Abstract
The pericarp of Capsicum fruit is a rich dietary source of carotenoids. Accumulation of these compounds may be controlled, in part, by gene transcription of biosynthetic enzymes. The carotenoid composition in a number of orange-coloured C. annuum cultivars was determined using HPLC and compared with transcript abundances for four carotenogenic enzymes, Psy, LcyB, CrtZ-2, and Ccs determined by qRT-PCR. There were unique carotenoid profiles as well as distinct patterns of transcription of carotenogenic enzymes within the seven orange-coloured cultivars. In one cultivar, ‘Fogo’, carrying the mutant ccs-3 allele, transcripts were detected for this gene, but no CCS protein accumulated. The premature stop termination in ccs-3 prevented expression of the biosynthetic activity to synthesize the capsanthin and capsorubin forms of carotenoids. In two other orange-coloured cultivars, ‘Orange Grande’ and ‘Oriole’, both with wild-type versions of all four carotenogenic enzymes, no transcripts for Ccs were detected and no red pigments accumulated. Finally, in a third case, the orange-coloured cultivar, Canary, transcripts for all four of the wild-type carotenogenic enzymes were readily detected yet no CCS protein appeared to accumulate and no red carotenoids were synthesized. In the past, mutations in Psy and Ccs have been identified as the loci controlling colour in the fruit. Now there is evidence that a non-structural gene may control colour development in Capsicum.
Keywords: Capsanthin, β-carotene, carotenoid biosynthesis, chromoplast, peppers, pericarp, qRT-PCR, xanthophylls
Introduction
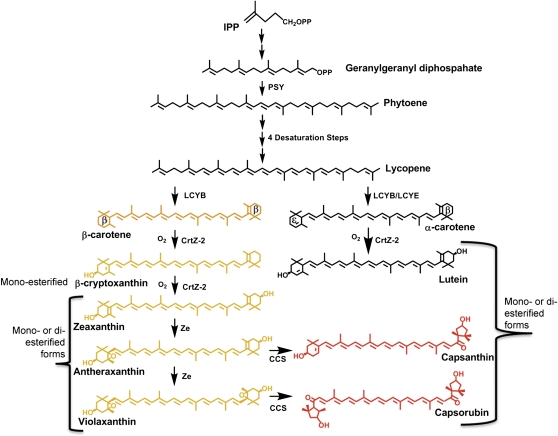
In plants, carotenoids are antenna pigments associated with the light-harvesting and reaction centres on the thylakoid membranes in chloroplasts (DellaPenna and Pogson, 2006). In ripening fruits and flower petals, carotenoids accumulate to even higher levels in chromoplasts. In animals, carotenoids provide antioxidant activity, macular pigments and specific carotenoids (β-carotene and β-cryptoxanthin) are essential dietary components as vitamin A precursors (von Lintig, 2010). Consumption of coloured peppers (Capsicum spp.) in the United States has become increasingly popular over the last two decades (USDA, 2011). As these fruits are rich sources of pro-Vitamin A carotenoids and anti-oxidant xanthophylls (Deli et al., 2001; Wall et al., 2001; Wahyuni et al., 2011), characterization of the carotenoid pathway (Fig. 1) in Capsicum spp, is important to the development of nutritious food. Ancestral peppers were hot, relatively small, and red; humans selected for bigger fruit with additional colours and with varying levels of heat from none to scorching (Paran and van der Knaap, 2007). Over the last 50 years, mutant lines allowed many of the loci controlling fruit quality to be identified. The simple locus controlling pungency, C or Pun1, maps to chromosome 2 (Blum et al., 2002, 2003) with a candidate gene at the locus (Stewart et al., 2005, 2007). By contrast, three loci are predicted to control the red, yellow, or orange fruit colour in pepper (Hurtado-Hernandez and Smith, 1985). Red colour is dominant over white and yellow in the F1 cross of a red pepper with a white and yellow pepper producing only red F1 progeny.
Fig. 1.
Carotenoid biosynthetic pathway.
A full understanding of the regulation of the carotenoid pathway, both structural and regulatory genes, is necessary in order to manipulate carotenoid levels in crops, particularly pro-Vitamin A carotenoids, through genetic engineering (Schaub et al., 2005; Diretto et al., 2010; Bai et al., 2011). Given the relatively high level of carotene and xanthophyll accumulation in Capsicum fruit, this system was and continues to be a very important source of carotenoid biosynthetic genes (Romer et al., 1993; Hugueney et al., 1995; Bouvier et al., 1996; Ha et al., 2007; Guzman et al., 2010). We have, among others, characterized the carotenoid pathway in orange-coloured fruit, with the intent to investigate ways to increase β-carotene accumulation in the fruit (Huh et al., 2001; Lang et al., 2004; Ha et al., 2007; Guzman et al., 2010). Expression studies on selected carotenoid structural genes show that pigment-related transcripts are detected as the fruit begins to ripen (Romer et al., 1993; Ha et al., 2007). Ha et al. (2007) used cDNAs for phytoene synthase (Psy), phytoene desaturase (Pds), β-carotene hydroxylase (CrtZ-2), and capsanthin-capsorubin synthase (Ccs) amplified from Korean red pepper as probes in Northern blot studies. Their results demonstrate high levels of transcripts in those peppers with high amounts of carotenoids.
Multiple distinct molecular explanations for the yellow or orange colour in pepper are proposed. A deletion of Ccs detected in yellow peppers is considered to be causal in the fruit phenotype (Lefebvre et al., 1998). A deletion of the amino terminus of Ccs in orange peppers is considered to be causal in that phenotype (Lang et al., 2004). A second site for mutation, the Psy locus on chromosome 4 is also associated with orange fruit colour in crosses between red-coloured C. annuum and orange-coloured C. chinense (Huh et al., 2001). As a result of these studies and additional Capsicum mapping data, the biochemical identities of two of the three loci proposed to control pepper fruit colour are known; Y is Ccs and C2 is Psy (Popovsky and Paran, 2000; Thorup et al., 2000).
In an earlier study it was demonstrated that there were orange-coloured peppers that did not accumulate β-carotene, rather a mixture of xanthophylls resulted in the apparent orange colour of the fruit (Guzman et al., 2010). Further, a novel gene mutation, ccs-3, was identified in one of the orange-coloured cultivars, Fogo, that generated a premature termination in the coding sequence of Ccs. In this report, gene expression patterns of carotenoid pathway genes are described in several orange-coloured C. annuum, L. fruit (Fig. 2). These results demonstrate that the control of this biosynthetic pathway is more complex than the currently accepted three-gene model.
