Abstract
Understanding the function of noncoding regions in the genome, such as introns, is of central importance to evolutionary biology. One approach is to assay for the targets of natural selection. On one hand, the sequence of introns, especially short introns, appears to evolve in an almost neutral manner. Whereas on the other hand, a large proportion of intronic sequence is under selective constraint. This discrepancy is largely dependent on intron length and differences in the methods used to infer selection. We have used a method based on DNA strand asymmetery that does not require comparison with any putatively neutrally evolving sequence, nor sequence conservation between species, to detect selection within introns. The strongest signal we identify is associated with short introns. This signal comes from a family of motifs that could act as cryptic 5′ splice sites during mRNA processing, suggesting a mechanistic justification underlying this signal of selection. Together with an analysis of intron length and splice site strength, we observe that the genomic signature of splicing-coupled selection differs between long and short introns.
Keywords: genome evolution, intron length, selection
Introns dominate the human genome, constituting ∼33% of DNA sequence and greater than 95% of the transcriptome. With only a few exceptions, all eukaryotic species contain a mix of both short and long introns. However, the amount of regulatory sequence necessary to identify and remove an intron will vary depending on intron length. The processing of long introns requires multiple sources of information including cis-regulatory motifs and trans-acting splicing factors, the chromatin landscape, and the kinetics of polymerase elongation (Berget 1995; Hertel 2008; Yu et al. 2008; Chen et al. 2010). The identification of short introns is thought to require less regulatory information, being largely dependent on the proximity of a 5′ and 3′ splice site, in a process termed intron definition (Talerico and Berget 1994; Lim and Burge 2001). Regardless of size, failure to correctly process an intron presents a significant cost to the cell (Jaillon et al. 2008; Ramani et al. 2009). Therefore, we have analyzed the targets of splicing-coupled selection that act to maintain efficient splicing, and how this selection pressure varies with intron length.
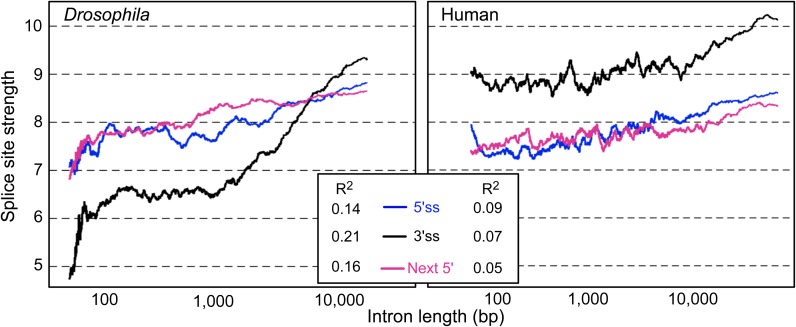
The 5′ and 3′ splice sites play a pivotal role during intron definition of short introns. Therefore, it is notable that in general short introns show weaker splice sites than longer introns. Our analysis of splice site strength (supplementary methods, Supplementary Material online) and intron length indicates that both in Drosophila (also see Fahey and Higgins 2007) and in humans, the 5′ and 3′ splice site strength increases with intron length (fig. 1).
FIG. 1.
Splice site strength increases with intron length. Splice site strength is positively correlated with intron length in Drosophila melanogaster (Fahey and Higgins 2007) and human. All introns were ranked according to length and mean splice site strength was measured within a sliding window of 1,000 introns (step size = 1 intron). All Pearson R2 values are significantly different from 0 (P < 0.0001).
