.1146/annurev.arplant.59.032607.092811. [DOI] [PubMed] [Google Scholar]
- Tarantino D, Vannini C, Bracale M, Campa M, Soave C, Murgia I. Antisense reduction of thylakoidal ascorbate peroxidase in Arabidopsis enhances paraquat-induced photooxidative stress and nitric oxide-induced cell death. Planta. 2005;221:757–765. doi: 10.1007/s00425-005-1485-9. [DOI] [PubMed] [Google Scholar]
- Uno Y, Rodriguez Milla MA, Maher E, Cushman JC. Identification of proteins that interact with catalytically active calcium-dependent protein kinases from Arabidopsis. Molecular Genetics and Genomics. 2009;281:375–390. doi: 10.1007/s00438-008-0419-1. [DOI] [PubMed] [Google Scholar]
- Xing S, Zachgo S. ROXY1 and ROXY2, two Arabidopsis glutaredoxin genes, are required for anther development. The Plant Journal. 2008;53:790–801. doi: 10.1111/j.1365-313X.2007.03375.x. [DOI] [PubMed] [Google Scholar]
- Xing SP, Rosso MG, Zachgo S. ROXY1, a member of the plant glutaredoxin family, is required for petal development in Arabidopsis thaliana. Development. 2005;132:1555–1565. doi: 10.1242/dev.01725. [DOI] [PubMed] [Google Scholar]
- Zaffagnini M, Michelet L, Massot V, Trost P, Lemaire SD. Biochemical characterization of glutaredoxins from Chlamydomonas reinhardtii reveals the unique properties of a chloroplastic CGFS-type glutaredoxin. Journal of Biological Chemistry. 2008;283:8868–8876. doi: 10.1074/jbc.M709567200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.