Fig. 2.
Capsicum annuum varieties: (A) NuMex Sunset; (B) Valencia; (C) Orange Grande; (D) Oriole; (E) NuMex Garnet; (F) Dove; (G) Canary; (H) Fogo.
Materials and methods
Plant material
Eight C. anuum cultivars: Valencia, Orange Grande, Oriole, Fogo, Dove, Canary, NuMex Sunset, and NuMex Garnet were grown from seed under greenhouse and field conditions at the Fabian Garcia Horticulture Farm, NMSU, Las Cruces, NM, USA. The cultivar Canary, a light orange pepper, was hybridized with Valencia as the pollen parent. Valencia is an example of a dominant orange pepper that produces orange not red F1 progeny in crosses with orange-yellow lines (Patent number 5 440 069). Thirty F1 progeny were planted and grown in greenhouse conditions. Fruits at different ripening stages, green, breaking, and fully mature, were collected and used for carotenoid, total RNA, and plastid protein extractions.
Carotenoid extractions and analysis
Harvested fruits were dissected and triplicate extractions of 5.0 g of pericarp samples were done as before (Guzman et al., 2010) except that the samples were lyophilized. Hexane-extracted carotenoids were dried under N2 gas flow, resuspended in isopropanol and 500 μl were saponified with 100 μl methanolic KOH for 30 min at 50 °C. Water (500 μl) and chloroform (300 μl) were added to the saponified extract, and the organic phase containing the saponified pigments was collected by centrifugation. Thin layer chromatography (TLC) plates were spotted with 0.5 μl of saponified or unsaponified carotenoid extracts along with the reference standards (violaxanthin, antheraxanthin, capsanthin, and capsorubin, β-cryptoxanthin, zeaxanthin, β-carotene, and lutein). The TLC plates were resolved with 5% butanol: 76% hexane: 9.5% ethyl acetate: 9.5% dichloromethane (v/v). Images of the plates were captured with a Seiko Epson scanner.
Extracts were analysed on a Waters HPLC system equipped with a 4.6×250 mm YMC carotenoid column. A linear gradient consisting of methanol:methyl-t-butyl ether:water:: 81:15:4 by vol. (solvent A) and methyl-t-butyl ether:methanol:water::88:8:4 by vol. (solvent B) was employed. The flow rate was 2 ml min−1 at 25 °C. Absorbance data were collected with a PDA from 400–600 nm. Calibration curves were generated at 450 nm with the reference standards: β-carotene, lutein, and lycopene, Sigma (St Louis, MO, USA); and capsanthin, capsorubin, zeaxanthin, antheraxanthin, violaxanthin, and β-cryptoxanthin, CaroteNature GmbH (Lupsingen, Switzerland). Triplicate extractions were performed on each genotype and the average concentration for each carotenoid was calculated following independent HPLC analyses.
Expression of gene transcripts
Total RNA extractions of fruit pericarp and Northern blot analyses were performed as previously described by Rodriguez-Uribe and O’Connell (2006). Transcripts were also detected by reverse-transcription PCR (RT-PCR) using the SuperScript™ First-Strand Synthesis System for RT-PCR (Invitrogen Life Technologies, Carlsbad CA) and primers for specific carotenoid biosynthetic genes as described earlier (Guzman et al., 2010).
For qRT-PCR, total RNA was treated with RNase free DNase1 (Invitrogen Life Technologies, Carlsbad, CA). The RNA concentration and integrity were determined in a nanodrop 2000 Spectrophotometer (Thermo Scientific) and by denaturing gel electrophoresis. Primers, Psy_535F: CCGAATGCATCACACATTACTCCG and Psy_719R: GACTTCCTCAAGTCCATACGCATTCC; CrtZ-2_4F: GTATCCTTCA CCGTACCGTACATG and CrtZ-2_180R: CTTTCTCCGAGACCG CAAAATTGC; Ccs_477F: GCTTCGGCTAGCTGAACAAGTTTC and Ccs_636R: CTCACAGGCCAC TTATGATCTAGAC were designed from published mRNA sequences to amplify regions of Psy, CrtZ-2, and Ccs. The amplicons ranged in size from 150 to 200 base pairs.
All qRT-PCR reactions were carried out in triplicate in a Master Cycler ep realplex (Eppendorf, Westbury, NY) with the Kapa SYBR Fast One-Step qRT-PCR Kit (Kapa Biosystems, Boston MA) as described by the manufacturer. Standard curves were generated using 10-fold dilutions of a template RNA of known concentration for each set of primer/template to determine the reaction optimization of each template. Absolute quantification of each gene was interpolated from the standard curves.
Isolation and purification of proteins from pericarp plastids
Crude extracts enriched for plastids were prepared from pericarp tissue of eight C. annuum cultivars at green, breaking, and orange ripening stages in GR buffer as described by Siddiqui et al. (2006) with subsequent purification of plastid integral membrane proteins. The protein patterns were analysed by SDS-PAGE. Finally, CCS protein identification was performed on ∼98% pure protein extracted from SDS-PAGE lanes, following trypsin digestion with the NanoLC-MS/MS peptide sequencing technology at ProTech, Inc. (Norristown, PA).
Ccs promoter cloning and analysis
Genomic DNA was extracted from young leaves of each cultivar using standard methods (Guzman et al., 2010). Primers were designed from a genomic DNA sequence of Ccs (YA4165) to amplify a 1000 bp region including the translation initiation of Ccs (Ccs_promo_F1: GACTCCGCCATTACTACAAG; Ccs_promo_F2: GACCTCCAACA ATCACCAATAC; Ccs_promo_R1: ATGGAAAGTAAAGGAGATGGAAAAGG; and Ccs_promo_R2 GGTATTGGTGATTGTTGGAGG). PCR amplification was carried out with the Advantage-HF 2 PCR Kit (Clontech, Mountain View, CA) with the following conditions: 94 °C for 1 min, followed by 35 cycles of 94 °C for 30 s, 57.5 °C for 35 s, 68 °C 1.4 min, with a final extension step of 68 °C for 7 min. The PCR products were cloned into the pGEM-T Easy Vector Systems (Promega, Madison, WI) and sequenced with the Li-Cor System. DNA sequences were aligned using DNASTAR software.