Interestingly, we also observe that the strength of the 5′ splice site of the next downstream intron also increases with the length of the upstream intron. To establish if this relationship was independent of the correlation between the length of adjacent introns, we fit a partial correlation between downstream 5′ splice site strength and the log10 length of the upstream intron, correcting for log10 length of downstream intron. This result remained significant (Pearson R2 = 0.10, P < 0.0001; Spearman R2 = 0.12, P < 0.0001 in Drosophila melanogaster). To double check this result, we fit a general linear model on downstream 5′ splice site strength as a response, with up and downstream intron length as explanatory variables. Both effects were significant (data not shown), indicating that intron length influences the selection pressure on the 5′, 3′, and next 5′ splice site of an intron. This finding is consistent with a model of exon definition in which the 5′ splice site of the next intron is required to validate an exon flanked by long introns (Berget 1995; Hicks et al. 2010). Notably, exon definition reduces the inclusion of false or pseudoexons during the splicing of very long introns (Sun and Chasin 2000).
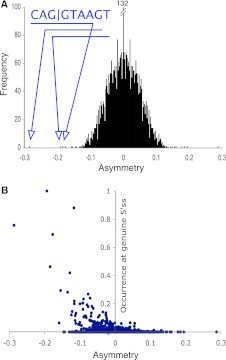
The ability of the spliceosome to identify and act upon a diverse set of 5′ splice site motifs offers a flexibility that is the basis of alternative splicing (Irimia et al. 2007). However, this also greatly expands the number of potential cryptic (or latent) splice sites that may disrupt splicing. If such motifs have phenotypic consequences they may be under selection. DNA sequence motifs under neutral evolution generally show a symmetric distribution between the forward and the reverse strand of a double-stranded genome (Mitchell and Bridge 2006). If splicing imposes a functional constraint upon a particular motif, it could lead to either an excess or depletion of that sequence on the coding strand of a gene. For the introns of D. melanogaster (and 18 further species, see supplementary methods, Supplementary Material online), we calculated the asymmetry of all possible motifs of length 5, 6, and 7 bp between the forward and the reverse strand (supplementary table 1, Supplementary Material online). The most highly asymmetric (underrepresented) motif at all three lengths matched perfectly to the consensus 5′ splice site (fig. 2A and supplementary fig. 1, Supplementary Material online). The significant under representation (Z = 28.4; P < 0.0001) of this motif, G|GTAAG (| denotes a potential exon–intron boundary), from introns is suggestive of purifying selection against a motif that could potentially be recognized as a spurious 5′ splice site. The trend toward negative asymmetry extends to all motifs that occur at high frequency within genuine 5′ splice site (fig. 2B), indicating that purifying selection acts upon a large family of sequence motifs that may compete with actual 5′ splice sites. These results are not due to any global bias in nucleotide composition between the coding and the noncoding strand nor any local asymmetry caused by the polypyrimidine tract or branch point (supplementary fig. 2, Supplementary Material online).
FIG. 2.
Cryptic 5' splice sites are underrepresented in introns. (A) The distribution of asymmetry values for all possible 4096 hexamers across the introns of Drosophila melanogaster. The distribution of asymmetry scores is symmetrical around 0 because each motif and its reverse complement have the same value but with opposite signs. The asymmetry against the motif G|GTAAG (−0.288) is six standard deviations off the mean of the distribution. Arrows indicate the top three values belong to 6mers that overlap the 9 bp of the consensus 5′ splice site, CAG|GTAAGT. Importantly, the next most significant value over laps the motif CAG|GTGAGT, indicating selection against both AA and GA cryptic splice sites (supplementary fig. 1, Supplementary Material online). (B) Motifs present at high frequency at actual 5′ splice sites show high asymmetry within D. melanogaster introns. This indicates that selection targets a large number of motifs that may compete during splicing, and it follows that introns with generally weak splice sites will be sensitive to competition from a larger number of potential cryptic splice sites. Number of motifs shown is all possible 4096 hexamers.
