Abstract
The sequences of the untranslated regions (UTRs) of mRNAs play important roles in posttranscriptional regulation, but whether a change in UTR length can significantly affect the regulation of gene expression is not clear. In this study, we examined the connection between UTR length and Expression Correlation with cytosolic ribosomal proteins (CRP) genes (ECC), which measures the level of expression similarity of a group of genes with CRP genes under various growth conditions. We used data from the aerobic fermentation yeast Saccharomyces cerevisiae and the aerobic respiration yeast Candida albicans. To reduce statistical fluctuations, we computed the ECC for the genes in a Gene Ontology (GO) functional group. We found that in both species, ECC is strongly correlated with the 5′ UTR length but not with the 3′ UTR length and that the 5′ UTR length is evolutionarily better conserved than the 3′ UTR length. Interestingly, we found 11 GO groups that have had a substantial increase in 5′ UTR length in the S. cerevisiae lineage and that the length increase was associated with a substantial decrease in ECC. Moreover, 9 of the 11 GO groups of genes are involved in mitochondrial respiration function, whose expression reprogramming has been shown to be a major factor for the evolution of aerobic fermentation. Finally, we found that an increase in 5′ UTR length may decrease the +1 nucleosome occupancy. This study provides a new angle to understand the role of 5′ UTR in gene expression regulation and evolution.
Keywords: UTR length, gene expression evolution, aerobic fermentation
Introduction
Gene transcriptional regulation is known to be mediated by transcription factors (TFs), RNA polymerase, chromatin structure and cis-acting elements, and gene expression is also subject to posttranscriptional regulation. The 5′ and 3′ untranslated regions (5′ UTR and 3′ UTR) have been recognized for their important roles in the regulation of gene expression at the posttranscriptional level by affecting the mRNA stability, localization, and translational efficiency (van der Velden and Thomas 1999; Jansen 2001; Mignone et al. 2002). The ability of UTRs to perform posttranscriptional regulation is mainly encoded by their sequences. For example, some 5′UTRs and 3′ UTRs contain regulatory sequences that may affect the mRNA’s stability, translational efficiency, or subcellular localization, such as selenocysteine insertion sequence elements, AU-rich elements, riboswitches, and microRNA-binding sites (van der Velden and Thomas 1999; Jansen 2001; Mignone et al. 2002).
In eukaryotes, the genome average of 5′ UTR lengths is remarkably similar across diverse taxonomic classes, ranging only from ∼100 to ∼200 base pair (bp) (Mignone et al. 2002; Nagalakshmi et al. 2008; Xu et al. 2009; Bruno et al. 2010). In sharp contrast, the 5′ UTR length varies considerably among the genes in a genome, ranging from a few base pair to several thousand base pair (Pesole et al. 2001; Mignone et al. 2002; Nagalakshmi et al. 2008). Large-scale studies of transcript boundaries in different yeasts showed that the 5′ transcript boundary of some genes is identical to the translation start site, indicating that 5′ UTR could be absent in these genes, though the possibility of errors in annotation of translation start sites or transcript boundaries could not be neglected (Nagalakshmi et al. 2008; Xu et al. 2009; Bruno et al. 2010). In fact, according to a study using a mammalian in vitro system, even a single nucleotide is a sufficient 5′ UTR for the initiation of translation (Hughes and Andrews 1997). In comparison, the genome average of 3′ UTR lengths is much more variable among eukaryotes, ranging from about 100 bp in fungi to 800 bp in human and other vertebrates (Pesole et al. 2001; Mignone et al. 2002; Nagalakshmi et al. 2008).
It has been noticed that genes with different functions show distinct UTR lengths. In a study of 669 vertebrate mRNAs, Kozak (Kozak 1987) found that genes with a long 5′ UTR, such as those involved in development or meiosis, are generally highly and finely regulated. Genome-wide surveys of transcript boundary with microarrays in Saccharomyces cerevisiae have revealed that genes that require fine regulation, such as those involved in the regulation of cellular processes, especially transcriptional regulation, tend to have longer UTRs, whereas genes with a reduced need for regulation, such as housekeeping genes and the ribosomal subunit genes, usually have shorter UTRs (Hurowitz and Brown 2003; David et al. 2006). Recent studies using RNA-seq further supported the uneven distribution of UTR lengths among genes with different functions in yeasts (Nagalakshmi et al. 2008; Bruno et al. 2010). These observations suggested that the regulation of gene expression might be in part mediated by different UTR lengths; this is especially true for 5′ UTR length because the 5′ UTR of a gene is the region immediately downstream of its transcription start site (TSS). To test this hypothesis, we have systematically examined the connection between UTR length and the pattern of gene expression across various conditions, and the correlation between evolutionary change in UTR length and gene expression divergence using data from two important model yeasts, S. cerevisiae and Candida albicans, which have diverged about 200 Ma (Berman and Sudbery 2002; Sudbery et al. 2004). As model yeast systems, abundant gene expression data under various growth or stress conditions have been accumulated in both species (Ihmels et al. 2005), and their genome-wide transcript length data have also been generated (Nagalakshmi et al. 2008; Xu et al. 2009; Bruno et al. 2010), providing an ideal opportunity to explore the role of UTR length in gene expression regulation.