Results
Carotenoid extractions
In previous analyses, the carotenoid content of orange-coloured pericarp was determined on samples using unsaponified hexane extracts (Guzman et al. 2010). This resulted in very accurate measurements of the β-carotene content of the fruit, but underestimated the abundance of the xanthophyll carotenoids. Saponification conditions were established that resulted in complete hydrolysis of the acyl side chains on the carotenoids, without any apparent reduction in carotenoid content. Using this method, pericarp samples were extracted and then saponified prior to chromatography. Table 1 lists the abundances of selected carotenoids that were detected in seven different orange-coloured C. annuum cultivars, and one red cultivar, NuMex Garnet.
Table 1.
Carotenoid accumulation in Capsicum fruit expressed as μg g−1 fresh wt pericarp ±SD, n=3, in immature fruit (turning) or mature fruit.
| β-carotene | β-cryptoxanthin | Lutein | Zeaxanthin | Violaxanthin | Capsanthin | Unidentified carotenoidsa | ||
| Canary | Turning | 1.94±0.20 | nd | nd | 3.43±0.48 | 11.48±1.13 | nd | 14.75±2.99 |
| Mature | nd | nd | 3.77±0.19 | 1.20±0.12 | 11.83±1.02 | nd | 6.50 ±0.55 | |
| Fogo | Turning | 4.74±0.72 | 1.11±0.17 | 10.50±0.52 | 5.68±0.68 | 11.79±1.08 | nd | 21.04±2.34 |
| Mature | 9.34±1.25 | 4.64±0.44 | 18.73±0.77 | 18.93±1.25 | 43.34±3.78 | nd | 66.59±5.99 | |
| Orange Grande | Turning | 2.04±0.55 | 1.29±0.35 | 10.66±1.56 | 20.90±3.53 | 1.65±0.21 | nd | 9.83±3.01 |
| Mature | 6.75.±3.11 | 2.16±1.77 | 14.41±4.65 | 44.44±16.50 | 2.16±0.34 | nd | 24.77±8.36 | |
| Oriole | Turning | 2.03±0.61 | 1.12±0.07 | 9.25±1.72 | 19.78±3.79 | 2.14±0.39 | nd | 11.88±3.55 |
| Mature | 4.94±1.47 | 2.67±0.66 | 10.18±1.87 | 39.14±6.98 | 1.64±0.22 | nd | 17.89±3.13 | |
| Valencia | Turning | 2.78±1.07 | nd | nd | 3.73±0.23 | 5.11±0.93 | nd | 16.17±3.36 |
| Mature | 5.48±0.27 | 1.11±0.08 | nd | 8.09±2.05 | 11.20±3.03 | 13.68±8.81 | 18.00±5.57 | |
| NuMex Sunset | Turning | nd | nd | nd | nd | 3.63±2.63 | 10.42±0.52 | 8.63±3.32 |
| Mature | nd | nd | nd | nd | nd | 8.18±0.51 | 2.42±0.09 | |
| Dove | Mature | nd | nd | nd | nd | 0.69±0.12 | 18.84±1.31 | 8.76±2.36 |
| NuMex Garnet | Mature | 36.00±6.00 | 19.02±3.44 | nd | 24.08±2.48 | 6.43±1.11 | 116.25±15.41 | 117.02±16.90 |
HPLC peaks were detected by absorbance at 450 nm, but not eluting with retentions times of any reference standards; concentrations calculated based on β-carotene calibration curves.
Among the orange-coloured fruit, there are varieties that accumulate β-carotene: Fogo, Orange Grande, Oriole, and Valencia; there are also varieties that do not appear to accumulate any β-carotene in their mature stage: Canary, NuMex Sunset, and Dove. Those varieties that accumulate the red pigment, capsanthin, Valencia, NuMex Sunset, Dove, and NuMex Garnet, do not appear to accumulate any appreciable quantities of lutein. There are some striking differences in the absolute amounts of specific carotenoids accumulated by these eight cultivars. Violaxanthin was detected in the mature fruit of all varieties, but the levels varied from a low of 0.7 μg g−1 fresh wt. in Dove to 43 μg g−1 fresh wt. in Fogo. Zeaxanthin was detected in six cultivars, and varied over a similar concentration range, 1.2 μg g−1 fresh wt. in Canary to 44 μg g−1 fresh wt. in Orange Grande. The concentrations of pro-Vitamin A carotenoids, β-carotene and β-cryptoxanthin, also varied in these fruit, with the highest concentrations detected in the red -coloured NuMex Garnet and no detectable levels, paradoxically, in the orange-coloured NuMex Sunset and Dove.
As described in the Materials and methods, the major peaks in the HPLC chromatograms were identified based on retentions times and spectra similarity to reference standards. In addition to the specific carotenoids listed in Table 1, the samples were characterized for the accumulation of lycopene, antheraxanthin, and capsorubin. None of the samples accumulated detectable levels of these carotenoids. There were additional unidentified carotenoid peaks in these chromatograms; the abundance of these carotenoids is indicated in the last column of Table 1. The unidentified carotenoid peaks represented between 17% and 35% of the total carotenoid profile, depending on cultivar. Most of the carotenoids accumulated by these cultivars were identified as one or more of the carotenoids listed in Table 1.
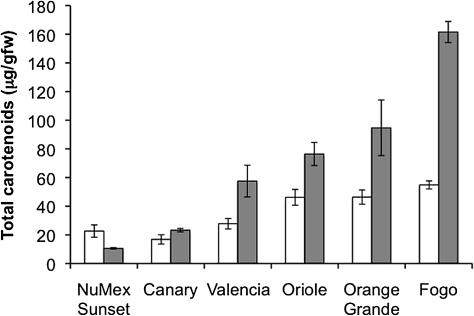
Most orange-coloured C. annuum varieties continue to accumulate carotenoids during development (Fig. 3). The total carotenoid content in the pericarp of turning or mature fruit are plotted for six different orange-coloured cultivars. Four of the six cultivars, double or triple the carotenoid content as the fruit develop from turning to mature. Canary increases carotenoid content by less than 50% and NuMex Sunset decreases the carotenoid content as the fruit mature. The two varieties with the lowest abundance of carotenoids are also the varieties that do not significantly increase carotenoid accumulation during development.
Fig. 3.