The cryptic splice site motif G|GTAAG also returns the strongest asymmetry in the introns of six other genomes (which are all dominated by short introns): the Dipterans Drosophila ananassae, Drosophila grimshawi, and Anopheles gambiae, the nematode Caenorhabditis elegans, and the yeast Aspergillus nidulans and Schizosaccharomyces pombe. Most species have an almost equal preference for A and G at position +3 of their actual 5′ splice site (Irimia et al. 2009), and our analysis indicates strong asymmetry against both motifs in these species (supplementary table 2, Supplementary Material online). However, several species, including A. gambiae and S. pombe, have a strong preference for only a single motif within genuine 5′ splice sites. In these two species, we only observe significant asymmetry against the single motif that is used in these species. This supports competition between the genuine 5′ splice site and similar motifs within an intron as the basis of this observed asymmetry.
Intron length has a major influence on the mode of action of the spliceosome and the choice between competing splice sites (Fox-Walsh et al. 2005; Kandul and Noor 2009). We therefore considered the relationship between asymmetry against cryptic splice sites and intron length. Both in Drosophila (R2 = 0.781, P < 0.0001) and in human (R2 = 0.806, P < 0.0001), we observed a highly significant correlation between asymmetry against the motif G|GTAAG and intron length (fig. 3A). This indicates that purifying selection against cryptic splice sites is stronger within short introns. This was confirmed when we considered 19 additional eukaryotic genomes (fig. 3B), with asymmetry against cryptic splice sites being highly dependent on the average intron length within a species (R2 = 0.696, P < 0.0001). While the relationship in figure 3A (and supplementary fig. 2B, Supplementary Material online) indicates that cryptic splice sites may be under selection even in long introns, the overwhelming signal of asymmetry against the motif G|GTAAG comes from introns < 1,000 bp (supplementary fig. 2C, Supplementary Material online). Considering the distribution of cryptic splice sites within introns, we observe a slight nonsignificant excess of cryptic splice sites within the first 44 bp of Drosophila introns relative to downstream sequence and a highly significant depletion within the last 30 bp (supplementary fig. 3, Supplementary Material online)
FIG. 3.
Selection targets cryptic splice sites in short but not long introns. (A) Asymmetry against the cryptic splice site motif (G|GTAAG) is stronger in the short introns of Drosophila melanogaster and human. Introns were partitioned into 20 nonoverlapping bins of increasing length such that each bin contains the same total amount of sequence (2.5 Mb for Drosophila and 47 Mb for human), and hence, a variable number of introns. Asymmetry was calculated for each bin. A general linear model of the raw data (not binned) was used to establish the significance of intercept and slope and Pearson correlation coefficients are reported. (B) Asymmetry against a cryptic splice sites is stronger in species with shorter introns. Asymmetry against the motif (G|GTAAG) versus the mean intron length for 19 eukaryotic genomes. Each data point represents the asymmetry for all concatenated intronic sequence of a species. In total, 69.6% of the variation in asymmetry between species is explained by intron length within that species. This indicates that cryptic splice sites within short introns are a common target of purifying selection across eukaryotes. R2 is the Pearson correlation coefficient calculated with a general linear model.
We note that this absence of strong asymmetry in long introns might in part result from the maintenance of alternative 5′ splice sites within introns or regulatory sequence that include cryptic splice sites. For example, very long introns in Drosophila (>20 kb) may contain recursive splicing sites that promote the stepwise processing of these introns (Burnette et al. 2005), however, such sites are in general rare.
A large proportion of intronic sequence is under selective constraint (Parsch 2003; Andolfatto 2005; Haddrill et al. 2005; Halligan and Keightley 2006; Sella et al. 2009). However, the traits that underlie this selection are largely unknown. Selection against spurious transcription factor binding sites produces significant constraint in the coding and noncoding sequence of both eubacterial and archaeal genomes (Hahn et al. 2003). Our data indicate that a proportion of the selective constraint observed within short introns of eukaryotes is associated with purifying selection against motifs that would otherwise disrupt the correct identification of 5′ splice sites. Despite the fact that cryptic splice sites greatly outnumber genuine splice sites in the genome, this effect is likely to account for only a small proportion of the inferred selective constraint within introns. However, our observation that most of this signal is associated with short introns should temper the view that this sequence is a good standard for neutrally evolving sequence within the genome (Parsch et al. 2010).