An important goal to study the regulatory mechanism of gene expression is to unravel how changes in genomic structure have contributed to organism’s gene expression divergence and phenotypic evolution. Saccharomyces cerevisiae has evolved a special ability to efficiently ferment glucose into ethanol even in the presence of oxygen, called aerobic fermentation (Pronk et al. 1996), whereas C. albicans still maintains aerobic respiration (Thomson et al. 2005; Piskur et al. 2006; Merico et al. 2007; Lin and Li 2011b). It has been shown that expression of the genes involved in mitochondrial respiration has been reprogrammed during the evolution of aerobic fermentation in the S. cerevisiae lineage (DeRisi et al. 1997; Ihmels et al. 2005; Field et al. 2009). Therefore, it is interesting to investigate whether UTR length change was associated with the evolution of gene expression and aerobic fermentation in the S. cerevisiae lineage. This study may increase our understanding of the genetic basis for the evolution of aerobic fermentation in the ancestor of S. cerevisiae.
Materials and Methods
Data Sources
The genome-wide 5′ UTR and 3′ UTR length data in S. cerevisiae were obtained from Xu et al. (2009), which are currently the most complete set of transcript boundary data in S. cerevisiae. The genome-wide 5′ UTR and 3′ UTR length data in C. albicans were retrieved from Bruno et al. (2010). The lengths of open reading frames (ORFs) of S. cerevisiae and C. albicans were downloaded from Saccharomyces Genome Database (http://www.yeastgenome.org/) and Candida Genome Database (http://www.candidagenome.org/). The 5′ UTR, 3′ UTR, and ORF length data in S. cerevisiae and C. albicans were compiled as supplementary table S1, Supplementary Material online. We used the yeast orthologous gene maps (Wapinski et al. 2007) to obtain 2,957 pairs of one-to-one orthologous genes between S. cerevisiae and C. albicans. Among them, 2,516 one-to-one orthologous pairs have UTR length data in both species (supplementary table S2, Supplementary Material online). We downloaded the large collections of compiled microarray data of S. cerevisiae (1,011 expression profiles) and C. albicans (198 expression profiles) from Ihmels et al. (2005). These expression data were obtained under a large variety of growth conditions, stress conditions, cell cycle stages, or genetic backgrounds. We used the Gene Ontology (GO) structure file (OBO v1.2) and GO annotation data of C. albicans (Revision: 1.587) and S. cerevisiae (Revision: 1.149), respectively (Ashburner et al. 2000). According to the GO term hierarchy structure, the genes of a GO term include: 1) all genes that are associated with this term in annotation and 2) all genes that are associated to any of its child terms. The complete list of genes under each GO term in the two species were compiled based on GO structure, and their annotation data using homemade perl scripts. We downloaded the gene lists for all the 86 transcription modules in S. cerevisiae which were clustered by the “signature algorithm” using all available genome-wide expression data (Ihmels et al. 2002). The in vivo nucleosome occupancy data of S. cerevisiae and C. albicans were downloaded from Field et al. (2009).
Computing Expression Correlations Between Gene Sets
The formation of cytosolic ribosomes is a response to demand of protein biosynthesis. In yeast, the rate of ribosome formation is perfectly tuned to the cellular growth rate (Mager and Planta 1991). At steady state growth conditions, the transcription of cytosolic ribosomal protein (CRP) genes is balanced and constitutive, and it produces a nearly constant amount of mRNA (Mager and Planta 1991). Under a rapid growth condition, the expression of CRP genes is coordinately adjusted to the new cellular demands. In short, the CRP genes have constitutive expression during steady growth but coordinately increase expression during rapid growth. Thus, they can be considered as growth-related housekeeping genes. This property can be used as a biologically meaningful proxy to evaluate gene expression pattern. If the expression of a group of genes is highly correlated with that of CRP genes, it means that these genes are expressed in a manner similar to growth-related genes; otherwise, these genes are expressed similar to nongrowth-related or stress-related genes. Therefore, using CRP genes as a proxy to compare gene expression profiles between different yeast species have been well accepted and used in several previous studies (Ihmels et al. 2005; Field et al. 2009; Lin and Li 2011a). To estimate the expression level changes under various conditions, we calculated Expression Correlation with CRP genes (ECC), which is defined as the average Pearson’s correlation coefficient of expression levels between genes in a GO group and the CRP genes:
where nG is the number of genes in the GO group under study, nC is the number of CRP genes, and Rji is the Pearson’s correlation coefficient of expression level between gene i in the GO group and CRP gene j. We obtained 132 and 78 CRP genes for S. cerevisiae and C. albicans, respectively, based on their genomic annotation information from Saccharomyces Genome Database and Candida Genome Database. To avoid the potential biases caused by different numbers of CRP genes between the two species, we run BlastP to obtain 74 pairs of CRP genes that were detected as reciprocal best hits (supplementary table S3, Supplementary Material online); thus, an equal number of CRP genes in the two species were used to calculate ECC. We also repeated our analyses using all CRP genes in both species and obtained essentially identical results (data not shown).
To obtain more accurate estimation of ECC, we only used the expression data that were obtained under similar conditions in the two species. That is, we excluded those expression data that were obtained from species-specific conditions, such as data from strains with deletion or overexpression of a certain gene. As a result, we used 593 and 162 expression profiles in S. cerevisiae and C. albicans, respectively. To determine if all genes in a GO group have coherent expression patterns, we computed the expression coherence score (ECS) by averaging Pearson’s correlation coefficient of expression profiles between every pair of genes in a GO group as
where n is the number of genes in the GO group under study, and Rji is the Pearson’s correlation coefficient of expression between gene i and gene j in the same GO group.