Accumulation of carotenoids in orange-coloured Capsicum during fruit development. Total carotenoids (total HPLC peak area detected at 450 nm) extracted from turning (open bars) or mature (grey bars) pericarp of the indicated cultivars. Carotenoid concentrations are expressed as μg g−1 fresh wt. pericarp, average ±SD, n=3.
Cultivar-specific gene expression patterns
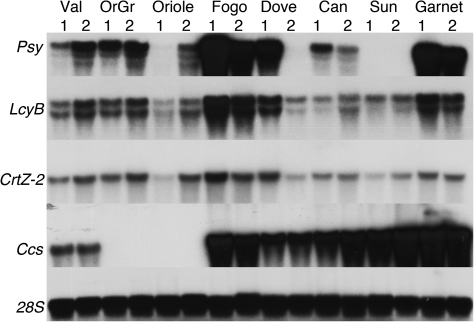
Given the variability in accumulation of specific carotenoids, the transcript abundance was investigated for four genes on the carotenoid biosynthetic pathway: Psy, LcyB, CrtZ-2, and Ccs. Total RNA from immature or turning as well as mature pericarp was used on Northern blots to monitor the expression level of these genes (Fig. 4). CrtZ-2 transcript accumulation was similar in all eight cultivars, with little or no differences between the immature and mature pericarp samples as well. LcyB transcripts were detected in all cultivars with similar levels in most cultivars. There were several unexpected patterns of expression for the other two genes, Psy and Ccs. Transcripts for Psy, an early step on the pathway, were readily detected in both developmental stages of five of the eight cultivars. Oriole accumulated transcripts for this gene only in mature fruit, while Dove accumulated Psy transcripts only in immature fruit; and no Psy transcripts were detected in either stage of pericarp for NuMex Sunset. In the case of Ccs, transcripts were readily detected in both developmental stages of pericarp from six of the cultivars; but no Ccs transcripts were detected in either stage of Orange Grande or Oriole. To confirm the absence of Psy transcripts in NuMex Sunset, RT-PCR reactions were run using primers designed for Psy. The expected 1255 bp amplicon was generated with the positive controls, cloned forms of the gene, and total RNA from Canary pericarp tissue, but no amplicon was detected with three different stages of NuMex Sunset: green, turning or mature. The earlier stage RNA was included in the RT-PCR screen in case this gene was only transcribed in immature fruit in this variety.
Fig. 4.
Transcript abundances for carotenoid pathway genes. Northern blots with total RNA from immature (1) or mature (2) pericarp from: Valencia (Val), Orange Grande (OrGr), Oriole, Fogo, Dove, Canary (Can), NuMex Sunset (Sun) or NuMex Garnet (Garnet) were hybridized with radiolabelled probes of the indicated carotenoid pathway genes or 28S rRNA.
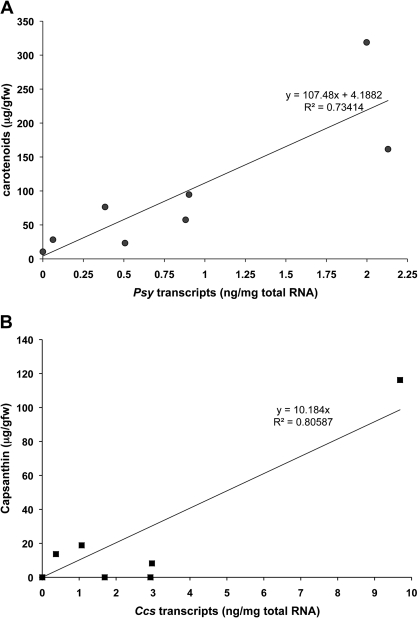
Quantitative RT-PCR (qRT-PCR) was conducted on the two developmental stages of fruit from eight different varieties using primers for Psy, Crtz-2, and Ccs (Table 2). This quantitative analysis clearly confirmed the visual images of the Northern blots in Fig. 4. The transcript abundances were used to investigate relationships with the accumulation of total carotenoids. Scatter plots of total carotenoid abundance in mature fruit versus Psy transcript abundance is presented in Fig. 5A; while the relationship between capsanthin accumulation in mature fruit versus Ccs transcript abundance is presented in Fig. 5B. There is a positive correlation between Psy transcript abundance and total carotenoid accumulation across the eight Capsicum cultivars. There is also a positive correlation between Ccs transcript abundance and capsanthin accumulation. These results support the description of PSY as the first committed step of carotenoid biosynthesis.
Table 2.
Transcript abundances for Psy, CrtZ-2, and Ccs detected by qRT-PCR in pericarp samples from immature or mature C. annuum cultivars (ng 100 ng−1 RNA, average ±SD, n=3)
| Psy | CrtZ-2 | Ccs | ||
| Canary | Turning | 0.56±0.02 | 0.19±0.0 | 2.24±0.07 |
| Mature | 0.51±0.05 | 0.19±0.0 | 1.69±0.23 | |
| Fogo | Turning | 5.6±0.24 | 0.41±0.0 | 1.93±0.00 |
| Mature | 2.13±0.17 | 0.98±0.0 | 2.93±0.00 | |
| Oro Grande | Turning | 0.64±0.05 | 0.26±0.0 | 0 |
| Mature | 0.90±0.08 | 0.56±0.0 | 0 | |
| Oriole | Turning | 0.12±0.0 | 0.14±0.0 | 0 |
| Mature | 0.38±0.02 | 0.23±0.0 | 0 | |
| Valencia | Turning | 0.18±0.03 | 0.19±0.0 | 0.21±0.03 |
| Mature | 0.88±0.03 | 0.40±0.0 | 0.37±0.01 | |
| NuMex Sunset | Turning | 0 | 0.11±0.0 | 1.62±0.16 |
| Mature | 0 | 0.18±0.0 | 2.97±0.06 | |
| Dove | Turning | 4.06±0.14 | 0.63±0.0 | 0.99±0.01 |
| Mature | 0.06±0.0 | 0.1±0.0 | 1.07±0.05 | |
| NuMex Garnet | Turning | 3.38±0.14 | 0.49±0.0 | 6.84±0.37 |
| Mature | 1.98±0.30 | 0.39±0.0 | 9.69±0.82 |
Fig. 5.
Relationship of carotenoid abundances to transcript abundance for carotenoid biosynthetic genes. (A) Psy versus total carotenoids; (B) Ccs versus capsanthin. The abundances of carotenoids expressed as microgram g−1 fresh wt. pericarp, transcripts as ng transcript mg−1 total RNA.