Intron length plays a crucial role during intron recognition and splicing. Various lines of evidence suggest that short introns evolve under selective constraint to maintain an optimal length (Carvalho and Clark 1999; Parsch 2003; Parsch et al. 2010). Long introns, on the other hand, utilize several sources of regulatory information, that include cis-regulatory motifs, chromatin structure, and exon length. However, the factors that govern intron length evolution are not well defined. Our data suggest that the frequency of cryptic splice sites might play a role in the evolution of intron length and the targets of splicing-coupled selection. As intron length increase, more potential cryptic splice sites can be generated by mutation. It is conceivable that within longer introns less purifying selection is needed to maintain the more elaborate signals associated with exon definition than to purge cryptic splice sites. Such a trade-off is indicative of an error threshold, the point at which a high mutation rate overwhelms selection (Bull et al. 2005; Wilke 2005).
It may be possible that the length at which intron definition ceases to function within a species is at least partly governed by the size range in which cryptic splice sites remain rare. The almost exclusive use of a single 5′ splice site in Saccharomyces cerevisiae (63% of introns begin with |GTAGT) (Irimia et al. 2007), considerably reduces the number of potential cryptic splice sites that may compete with the genuine splice site, consistent with this species having an unusually high cutoff between short and long introns (Lim and Burge 2001).
Supplementary Material
Supplementary methods, figures 1–3, and tables 1 and 2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Roy W. Scott for kindly providing the Cryptococcus intronic sequence. We are grateful to Claus Wilke, Thomas Flatt, Nico Posnien, and Jacki Heraud for comments on this manuscript. This work was supported by grants of the Fonds zur Förderung der wissenschaftlichen Forschung (P19832, L403) awarded to C.S. and Eurasia-Pacific Uninet PhD scholarship awarded to L.H.
References
- Andolfatto P. Adaptive evolution of non-coding DNA in Drosophila. Nature. 2005;437:1149–1152. doi: 10.1038/nature04107. [DOI] [PubMed] [Google Scholar]
- Berget SM. Exon recognition in vertebrate splicing. J Biol Chem. 1995;270:2411–2414. doi: 10.1074/jbc.270.6.2411. [DOI] [PubMed] [Google Scholar]
- Bull JJ, Meyers LA, Lachmann M. Quasispecies made simple. PLoS Comput Biol. 2005;1:e61. doi: 10.1371/journal.pcbi.0010061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnette JM, Miyamoto-Sato E, Schaub MA, Conklin J, Lopez AJ. Subdivision of large introns in Drosophila by recursive splicing at nonexonic elements. Genetics. 2005;170:661–674. doi: 10.1534/genetics.104.039701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho AB, Clark AG. Intron size and natural selection. Nature. 1999;401:344. doi: 10.1038/43827. [DOI] [PubMed] [Google Scholar]
- Chen W, Luo L, Zhang L. The organization of nucleosomes around splice sites. Nucleic Acids Res. 2010;38:2788–2798. doi: 10.1093/nar/gkq007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahey M, Higgins D. Gene expression, intron density, and splice site strength in Drosophila and Caenorhabditis. J Mol Evol. 2007;65(3):349–357. doi: 10.1007/s00239-007-9015-y. [DOI] [PubMed] [Google Scholar]
- Fox-Walsh KL, Dou Y, Lam BJ, Hung SP, Baldi PF, Hertel KJ. The architecture of pre-mRNAs affects mechanisms of splice-site pairing. Proc Natl Acad Sci U S A. 2005;102:16176–16181. doi: 10.1073/pnas.0508489102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haddrill PR, Charlesworth B, Halligan DL, Andolfatto P. Patterns of intron sequence evolution in Drosophila are dependent upon length and GC content. Genome Biol. 2005;6:R67. doi: 10.1186/gb-2005-6-8-r67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn MW, Stajich JE, Wray GA. The effects of selection against spurious transcription factor binding sites. Mol Biol Evol. 2003;20:901–906. doi: 10.1093/molbev/msg096. [DOI] [PubMed] [Google Scholar]