Calculating In vivo +1 Nucleosome Occupancy of a GO Group
We used the in vivo nucleosome occupancy data in S. cerevisiae and C. albicans to study the average +1 nucleosome occupancy for each GO group (Field et al. 2009). Because 85% of genes in S. cerevisiae and 78% of genes in C. albicans have 5′ UTR length ≤200 bp (supplementary table S1, Supplementary Material online) and approximately 147 bp of DNA wrap around a histone octamer (Luger et al. 1997), we used a sliding window of 100 bp to scan nucleosome occupancy in the 200 bp region immediately downstream of TSS for each gene. The window with the peak nucleosome occupancy is likely to be where the +1 nucleosome positioned in the 5′ UTR. The +1 nucleosome occupancy of a GO group was calculated as the average of nucleosome occupancies of peak windows in all genes of the group.
All statistical analyses in this work were conducted in R (http://www.R-project.org). The graphs were originally created with R and further edited using Adobe Illustrator CS2.
Results
The 5′ UTR Length Is Evolutionarily Better Conserved Than the 3′ UTR Length
To obtain an overall picture of UTR length change in evolution, we examined the length differences for one-to-one orthologous pairs between S. cerevisiae and C. albicans using the 5′ UTR and 3′ UTR length data from Xu et al. (2009) and Bruno et al. (2010). We obtained 2,516 one-to-one orthologous pairs with available UTR length data (supplementary table S2, Supplementary Material online). The ORF lengths in the two species were also studied for comparison. The mean lengths and standard deviations for 5′ UTR, 3′ UTR, and ORF are 96.5 ± 116.8 bp, 146.7 ± 138.4 bp, and 1537.1 ± 1164.8 bp in S. cerevisiae and 120.9 ± 147.2 bp, 141.2 ± 126.7 bp, and 1566.0 ± 1179.8 bp in C. albicans. The ORF lengths are highly conserved between the two species with a Spearman’s rank correlation rho = 0.97, which is consistent with a previous study on the protein length evolution in eukaryotes (Wang et al. 2005). In contrast, Spearman’s rank correlation is only 0.20 (P value < 2.20 × 10−16) for 5′ UTR length and only 0.08 (P value = 7.55 × 10−5) for 3′ UTR length between the two species (supplementary fig. S1, Supplementary Material online), suggesting that 5′ UTR and 3′ UTR lengths are much more variable than ORF lengths. However, we should consider the fact that the measurements of 5′ UTR and 3′ UTR lengths of a gene are much less accurate than the measurement of ORF lengths. Currently, the most common methods used to measure the UTR lengths are rapid amplification of cDNA ends, microarray, and RNA-seq. For technical and sample preparation issues, discrepancy of UTR lengths between different measurements is very common, although most of UTR lengths are differentiated within 50 bp (Nagalakshmi et al. 2008; Bruno et al. 2010). Furthermore, some genes might use alternative TSSs under different conditions or in different tissues, which creates additional uncertainty of 5′ UTR length (Hake et al. 1990; Yiu et al. 1994). Therefore, we might underestimate the conservation level of UTR lengths using current UTR length data.
It is worth noting that the Spearman’s correlation coefficient of 5′ UTR length is considerably higher than that of 3′ UTR length, suggesting that the 5′ UTR length is less variable during evolution than 3′ UTR length. The mean differences for 5′ UTR and 3′ UTR lengths between S. cerevisiae and C. albicans are −24.5 ± 172.36 bp and −5.4 ± 182.3 bp, respectively. The length differences for 5′ UTR and 3′ UTR between the two species are both nearly normally distributed around 0 bp (supplementary fig. S2, Supplementary Material online), but the distribution of 5′ UTR length differences has a higher peak than that of 3′ UTR, indicating the evolutionary change in 5′ UTR length is not as large as 3′ UTR length change. To quantitatively measure the variability of UTR length change in S. cerevisiae since its divergence from C. albicans, we calculated the median absolute deviation (MAD) for 5′ UTR length differences (MAD = 63.8) and for 3′ UTR length differences (MAD = 105.3), respectively. Note that the MAD of 3′ UTR length differences is 65% higher than that of 5′ UTR length differences, further supporting a better evolutionary conservation of 5′ UTR length. This observation is consistent with the fact that average 3′ UTR lengths among different eukaryotes are much more variable than 5′ UTR lengths (Mignone et al. 2002).
ECC Is Correlated With 5′ UTR Length But Not 3′ UTR Length
As mentioned above, current measurement of UTR lengths of a gene may be inaccurate. Therefore, if we study the correlation between UTR length and gene expression pattern of individual genes, the signal could be diluted by potential noises due to data inaccuracy. To mitigate this effect, we used GO functional groups as units to study the correlation between UTR lengths and gene expression pattern (see Materials and Methods). To reduce the potential biases due to a small sample size, we only considered GO groups that contain ≥10 genes with UTR length data. We also calculated the coefficient of variation (CV) of GO transcript lengths, which is measured as the ratio of the standard deviation of the gene transcript lengths to the mean transcript length in a GO group. If CV > 1, it indicates that the transcript lengths within a GO group are very inconsistent or highly skewed, and thus, these GO groups were excluded from our subsequent analysis.