Expression of proteins in pepper fruit chromoplasts
In previous work (Guzman et al., 2010) it had been demonstrated that there were no DNA sequence differences among the cultivars under investigation within the coding regions of three of the four genes (Psy, LcyB, and CrtZ-2). Only Ccs had a variant sequence and that was found in only one cultivar ‘Fogo’. This variant, ccs-3, generated a premature stop codon and the predicted gene product was expected to be inactive. Since transcripts for ccs-3 were readily detected in Fogo (Fig. 4; Table 2) it was examined whether proteins for Ccs and ccs-3 accumulated in the fruit of cultivars carrying these genes, Valencia and Fogo.
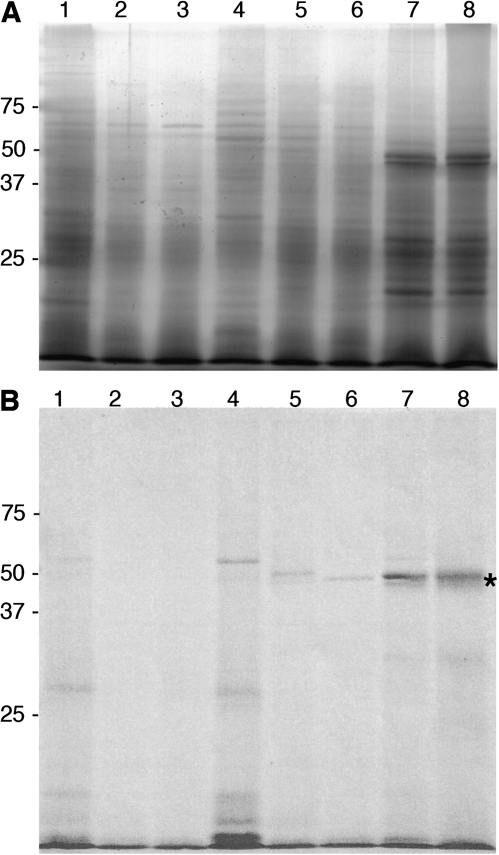
Chromoplasts were isolated from pericarp of mature fruit of all eight varieties, and the integral membrane proteins were resolved on SDS-PAGE gels (Fig. 6). A prominent band of approximately the correct size for CCS, is present in the two varieties that accumulate significant amounts of capsanthin, NuMex Sunset and NuMex Garnet (Fig. 6B). To determine if this band was CCS, gel fragments containing this protein were collected and submitted for tryptic peptide analysis by LC/MS. An equivalent region from the lane containing the Fogo sample was also prepared for LC/MS analysis.
Fig. 6.
Chromoplast proteins. Proteins extracted from (A) whole chromoplasts or (B) inner membranes from: 1, Valencia; 2, Orange Grande; 3, Oriole; 4, Fogo; 5, Dove; 6, Canary; 7, NuMex Sunset; 8, NuMex Garnet. Migration of molecular weight standards in kDa is indicated; * marks the CCS band.
All of the predicted tryptic peptides for CCS were present in the NuMex Garnet sample. The major band in the integral membrane protein preparation with apparent 55 kDa MW, was in fact CCS. The tryptic peptides in the Fogo sample did not match CCS peptides. So while there were transcripts for ccs-3, no protein accumulated. Bands of the correct size in the integral membrane fraction were observed in the samples prepared from Dove and Valencia; while no bands of this size were observed in Canary, Fogo, Orange Grande or Oriole (Fig. 6B).
Ccs promoter analysis
As transcript accumulation for Ccs was distinct and variable across the orange varieties, the DNA sequences of the promoter of this gene were obtained following PCR amplification from selected cultivars. Alignment of ∼860 bp upstream of the ATG translation start of the Ccs gene, revealed only three single nucleotide polymorphisms (Table 3; see Supplementary Fig. S1 at JXB online). The remainder of the sequence for this promoter region was identical in all six orange varieties and identical to the published promoter sequence of a red C. annuum cultivar (Ha et al., 2007). Interestingly, the hybrid nature of some of these cultivars (Valencia, Orange Grande, and Oriole) was observed as two distinct promoter sequences were cloned. However, none of these sequence differences in the Ccs promoter are likely to be associated with the differences in Ccs transcription. There were no sequence differences that distinguished the two varieties that do not accumulate any Ccs transcripts, Orange Grande and Oriole, from those varieties that do accumulate Ccs transcripts.
Table 3.
Nucleotide sequence polymorphisms in the Ccs promoter Positions of DNA sequence polymorphisms in the Ccs promoter of the orange C. annuum cultivars relative to the reference sequence of the Ccs promoter in the red C. annuum cultivar (GenBank Y14165; Ha et al., 2007). Numbers indicate the position relative to ATG codon.
| Variety | Position –711 | Position –483 | Position –312 |
| GenBank Y14165 | C | C | G |
| Canary | T | C | A |
| Fogo | T | C | A |
| NuMex Sunset | C | T | G |
| Valencia | T or C | T or C | A or G |
| Oriole | T or C | T or C | A or G |
| Orange Grande | C | T or C | A or G |
Carotenoid profiles and gene expression in segregating population
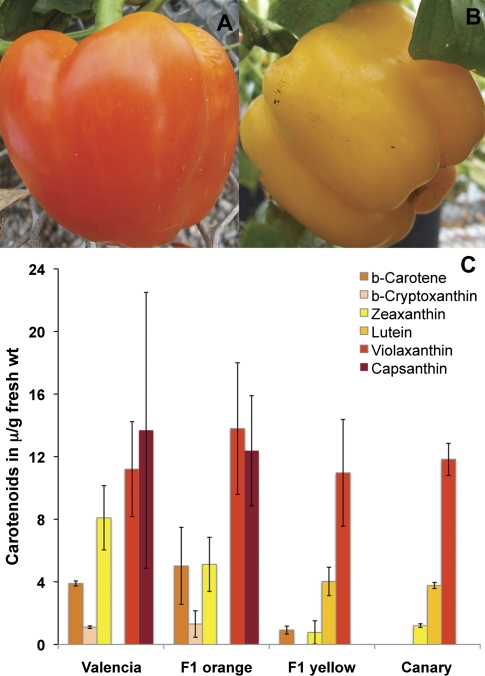
Valencia is considered to be a source of the unusual dominant orange phenotype. The F1 progeny of the Canary×Valencia cross were examined for segregation of fruit colour. Only 17 viable progeny grew to produce fruit; of those 10 produced orange fruit, while seven produced yellow fruit. Representative images of these fruit are shown in Fig. 7A and B.