- Halligan DL, Keightley PD. Ubiquitous selective constraints in the Drosophila genome revealed by a genome-wide interspecies comparison. Genome Res. 2006;16:875–884. doi: 10.1101/gr.5022906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertel KJ. Combinatorial control of exon recognition. J Biol Chem. 2008;283:1211–1215. doi: 10.1074/jbc.R700035200. [DOI] [PubMed] [Google Scholar]
- Hicks MJ, Mueller WF, Shepard PJ, Hertel KJ. Competing upstream 5′ splice sites enhance the rate of proximal splicing. Mol Cell Biol. 2010;30(8):1878–1886. doi: 10.1128/MCB.01071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Penny D, Roy SW. Coevolution of genomic intron number and splice sites. Trends Genet. 2007;23:321–325. doi: 10.1016/j.tig.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Irimia M, Roy SW, Neafsey DE, Abril JF, Garcia-Fernandez J, Koonin EV. Complex selection on 5′ splice sites in intron-rich organisms. Genome Res. 2009;19:2021–2027. doi: 10.1101/gr.089276.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillon O, Bouhouche K, Gout J, et al. (19 co-authors) Translational control of intron splicing in eukaryotes. Nature. 2008;451:359–362. doi: 10.1038/nature06495. [DOI] [PubMed] [Google Scholar]
- Kandul NP, Noor MA. Large introns in relation to alternative splicing and gene evolution: a case study of Drosophila bruno-3. BMC Genet. 2009;10:67. doi: 10.1186/1471-2156-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim LP, Burge CB. A computational analysis of sequence features involved in recognition of short introns. Proc Natl Acad Sci U S A. 2001;98:11193–11198. doi: 10.1073/pnas.201407298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell D, Bridge R. A test of Chargaff's second rule. Biochem Biophys Res Commun. 2006;340:90–94. doi: 10.1016/j.bbrc.2005.11.160. [DOI] [PubMed] [Google Scholar]
- Parsch J. Selective constraints on intron evolution in Drosophila. Genetics. 2003;165:1843–1851. doi: 10.1093/genetics/165.4.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsch J, Novozhilov S, Saminadin-Peter SS, Wong KM, Andolfatto P. On the utility of short intron sequences as a reference for the detection of positive and negative selection in Drosophila. Mol Biol Evol. 2010;27(6):1226–1234. doi: 10.1093/molbev/msq046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramani AK, Nelson AC, Kapranov P, Bell I, Gingeras TR, Fraser AG. High resolution transcriptome maps for wild-type and NMD mutant C. elegans through development. Genome Biol. 2009;10:R101. doi: 10.1186/gb-2009-10-9-r101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G, Petrov DA, Przeworski M, Andolfatto P. Pervasive natural selection in the Drosophila genome? PLoS Genet. 2009;5:e1000495. doi: 10.1371/journal.pgen.1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Chasin LA. Multiple splicing defects in an intronic false exon. Mol Cell Biol. 2000;20:6414–6425. doi: 10.1128/mcb.20.17.6414-6425.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talerico M, Berget SM. Intron definition in splicing of small Drosophila introns. Mol Cell Biol. 1994;14:3434–3445. doi: 10.1128/mcb.14.5.3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilke CO. Quasispecies theory in the context of population genetics. BMC Evol Biol. 2005;5:44. doi: 10.1186/1471-2148-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Maroney P, Denker J, Zhang X, Dybkov O, Luhrmann R, Jankowsky E, Chasin L, Nilsen T. Dynamic regulation of alternative splicing by silencers that modulate 5 splice site competition. Cell. 2008;135:1224–1236. doi: 10.1016/j.cell.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.