To assess whether the genes of a GO group have expression patterns similar to CRP genes under various conditions, we calculated their ECC. ECC ranges from −1 to 1; ECC = 1 indicates that all genes in a GO group have a perfectly positive ECC under all examined conditions. It has been found that genes with a high ECC are usually involved in growth (“growth”-related genes), whereas genes with a low ECC tend to have functions related to stress response (“stress”-related genes) (Tirosh and Barkai 2008; Field et al. 2008, 2009; Tsankov et al. 2010). To determine if the genes in a GO group have coherent expression patterns, we calculated the ECS for each GO group (see Materials and Methods).
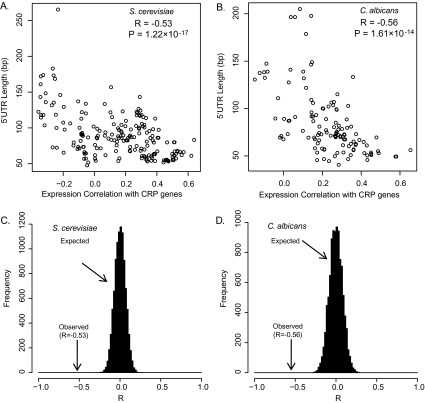
To ensure the robustness of our conclusion, we used five different ECS cutoffs (ECS ≥ 0.10, 0.15, 0.20, 0.25, and 0.30) to select the coherent expressed GO groups and calculated the correlations between their UTR lengths and ECC values. We obtained 844, 440, 229, 139, and 110 coherent GO groups, respectively, in S. cerevisiae (supplementary table S4, Supplementary Material online). By plotting the average 5′ UTR lengths of these GO groups against their ECC values, we observed significant negative correlations (R ranging from −0.38 to −0.60, P < 0.01) with all ECS cutoffs (supplementary table S4, Supplementary Material online). Similarly, the average 5′ UTR lengths and ECCs of C. albicans coherent GO groups are negatively correlated (R ranging from −0.43 to −0.58, P < 0.01) (supplementary table S4, Supplementary Material online). These results reveal a robust correlation between 5′ UTR length and ECC. Specifically, genes with a shorter 5′ UTR tend to have a higher ECC, indicating more stable expression levels under different conditions. We then used the median ECS cutoff (ECS ≥ 0.20) for subsequent analysis, unless otherwise indicated (fig. 1). We also conducted a random permutation test. We randomly assigned an observed 5′ UTR length to a gene in a species while keeping the ECC value and gene number in each GO group unchanged to estimate the correlations expected by chance between 5′ UTR length and ECC. The simulation was repeated 10,000 times, and the simulated correlation coefficients were normally distributed around 0 with a standard deviation σ = 0.067 in S. cerevisiae and σ = 0.080 in C. albicans (fig. 1C and D). The observed correlations in S. cerevisiae (R = −0.53) and C. albicans (R = −0.56) are at least seven standard deviations away from the simulation means, and thus, they are highly unlikely to have occurred by chance (P < 0.0001, fig. 1C and D).
FIG. 1.
Negative correlation between 5′ UTR length and ECC in Saccharomyces cerevisiae and Candida albicans. (A–B) Scatter plot of 5′ UTR lengths and ECCs of 229 GO groups in S. cerevisiae (A) and 158 GO groups in C. albicans (B) with a cutoff of ECS ≥ 0.20. The 5′ UTR length is negatively correlated with ECC in both S. cerevisiae and C. albicans. (C−D) The observed negative correlation between 5′ UTR length and ECC is statistically significant in both S. cerevisiae (C) and C. albicans (D). The x axis shows the Pearson’s correlation coefficients (R) between 5′ UTR length and ECC in 10,000 random simulations. The y axis indicates the number of simulated Rs in a bin of width 0.02. In both S. cerevisiae and C. albicans, the observed Pearson’s correlation between 5′ UTR length and ECC is negative, and its absolute value is larger than all of the 10,000 simulated correlations, that is, P value < 0.0001.
Unlike 5′ UTR, the variation of 3′ UTR length appears to have very limited impacts on gene expression. As can be seen in supplementary figure S3, Supplementary Material online, no significant correlation at the P value ≤ 0.01 level is observed between 3′ UTR length and ECC in S. cerevisiae (R = −0.07, P value = 0.23) and in C. albicans (R = −0.16, P value = 0.02). Similar to 3′ UTR, the ORF length also has no significant correlation with ECC in either species (supplementary fig. S3C and D, Supplementary Material online). Therefore, our results reveal that among the three different parts of an mRNA, only the 5′ UTR length is involved in the regulation of gene expression in yeasts.