Fig. 7.
Fruit colours and carotenoid accumulation in a segregating Valencia×Canary F1 population. (A) Orange F1; (B) yellow F1; (C) average carotenoid accumulation (μg g−1 fresh wt. pericarp) in parents (n=3), orange-coloured F1 (n=4), and yellow-coloured F1 (n=4) determined following separation by HPLC.
The carotenoid abundances in the fruit from four orange F1 and from four yellow F1 individuals were determined (Fig. 7C). There were clear differences in carotenoid accumulation in the two different F1 phenotypes with β-cryptoxanthin and capsanthin only accumulating in the orange F1 progeny, and lutein only accumulating in the yellow F1 progeny. There were also significant differences in the abundances of zeaxanthin and β-carotene between the two progeny types, with these carotenoids accumulating to higher levels in the orange F1 progeny.
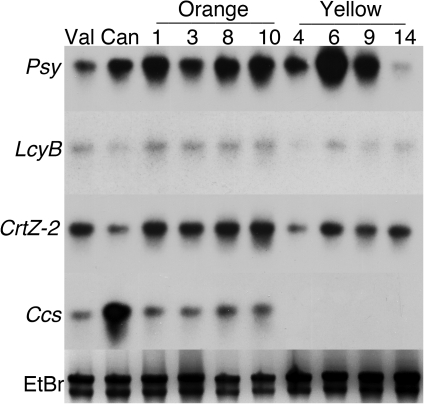
Transcript abundances for the carotenoid biosynthetic genes were also measured in the two classes of F1, orange and yellow (Fig. 8). Transcript levels for three of the four genes were similar in both parents and F1 progeny. Transcripts for Psy, LcyB, and CrtZ-2 all accumulated to similar levels as detected by a Northern blot. Ccs transcripts, however, were not detected in any of the yellow F1 progeny. Both parents accumulated easily detected levels of this transcript, so this pattern of gene expression was not expected.
Fig. 8.
Transcript abundances for carotenoid pathway genes in a segregating Valencia×Canary F1 populations. Northern blots with total RNA from turning stage pericarp from each parent, Valencia (Val) or Canary (Can), and individual F1 progeny sorted by fruit colour into orange (No. 1, 3, 8, or 10) and yellow (No. 4, 6, 9, or 14) were hybridized with radiolabelled probes for the indicated genes. A representative image of the ethidium-bromide-stained gel is shown in the lowest panel as a loading control.
Discussion
Investigation into the molecular basis for the control of carotenoid accumulation is an active area of research for a number of agronomic fruits, vegetables, and spices (Clotault et al., 2008; Ampomah-Dwamena et al., 2009; Guzman et al., 2010; Ahrazem et al., 2010). Genetic variation in specific alleles of carotenoid biosynthetic enzymes have been associated with altered carotenoid accumulation patterns (Welsch et al., 2010; Yan et al., 2010). The results in this report identify varietal specific patterns of expression of carotenoid pathway genes in cultivars with seemingly similar phenotypes, i.e. orange-coloured fruit.
Carotenoid accumulation in Capsicum fruit is a key trait for nutritional value. Early work suggested that genetic control on this trait might be explained by relatively few genes, the three gene model of Hurtado-Hernandez and Smith (1985). Later investigators have identified candidate genes with biochemical functions on the carotenoid pathway for these genetic loci (Thorup et al., 2000). Our work suggests that there are additional mechanisms working to control carotenoid accumulation and categorizing varieties by overall fruit colour may obscure unique genetic and biochemical mechanisms. In one report, yellow and orange peppers did not accumulate carotenoids during fruit development, while red peppers did increase carotenoid accumulation during fruit ripening (Ha et al., 2007). Our results (Fig. 3) clearly show that most of the orange varieties characterized did increase carotenoid accumulation during fruit maturation. Only those varieties with very low carotenoid concentrations did not increase carotenoid accumulation over time.
Among the seven varieties with orange-coloured fruit in this study, the unique distributions of β-carotene was confirmed in those varieties described earlier (Guzman et al., 2010) and the characterization extended to document additional distinct patterns of xanthophyll accumulation. Specifically, varieties that accumulate lutein do not accumulate any capsanthin. This correlation was observed in the cultivars as well as in the segregating F1 population from the Canary×Valencia cross.
The metabolite accumulation data needs to be interpreted using the molecular data also available. Differences in metabolite accumulation could be explained by differences in carotenoid biosynthetic enzymes catalytic rates, or differences in enzyme abundance. With the exception of Fogo, all of the orange-fruited varieties have exactly the same predicted amino acid sequences for all four of the carotenoid pathway genes, making inherent enzyme efficiency differences difficult to postulate.
In Valencia, Orange Grande, and Oriole, the abundance of Psy transcripts increased as the fruit ripened while in Fogo, Dove, Canary, and NuMex Garnet the transcript abundance decreased during ripening. The most dramatic examples of these two classes of responses are seen in Oriole and Dove. It has been recognized that these are RNA transcript levels and protein accumulation may be more stable or persistent.
One of the more surprising results observed here was the absence of detectable Psy transcripts in NuMex Sunset. Is there an alternate gene family member expressed in the fruit of this variety? In tomato, Psy1 and Psy2 are very similar, with most of the differences occurring in the first two exons in the region predicted to be processed during chloroplast import (Giorio et al., 2008). Capsicum is expected to have two Psy genes, although the second gene has not been cloned. Evidence in support of a second Psy, is the detection of two different transcript sizes on Northern blots (Romer et al., 1993) and presumably two bands on a genomic Southern (Thorup et al., 2000). The conditions of hybridization for the Northern blots used to monitor Psy transcripts in NuMex Sunset pericarp samples would have detected transcripts that had ≥85% sequence complementarity. Perhaps the second Psy gene in Capsicum is more divergent than seen in tomato, in that case, the transcripts might not be detected by hybridization or the PCR primers might not be able to support amplification of divergent sequences. Identification and sequencing of the Capsicum version of the tomato Psy2 would be informative.
PSY is considered to be one of the key biosynthetic enzymes in carotenoid accumulation (Fraser et al., 2007). Despite the variable expression patterns for Psy, our results comparing the Psy transcript abundance with total carotenoid accumulation (Fig. 5) in eight different Capsicum varieties support this key role for the enzyme. It was also demonstrated that transcript abundance for Ccs was proportional to the capsanthin accumulation in six fruit. Together these results suggest that transcriptional coordination of selected genes on the carotenoid pathway is likely.
To date, two of the loci controlling fruit colour in Capsicum have been annotated with carotenoid biosynthetic activities, Y is Ccs and C2 is Psy (Lefebvre et al., 1998; Popovsky and Paran, 2000; Thorup et al., 2000; Huh et al., 2001). The nature of the gene(s) at C1 is still unknown. Our studies with orange-coloured Capsicum cultivars have revealed that the control of colour development is more complex than a deletion in a structural gene for a biosynthetic step. Two cultivars are documented (Orange Grande and Oriole) with intact forms of all four genes including Ccs, yet no transcripts for Ccs accumulate in either variety and, as expected, there is no accumulation of capsanthin or capsorubin xanthophylls and the fruit appears orange. How expression of Ccs is controlled at a transcriptional level has not been characterized. Our data with the F1 progeny between Valencia and Canary (Figs 7, 8), demonstrate that transcription of Ccs was repressed in all the yellow-orange progeny but not in the orange progeny. Clearly a regulatory gene segregated in this population. Transcriptional control of the genes on the carotenoid pathway appears to play a significant role in the expression of fruit colour. Finally, translational control, in the case again of Ccs appears to play a significant role in fruit colour. Canary, is an orange-yellow coloured cultivar, that does not accumulate any red carotenoids (Table 1), but has wild type Ccs (Guzman et al., 2010), and readily detectable transcripts for Ccs (Fig. 4; Table 2). Very low to undetectable levels of CCS protein are observed (Fig. 6), suggesting that translational control of some sort is effective in preventing the expression of Ccs in this cultivar and resulting in the orange fruit colour.
Complex regulation of carotenoid accumulation in Capsicum fruit is predicted based on our comparisons of gene sequence and expression with chemical profiles for carotenoids and physical appearance of the fruit. This system for studying carotenogeneis will continue to reveal exciting mechanisms in the future.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Alignment of Ccs promoter DNA sequences cloned from C. annuum cultivars: Canary, Fogo, NuMex Sunset, Valencia, Oriole, and Orange Grande.
Acknowledgments
We thank Paul Bosland and The Chile Pepper Institute for seeds of Capsicum varieties used in this study. This work was supported in part by the NM Agricultural Experiment Station, USDA CSREES grant 2009-34604-19939, USDA NIFA 2010-34604-20886 and NIH R25 GM61222.
References
- Ahrazem O, Rubio-Moraga A, López R, Gómez-Gómez L. The expression of a chromoplast-specific lycopene beta cyclase gene is involved in the high production of saffron's apocarotenoid precursors. Journal of Experimental Botany. 2010;61:105–119. doi: 10.1093/jxb/erp283. [DOI] [PubMed] [Google Scholar]
- Ampomah-Dwamena C, McGhie T, Wibisono R, Montefiori M, Hellens R, Allan A. The kiwifruit lycopene beta-cyclase plays a significant role in carotenoid accumulation in fruit. Journal of Experimental Botany. 2009;60:3765–3779. doi: 10.1093/jxb/erp218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai C, Twyman RM, Farre G, Sanahuja G, Christou P, Capell T, Zhu C. A golden era: pro-vitamin A enhancement in diverse crops. In vitro. Cell and Developmental Biology—Plant. 2011;47:205–221. [Google Scholar]
- Blum E, Liu K, Mazourek M, Yoo E, Jahn M, Paran I. Molecular mapping of the C locus for presence of pungency in Capsicum. Genome. 2002;45:702–705. doi: 10.1139/g02-031. [DOI] [PubMed] [Google Scholar]
- Blum E, Mazourek M, O’Connell M, Curry J, Liu K, Jahn M, Paran I. Molecular mapping of capsaicinoid biosynthetic genes and QTL analysis for capsaicinoid content in Capsicum. Theoretical and Applied Genetics. 2003;108:79–86. doi: 10.1007/s00122-003-1405-y. [DOI] [PubMed] [Google Scholar]
- Bouvier F, d’Harlingue A, Hugueney P, Marin E, Marion-Poll A, Camara B. Xanthophyll biosynthesis: cloning, expression, functional reconstitution, and regulation of β-cyclohexenyl carotenoid epoxidase from pepper (Capsicum annuum) Journal of Biological Chemistry. 1996;271:28861–22867. doi: 10.1074/jbc.271.46.28861. [DOI] [PubMed] [Google Scholar]
- Clotault J, Peltier D, Berruyer R, Thomas M, Briard M, Geoffriau E. Expression of carotenoid biosynthesis genes during carrot root development. Journal of Experimental Botany. 2008;59:3563–3573. doi: 10.1093/jxb/ern210. [DOI] [PubMed] [Google Scholar]
- Deli J, Molnar P, Matus Z, Toth G. Carotenoid composition in the fruits of red paprika (Capsicum annuum var. lycopersiciforme rubrum) during ripening: biosynthesis of carotenoids in red paprika. Journal of Agricultural and Food Chemistry. 2001;49:1517–1523. doi: 10.1021/jf000958d. [DOI] [PubMed] [Google Scholar]
- DellaPenna D, Pogson BJ. Vitamin synthesis in plants: tocopherols and carotenoids. Annual Review of Plant Biology. 2006;57:711–738. doi: 10.1146/annurev.arplant.56.032604.144301. [DOI] [PubMed] [Google Scholar]