Association Between Evolutionary Change in 5′ UTR Length and Reprogramming of Gene Expression
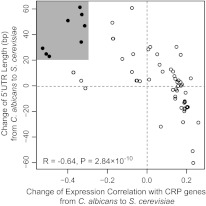
If 5′ UTR length is important for fine regulation of gene expression, it is reasonable to speculate that evolutionary change in 5′ UTR length has contributed to the divergence of gene expression. To test this hypothesis, we calculated the differences in 5′ UTR length and in ECC values between S. cerevisiae and C. albicans for 78 coherent GO groups that are shared by the two species. As shown in figure 2, the increase in 5′ UTR length is strongly correlated with the decrease in ECC (R = −0.64, P value = 2.84×10−10), suggesting that changes in 5′ UTR length have contributed to reprogramming of gene expression. Because the differences in 5′ UTR length between the two species in most GO groups are <20 bp and most ECC changes are less than 0.3, we focused on those GO groups with larger changes in 5′ UTR length (>20 bp) and ECC (>0.3). The distribution of GO groups with large ECC changes in S. cerevisiae are negatively skewed (reduced ECC), suggesting that the expression of these genes in S. cerevisiae has become more variable over different conditions compared with C. albicans, similar to a switch from growth-related genes to stress-related genes. Interestingly, all these GO groups with a reduced ECC are associated with a 5′ UTR length increase (fig. 2). Among the 11 GO groups with larger increases in 5′ UTR length (>20 bp) and more decreases in ECC (<−0.3) in S. cerevisiae, nine are involved in S. cerevisiae mitochondrial respiration and energy production (table 1 and supplementary table S5, Supplementary Material online). The average 5′ UTR lengths of some GO groups have even increased >100% in S. cerevisiae, such as GO:0046933 (124.8%) and GO:0005753 (109%).
FIG. 2.
Association of reduced ECC with 5′ UTR length increase. The changes in 5′ UTR length from Candida albicans to Saccharomyces cerevisiae in 78 shared GO groups are plotted against ECC changes. The increase in 5′ UTR length in S. cerevisiae is strongly correlated with the decrease in ECC. The 11 GO groups with the most significant changes in 5′ UTR length (>20 bp) and in ECC (<−0.3) since the divergence between the S. cerevisiae and C. albicans lineages are shaded in gray.
Table 1.
The 11 GO Groups That Show a Large Increase in 5′ UTR Length and a Reduced ECC in Saccharomyces cerevisiae Since Its Divergence from Candida albicans.
| GO ID | GO Annotation | ECC Changea | 5′ UTR Length Change (bp) | % Length Changeb |
| GO:0005746c | Mitochondrial respiratory chain | −0.53 | 29.30 | 31.5 |
| GO:0070469c | Respiratory chain | −0.51 | 24.85 | 26.7 |
| GO:0022904c | Respiratory electron transport chain | −0.49 | 23.01 | 25.2 |
| GO:0042773c | ATP synthesis coupled electron transport | −0.49 | 23.01 | 25.2 |
| GO:0042775c | Mitochondrial ATP synthesis coupled electron transport | −0.49 | 23.01 | 25.2 |
| GO:0005753c | Mitochondrial proton-transporting ATP synthase complex | −0.40 | 50.91 | 109.0 |
| GO:0045259c | Proton-transporting ATP synthase complex | −0.40 | 50.91 | 109.0 |
| GO:0051187 | Cofactor catabolic process | −0.34 | 61.35 | 70.5 |
| GO:0006119c | Oxidative phosphorylation | −0.33 | 34.17 | 47.4 |
| GO:0046933c | Hydrogen ion transporting ATP synthase activity, rotational mechanism | −0.33 | 55.61 | 124.8 |
| GO:0009109 | Coenzyme catabolic process | −0.31 | 46.85 | 52.6 |
The GO groups are arranged in the decreasing order of the change in ECC.
The percent change in length is with respect to the 5′ UTR length of the gene in C. albicans.
GO groups involved in the mitochondrial respiration process.
To provide a test of the robustness of our results, we repeated our analysis using gene clusters binned by a different method. Ihmels et al. (2002) applied the signature algorithm to cluster S. cerevisiae genes into 86 transcriptional modules, using all available genome-wide expression data. The genes in the same module were shown to share a common TF–binding motif and were postulated to be involved in the same cellular pathway. Under the same cutoff (≥10 genes and CV < 1), we obtained 41 and 39 eligible modules in S. cerevisiae and in C. albicans, respectively. The correlations between the 5′ UTR length and ECC are −0.34 and −0.59 in the two species (supplementary fig. S4, Supplementary Material online). The correlation between the 5′ UTR length difference and ECC difference is −0.55 (P = 0.003). These results are consistent with that based on GO groups (figs. 1 and 2). In addition, there are three modules (module 5: 50 genes, module 55: 76 genes, and module 66: 24 genes) with the most significant changes in 5′ UTR length (>20 bp) and in ECC (<−0.3) since the divergence between the S. cerevisiae and C. albicans lineages (supplementary fig. S4, Supplementary Material online). Two of them (modules 5 and 55) are involved in the cellular respiration process, providing further support to our conclusion (supplementary table S6, Supplementary Material online).
Therefore, our results are consistent with previous studies showing that mitochondrial respiration-related genes have experienced expression reprogramming during the evolution of aerobic fermentation in the S. cerevisiae lineage (Ihmels et al. 2005; Field et al. 2009). It has been shown that changes in cis regulatory elements and promoter nucleosome organization were associated with the divergence of gene expression (Ihmels et al. 2005; Field et al. 2009). We showed here that increases in 5′ UTR length have also contributed to expression reprogramming of respiration-related genes and the evolution of aerobic fermentation in S. cerevisiae.
Lengthening of 5′ UTR and Reduced +1 Nucleosome Occupancy
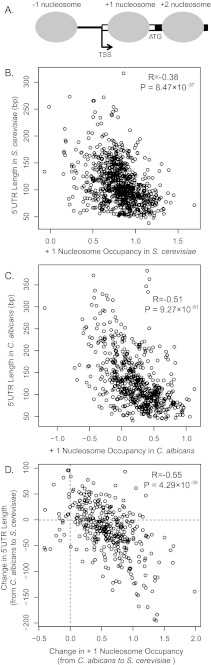
The 5′ UTR length of a gene is determined by where TSS is located (fig. 3A). It was found that TSSs are tightly distributed ∼10–15 bp upstream of the border of the +1 nucleosome (Albert et al. 2007). Because the preinitiation complex contains nucleosome-binding subunits, and because it is extremely unlikely that the positioning of the TSS and the +1 nucleosome have arisen independently and maintained a fixed distance at thousands of genes in yeast, it has been speculated that the selection of TSS is defined by the positioning of the +1 nucleosome (Jiang and Pugh 2009). Therefore, to obtain a better understanding about the underlying molecular basis for evolutionary change of 5′ UTR length, it is necessary to elucidate the connection between 5′ UTR length and +1 nucleosome positioning pattern. To estimate the +1 nucleosome positioning for a gene, we searched for the peak nucleosome occupancy in the 200 bp region downstream of the TSS using a sliding window of 100 bp because the peak window is likely to be where the +1 nucleosome positioned (see Materials and Methods). The +1 nucleosome occupancy of a GO group was defined as the average of peak nucleosome occupancies in a 100 bp window of all genes in the group.
FIG. 3.
Evolutionary change in 5′ UTR length was linked to variation in +1 nucleosome occupancy. (A) A typical nucleosome occupancy pattern in the promoter region of a gene in yeast. TSS is tightly located ∼10–15 bp upstream of the +1 nucleosome. (B–C) Scatter plot of average 5′ UTR lengths and average +1 nucleosome occupancies of GO groups in Saccharomyces cerevisiae (B) and in Candida albicans (C). The 5′ UTR length and the +1 nucleosome occupancy are negatively correlated in both species. (D) Increases in the 5′ UTR length and decreases in the +1 nucleosome occupancy are correlated during the evolution of S. cerevisiae. The unit of nucleosome occupancy in x axis is defined as the log ratio of the number of sequencing reads at a base pair and the median number of reads per base pair across entire genome.
In S. cerevisiae, we obtained 1,175 GO groups with coherent 5′ UTR lengths (CV < 1). By plotting the average 5′ UTR lengths of these GO groups against their +1 nucleosome occupancies, we observed a negative correlation between 5′ UTR length and +1 nucleosome occupancy (R = −0.38, P value 8.5×10−37). We also observed a similar pattern (R = −0.51, P value = 9.3×10−51) in C. albicans based on 758 GO groups with coherent 5′ UTR lengths (fig. 3). Therefore, a well-positioned +1 nucleosome is usually found in genes with a short 5′ UTR but tends to be absent in genes with a long 5′ UTRs, and this pattern is evolutionarily conserved. If the strongly positioned +1 nucleosome tends to be associated with a short 5′ UTR, we ask if reduced +1 nucleosome occupancy tends to be coupled with lengthening of 5′ UTR in evolution. By plotting evolutionary changes in average 5′ UTR length against the changes in +1 nucleosome occupancies for 422 GO groups shared by the two species, we found that the decrease in the +1 nucleosome occupancy is strongly correlated with lengthening of 5′ UTR (R = −0.55, P value = 4.3×10−35, fig. 3D). Therefore, change in 5′ UTR length during evolution is negatively correlated with change in the strength of +1 nucleosome positioning.
Discussion
In the present study, we showed that ECC and 5′ UTR length are strongly negatively correlated in both S. cerevisiae and C. albicans (fig. 1). In contrast, ECC is not correlated with 3′ UTR length or ORF length (supplementary fig. S3, Supplementary Material online). Moreover, 5′ UTR lengths tend to be better conserved than 3′ UTR lengths in evolution. These results suggest an important role of 5′ UTR length in gene expression regulation, especially at the transcriptional level. We also found an association between evolutionary change in 5′ UTR length and reprogramming of gene expression in the S. cerevisiae lineage. We showed here that genes with more extreme expression reprogramming in S. cerevisiae are mainly involved in mitochondrial respiration functions, which was consistent with previous studies and was believed as a major factor for the evolution of aerobic fermentation (Ihmels et al. 2005; Field et al. 2009). Those genes are also associated with large increase in 5′ UTR length in S. cerevisiae. Therefore, 5′ UTR length changes might have contributed to gene expression divergence and phenotypic evolution in yeasts. To our knowledge, this is the first genome-wide study that unveils the connection between 5′ UTR length and gene transcriptional regulation. Gene expression regulation has been regarded as a highly complex process, and our results suggest that 5′ UTR length plays a significant role in this process.
Lynch et al. have proposed a null model that 5′ UTR length is determined by several stochastic processes, and thus, its evolution is selectively neutral (Lynch et al. 2005). Reuter et al. reasoned that under this null model, a positive correlation between UTR length and GC content can be predicted, but they observed large quantitative discrepancies between empirical data and model prediction, so they argued that the evolution of 5′ UTR length might be under some selection pressure (Reuter et al. 2008). In addition, genome-wide UTR length surveys have revealed that finely regulated genes tend to have long 5′ UTR in different yeasts, which is not consistent with the model of neutral evolution of 5′ UTR length (Hurowitz and Brown 2003; Miura et al. 2006; Nagalakshmi et al. 2008; Bruno et al. 2010). By using GO groups and transcriptional modules as units to mitigate the possibility of inaccurate measurement of 5′ UTR length, we showed here that lengthening of 5′ UTR was associated with gene expression divergence and evolution of aerobic fermentation, further supporting the view that evolution of 5′ UTR length has been subjected to natural selection.