- Diretto G, Al-Babili S, Tavazza R, Scossa F, Papacchioli V, Migliore M, Beyer P, Giuliano G. Transcriptional-metabolic networks in β-carotene-enriched potato tubers: the long and winding road to the golden phenotype. Plant Physiology. 2010;154:899–912. doi: 10.1104/pp.110.159368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fraser PD, Enfissi EMA, Halket JM, Truesdale MR, Yu D, Gerrish C, Bramley PM. Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids, and intermediary metabolism. The Plant Cell. 2007;19:3194–3211. doi: 10.1105/tpc.106.049817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorio G, Stigliani A, D’Ambrosio C. Phytoene synthase genes in tomato (Solanum lycopersicum L.): new data on the structures, the deduced amino acid sequences and the expression patterns. FEBS Letters. 2008;275:527–535. doi: 10.1111/j.1742-4658.2007.06219.x. [DOI] [PubMed] [Google Scholar]
- Guzman I, Hambly S, Romero J, Bosland P, O’Connell M. Variability of carotenoid biosynthesis in orange coloured Capsicum spp. Plant Science. 2010;179:49–59. doi: 10.1016/j.plantsci.2010.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha S-H, Kim J-B, Park J-S, Lee S-W, Cho K- J. A comparison of the carotenoid accumulation in Capsicum varieties that show different ripening colours: deletion of the capsanthin-capsorubin synthase gene is not a prerequisite for the formation of a yellow pepper. Journal of Experimental Botany. 2007;58:3135–3144. doi: 10.1093/jxb/erm132. [DOI] [PubMed] [Google Scholar]
- Hugueney P, Badillo A, Chen H-C, Klein A, Hirschberg J, Camara B, Kuntz M. Metabolism of cyclic carotenoids: a model for the alteration of this biosynthetic pathway in Capsicum annuum chromoplasts. The Plant Journal. 1995;8:417–424. doi: 10.1046/j.1365-313x.1995.08030417.x. [DOI] [PubMed] [Google Scholar]
- Huh J, Kang B, Nahm S, Kim S, Ha K, Lee M, Kim B. A candidate gene approach identified phytoene synthase as the locus for mature fruit colour in red pepper (Capsicum spp.) Theoretical and Applied Genetics. 2001;102:524–530. [Google Scholar]
- Hurtado-Hernandez H, Smith P. Inheritance of mature fruit colour in Capsicum annuum L. Journal of Heredity. 1985;76:211–213. [Google Scholar]
- Lang Y, Yanagawa S, Sasanuma T, Sasakuma T. Orange fruit colour in Capsicum due to deletion of capsanthin-capsorubin synthesis gene. Breeding Science. 2004;54:33–39. [Google Scholar]
- Lefebvre V, Kuntz M, Camara B, Palloix A. The capsanthin-capsorubin synthase gene: a candidate gene for the y locus controlling the red fruit colour in pepper. Plant Molecular Biology. 1998;36:785–789. doi: 10.1023/a:1005966313415. [DOI] [PubMed] [Google Scholar]
- Paran I, van der Knaap E. Genetic and molecular regulation of fruit and plant domestication traits in tomato and pepper. Journal of Experimental Botany. 2007;58:3841–3852. doi: 10.1093/jxb/erm257. [DOI] [PubMed] [Google Scholar]
- Popovsky S, Paran I. Molecular genetics of the y locus in pepper: its relation to capsanthin-capsorubin synthase and to fruit colour. Theoretical and Applied Genetics. 2000;101:86–89. [Google Scholar]
- Rodriguez-Uribe L, O’Connell M. A root-specific bZIP transcription factor is drought responsive in tepary bean (Phaseolus acutifolius) and common bean (P. vulgaris) Journal of Experimental Botany. 2006;57:1391–1398. doi: 10.1093/jxb/erj118. [DOI] [PubMed] [Google Scholar]
- Romer S, Hugueney P, Bouvier F, Camara B, Kuntz M. Expression of the genes encoding the early carotenoid biosynthetic enzymes in Capsicum annuum. Biochemical and Biophysical Research Communications. 1993;196:1414–1421. doi: 10.1006/bbrc.1993.2410. [DOI] [PubMed] [Google Scholar]
- Schaub P, Al-Babili S, Drake R, Beyer P. Why is golden rice golden (yellow) instead of red? Plant Physiology. 2005;138:441–450. doi: 10.1104/pp.104.057927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siddiqui MA, Grossmann J, Gruissem W, Baginsky S. Proteome analysis of bell pepper (Capsicum annuum L.) chromoplasts. Plant and Cell Physiology. 2006;47:1663–1673. doi: 10.1093/pcp/pcl033. [DOI] [PubMed] [Google Scholar]
- Stewart C, Kang B-C, Liu K, Mazourek M, Moore S, Yoo E, Kim B-D, Paran I, Jahn M. The Pun1 gene for pungency in pepper encodes a putative acyltransferase. The Plant Journal. 2005;42:675–688. doi: 10.1111/j.1365-313X.2005.02410.x. [DOI] [PubMed] [Google Scholar]
- Stewart C, Mazourek M, Stellari G, O’Connell M, Jahn M. Genetic control of pungency in C. chinense via the Pun 1 locus. Journal of Experimental Botany. 2007;58:979–991. doi: 10.1093/jxb/erl243. [DOI] [PubMed] [Google Scholar]
- Thorup T, Tanyolac B, Livingstone K, Popovsky S, Paran I, Jahn M. Candidate gene analysis of organ pigmentation loci in the Solanaceae. Proceedings of the National Academy of Sciences, USA. 2000;97:11192–11197. doi: 10.1073/pnas.97.21.11192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- USDA. Vegetables and melons yearbook: dataset. 2011 http://usda.mannlib.cornell.edu/MannUsda/viewDocumentInfo.do?documentID=1212. [Google Scholar]
- von Lintig J. Colours with functions: elucidating the biochemical and molecular basis of carotenoid metabolism. Annual Review of Nutrition. 2010;30:5.1–5.22. doi: 10.1146/annurev-nutr-080508-141027. [DOI] [PubMed] [Google Scholar]
- Wahyuni Y, Ballester A-R, Sudarmonowati E, Bino RJ, Bovy AG. Metabolite diversity in pepper (Capsicum) fruits of thirty-two diverse accessions: variation in health-related compounds and implications for breeding. Phytochemistry. 2011;72:1358–1370. doi: 10.1016/j.phytochem.2011.03.016. [DOI] [PubMed] [Google Scholar]
- Wall M, Waddell C, Bosland P. Variation in β-carotene and total carotenoid content in fruits of. Capsicum. HortScience. 2001;36:746–749. [Google Scholar]
- Welsch R, Arango J, Bar C, Salazar B, Al-Babili S, Beltran J, Chavarriaga R, Ceballos H, Tohme J, Beyer P. Provitamin A accumulation in cassava (Manihot esculenta) roots driven by a single nucleotide polymorphism in a phytoene synthase gene. The Plant Cell. 2010;22:3348–3356. doi: 10.1105/tpc.110.077560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Kandianis CB, Harjes CE, et al. Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nature Genetics. 2010;42:322–327. doi: 10.1038/ng.551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.