Deciphering the mechanism of 5′ UTR length in gene regulation is important for understanding the regulation of gene expression. Because our study suggests that a change in 5′ UTR length can affect the +1 nucleosome occupancy pattern (fig. 3), and because it has been shown that gene expression can be regulated by promoter nucleosome organization (Tirosh and Barkai 2008; Field et al. 2009), one may speculate that the impact of 5′ UTR length on gene expression is due to its effect on +1 nucleosome occupancy. However, we did not observe a significant correlation between the +1 nucleosome occupancy and ECC in either of the two species (supplementary fig. S5, Supplementary Material online). Therefore, the role of 5′ UTR length on the gene expression regulation may not be explained by different +1 nucleosome occupancies. Castillo-Davis et al. (2002) found that highly expressed genes have substantially shorter introns than those in lowly expressed genes and speculated that short introns could minimize the cost of transcription. However, no significant correlation was found between 5′ UTR length and expression rate which was measured by the codon adaptation index (Reuter et al. 2008) or between 5′ UTR length and transcript level (David et al. 2006). Another possible factor is that a long 5′ UTR may contain more regulatory elements than short ones. For example, some eukaryotic 5′ UTRs may contain upstream ORFs (uORFs), which generally inhibit the translation of the main ORF by interfering with reassembly of the translation initiation complex at the main start codon (Morris and Geballe 2000; Vilela and McCarthy 2003). The average 5′ UTR length of 256 uORF-containing genes (737 bp) in S. cerevisiae is much longer than 115 bp, the genome average 5′ UTR length (Cvijovic et al. 2007). However, the uORF affects gene expression at the translation level, so that it still cannot directly explain the transcription regulatory effects of 5′ UTR length.
In summary, our study unraveled a strong correlation between 5′ UTR length and gene expression pattern in both S. cerevisiae and C. albicans and an association between evolutionary change in 5′ UTR length and gene expression reprogramming in the S. cerevisiae lineage. However, it is still unclear as to how the regulation of gene transcription is affected by 5′ UTR length. This should be an interesting topic for future study.
Supplementary Material
Supplementary tables S1–S6 and figures S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Dr. Yong Woo for his comments on the manuscript and helpful advice on statistical inferences. We also gratefully thank two anonymous reviewers for their valuable comments. This study was funded by NIH grant GM30998.
References
- Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, et al. (20 co-authors) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman J, Sudbery PE. Candida Albicans: a molecular revolution built on lessons from budding yeast. Nat Rev Genet. 2002;3:918–930. doi: 10.1038/nrg948. [DOI] [PubMed] [Google Scholar]
- Bruno VM, Wang Z, Marjani SL, Euskirchen GM, Martin J, Sherlock G, Snyder M. Comprehensive annotation of the transcriptome of the human fungal pathogen Candida albicans using RNA-seq. Genome Res. 2010;20:1451–1458. doi: 10.1101/gr.109553.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castillo-Davis CI, Mekhedov SL, Hartl DL, Koonin EV, Kondrashov FA. Selection for short introns in highly expressed genes. Nat Genet. 2002;31:415–418. doi: 10.1038/ng940. [DOI] [PubMed] [Google Scholar]
- Cvijovic M, Dalevi D, Bilsland E, Kemp GJ, Sunnerhagen P. Identification of putative regulatory upstream ORFs in the yeast genome using heuristics and evolutionary conservation. BMC Bioinformatics. 2007;8:295. doi: 10.1186/1471-2105-8-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David L, Huber W, Granovskaia M, Toedling J, Palm CJ, Bofkin L, Jones T, Davis RW, Steinmetz LM. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci U S A. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997;278:680–686. doi: 10.1126/science.278.5338.680. [DOI] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Fondufe-Mittendorf Y, Moore IK, Mieczkowski P, Kaplan N, Lubling Y, Lieb JD, Widom J, Segal E. Gene expression divergence in yeast is coupled to evolution of DNA-encoded nucleosome organization. Nat Genet. 2009;41:438–445. doi: 10.1038/ng.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hake LE, Alcivar AA, Hecht NB. Changes in mRNA length accompany translational regulation of the somatic and testis-specific cytochrome c genes during spermatogenesis in the mouse. Development. 1990;110:249–257. doi: 10.1242/dev.110.1.249. [DOI] [PubMed] [Google Scholar]
- Hughes MJ, Andrews DW. A single nucleotide is a sufficient 5' untranslated region for translation in an eukaryotic in vitro system. FEBS Lett. 1997;414:19–22. doi: 10.1016/s0014-5793(97)00965-4. [DOI] [PubMed] [Google Scholar]
- Hurowitz EH, Brown PO. Genome-wide analysis of mRNA lengths in Saccharomyces cerevisiae. Genome Biol. 2003;5:R2. doi: 10.1186/gb-2003-5-1-r2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ihmels J, Bergmann S, Gerami-Nejad M, Yanai I, McClellan M, Berman J, Barkai N. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science. 2005;309:938–940. doi: 10.1126/science.1113833. [DOI] [PubMed] [Google Scholar]
- Ihmels J, Friedlander G, Bergmann S, Sarig O, Ziv Y, Barkai N. Revealing modular organization in the yeast transcriptional network. Nat Genet. 2002;31:370–377. doi: 10.1038/ng941. [DOI] [PubMed] [Google Scholar]
- Jansen RP. mRNA localization: message on the move. Nat Rev Mol Cell Biol. 2001;2:247–256. doi: 10.1038/35067016. [DOI] [PubMed] [Google Scholar]
- Jiang C, Pugh BF. Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet. 2009;10:161–172. doi: 10.1038/nrg2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Li WH. The evolution of aerobic fermentation in Schizosaccharomyces pombe was associated with regulatory reprogramming but not nucleosome reorganization. Mol Biol Evol. 2011a;28:1407–1413. doi: 10.1093/molbev/msq324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Li WH. Expansion of hexose transporter genes was associated with the evolution of aerobic fermentation in yeasts. Mol Biol Evol. 2011b;28:131–142. doi: 10.1093/molbev/msq184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- Lynch M, Scofield DG, Hong X. The evolution of transcription-initiation sites. Mol Biol Evol. 2005;22:1137–1146. doi: 10.1093/molbev/msi100. [DOI] [PubMed] [Google Scholar]
- Mager WH, Planta RJ. Coordinate expression of ribosomal protein genes in yeast as a function of cellular growth rate. Mol Cell Biochem. 1991;104:181–187. doi: 10.1007/BF00229818. [DOI] [PubMed] [Google Scholar]
- Merico A, Sulo P, Piskur J, Compagno C. Fermentative lifestyle in yeasts belonging to the Saccharomyces complex. FEBS J. 2007;274:976–989. doi: 10.1111/j.1742-4658.2007.05645.x. [DOI] [PubMed] [Google Scholar]
- Mignone F, Gissi C, Liuni S, Pesole G. Untranslated regions of mRNAs. Genome Biol. 2002;3:REVIEWS0004. doi: 10.1186/gb-2002-3-3-reviews0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura F, Kawaguchi N, Sese J, Toyoda A, Hattori M, Morishita S, Ito T. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc Natl Acad Sci U S A. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DR, Geballe AP. Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol. 2000;20:8635–8642. doi: 10.1128/mcb.20.23.8635-8642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science. 2008;320:1344–1349. doi: 10.1126/science.1158441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesole G, Mignone F, Gissi C, Grillo G, Licciulli F, Liuni S. Structural and functional features of eukaryotic mRNA untranslated regions. Gene. 2001;276:73–81. doi: 10.1016/s0378-1119(01)00674-6. [DOI] [PubMed] [Google Scholar]
- Piskur J, Rozpedowska E, Polakova S, Merico A, Compagno C. How did Saccharomyces evolve to become a good brewer? Trends Genet. 2006;22:183–186. doi: 10.1016/j.tig.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Pronk JT, Yde Steensma H, Van Dijken JP. Pyruvate metabolism in Saccharomyces cerevisiae. Yeast. 1996;12:1607–1633. doi: 10.1002/(sici)1097-0061(199612)12:16<1607::aid-yea70>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- Reuter M, Engelstadter J, Fontanillas P, Hurst LD. A test of the null model for 5' UTR evolution based on GC content. Mol Biol Evol. 2008;25:801–804. doi: 10.1093/molbev/msn044. [DOI] [PubMed] [Google Scholar]
- Sudbery P, Gow N, Berman J. The distinct morphogenic states of Candida albicans. Trends Microbiol. 2004;12:317–324. doi: 10.1016/j.tim.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Thomson JM, Gaucher EA, Burgan MF, De Kee DW, Li T, Aris JP, Benner SA. Resurrecting ancestral alcohol dehydrogenases from yeast. Nat Genet. 2005;37:630–635. doi: 10.1038/ng1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh I, Barkai N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 2008;18:1084–1091. doi: 10.1101/gr.076059.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsankov AM, Thompson DA, Socha A, Regev A, Rando OJ. The role of nucleosome positioning in the evolution of gene regulation. PLoS Biol. 2010;8:e1000414. doi: 10.1371/journal.pbio.1000414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Velden AW, Thomas AA. The role of the 5' untranslated region of an mRNA in translation regulation during development. Int J Biochem Cell Biol. 1999;31:87–106. doi: 10.1016/s1357-2725(98)00134-4. [DOI] [PubMed] [Google Scholar]
- Vilela C, McCarthy JE. Regulation of fungal gene expression via short open reading frames in the mRNA 5'untranslated region. Mol Microbiol. 2003;49:859–867. doi: 10.1046/j.1365-2958.2003.03622.x. [DOI] [PubMed] [Google Scholar]
- Wang D, Hsieh M, Li WH. A general tendency for conservation of protein length across eukaryotic kingdoms. Mol Biol Evol. 2005;22:142–147. doi: 10.1093/molbev/msh263. [DOI] [PubMed] [Google Scholar]
- Wapinski I, Pfeffer A, Friedman N, Regev A. Natural history and evolutionary principles of gene duplication in fungi. Nature. 2007;449:54–61. doi: 10.1038/nature06107. [DOI] [PubMed] [Google Scholar]
- Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yiu GK, Gu W, Hecht NB. Heterogeneity in the 5' untranslated region of mouse cytochrome cT mRNAs leads to altered translational status of the mRNAs. Nucleic Acids Res. 1994;22:4599–4606. doi: 10.1093/nar/22.22.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.