Abstract
Inflammatory bowel disease 5 (IBD5) is a 250 kb haplotype on chromosome 5 that is associated with an increased risk of Crohn’s disease in Europeans. The OCTN1 gene is centrally located on IBD5 and encodes a transporter of the antioxidant ergothioneine (ET). The 503F variant of OCTN1 is strongly associated with IBD5 and is a gain-of-function mutation that increases absorption of ET. Although 503F has been implicated as the variant potentially responsible for Crohn’s disease susceptibility at IBD5, there is little evidence beyond statistical association to support its role in disease causation. We hypothesize that 503F is a recent adaptation in Europeans that swept to relatively high frequency and that disease association at IBD5 results not from 503F itself, but from one or more nearby hitchhiking variants, in the genes IRF1 or IL5. To test for evidence of recent positive selection on the 503F allele, we employed the iHS statistic, which was significant in the European CEU HapMap population (P = 0.0007) and European Human Genome Diversity Panel populations (P ≤ 0.01). To evaluate the hypothesis of disease-variant hitchhiking, we performed haplotype association tests on high-density microarray data in a sample of 1,868 Crohn’s disease cases and 5,550 controls. We found that 503F haplotypes with recombination breakpoints between OCTN1 and IRF1 or IL5 were not associated with disease (odds ratio [OR]: 1.05, P = 0.21). In contrast, we observed strong disease association for 503F haplotypes with no recombination between these three genes (OR: 1.24, P = 2.6 × 10−8), as expected if the sweeping haplotype harbored one or more disease-causing mutations in IRF1 or IL5. To further evaluate these disease-gene candidates, we obtained expression data from lower gastrointestinal biopsies of healthy individuals and Crohn’s disease patients. We observed a 72% increase in gene expression of IRF1 among Crohn’s disease patients (P = 0.0006) and no significant difference in expression of OCTN1. Collectively, these data indicate that the 503F variant has increased in frequency due to recent positive selection and that disease-causing variants in linkage disequilibrium with 503F have hitchhiked to relatively high frequency, thus forming the IBD5 risk haplotype. Finally, our association results and expression data support IRF1 as a strong candidate for Crohn’s disease causation.
Keywords: positive selection, genetic hitchhiking, Crohn's disease, IBD5, IRF1
Introduction
As an advantageous allele spreads through a population during a selective sweep, alleles in linkage disequilibrium (LD) with the advantageous allele can rapidly increase in frequency as a result of genetic hitchhiking. Under some conditions, genetic hitchhiking can also drive deleterious disease-causing alleles to high frequency (Wagener and Cavalli-Sforza 1975; Rice 1987). However, the study of this phenomenon has been limited to complete selective sweeps, primarily in nonrecombining genomes (Wagener and Cavalli-Sforza 1975; Rice 1987; Charlesworth B and Charlesworth D 2000; Seger et al. 2010). As a consequence, earlier studies of genetic hitchhiking are not directly relevant to the appreciable number of incomplete selective sweeps that have recently been identified in various human populations (Voight et al. 2006; Hawks et al. 2007; Pickrell et al. 2009; Simonson et al. 2010), and thus, the role of genetic hitchhiking in human disease remains unclear. Here, we analyze the disease implications of incomplete selective sweeps in large recombining genomes and identify the conditions that can lead to an increase in the frequency of disease-causing alleles. We then apply these results to the analysis of the inflammatory bowel disease 5 (IBD5) haplotype on chromosome 5.
Among Europeans, the IBD5 haplotype is associated with an increased risk of developing Crohn’s disease (Ma et al. 1999; Rioux et al. 2000, 2001; Burton et al. 2007), a chronic inflammatory disorder of the gastrointestinal tract. IBD5 has a frequency of approximately 40% in healthy Europeans but has a very low frequency (<5%) in African and East Asian populations (Fisher et al. 2006; Tosa et al. 2006; Silverberg et al. 2007). Due to extensive LD extending across 250 kb, multiple single nucleotide polymorphisms (SNPs) in this haplotype have equivalent statistical association with Crohn’s disease, including SNPs in the genes P4HA2, PDLIM4, OCTN1, OCTN2, and IRF1 (Onnie et al. 2006; Silverberg et al. 2007; Franke et al. 2010). We hypothesize that the disease association and extensive LD at IBD5 are the results of a recent selective sweep (fig. 1). Here, we evaluate the case for positive selection at IBD5 with statistical tests for selection in European populations. We then identify patterns of Crohn’s disease association at IBD5 that are consistent with genetic hitchhiking of disease-causing alleles on a sweeping haplotype.
FIG. 1.
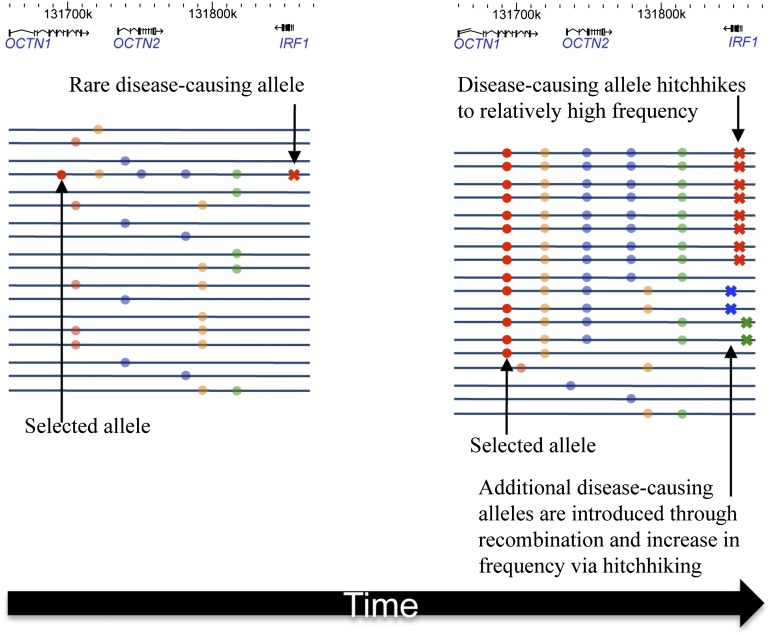
The disease-hitchhiking hypothesis. A new advantageous mutation arises on a chromosome with a rare disease-causing mutation. As the advantageous mutation (red dot) spreads rapidly through the population, the disease-causing mutation (red X) hitchhikes to relatively high frequency, and additional disease-causing mutations (green and blue Xs) are introduced into the sweeping haplotype block by recombination. Neutral and advantageous mutations are shown as dots, and disease-causing mutations are shown as crosses.
Materials and Methods
Simulations
The simulations in figure 2 model the interaction between a strongly favored allele and mildly deleterious alleles at a neighboring locus, as a special case of asymmetric Hill–Robertson interference (Hill and Robertson 1966). All simulations begin with a 5N generation burn-in time, where N is the effective population size. The simulations are conditioned on reaching the desired frequency of the favored allele by rejection sampling. The deleterious mutation rate is 1.1 × 10−8 per site per generation (Roach et al. 2010). The segregating sites within the disease susceptibility locus are nonrecombining. The fitness effects of multiple alleles are assumed to combine multiplicatively, although given the asymmetric relationship between the advantageous allele and deleterious alleles (s = 0.02 vs. s < = −0.005), the specific fitness model is of little consequence.
FIG. 2.
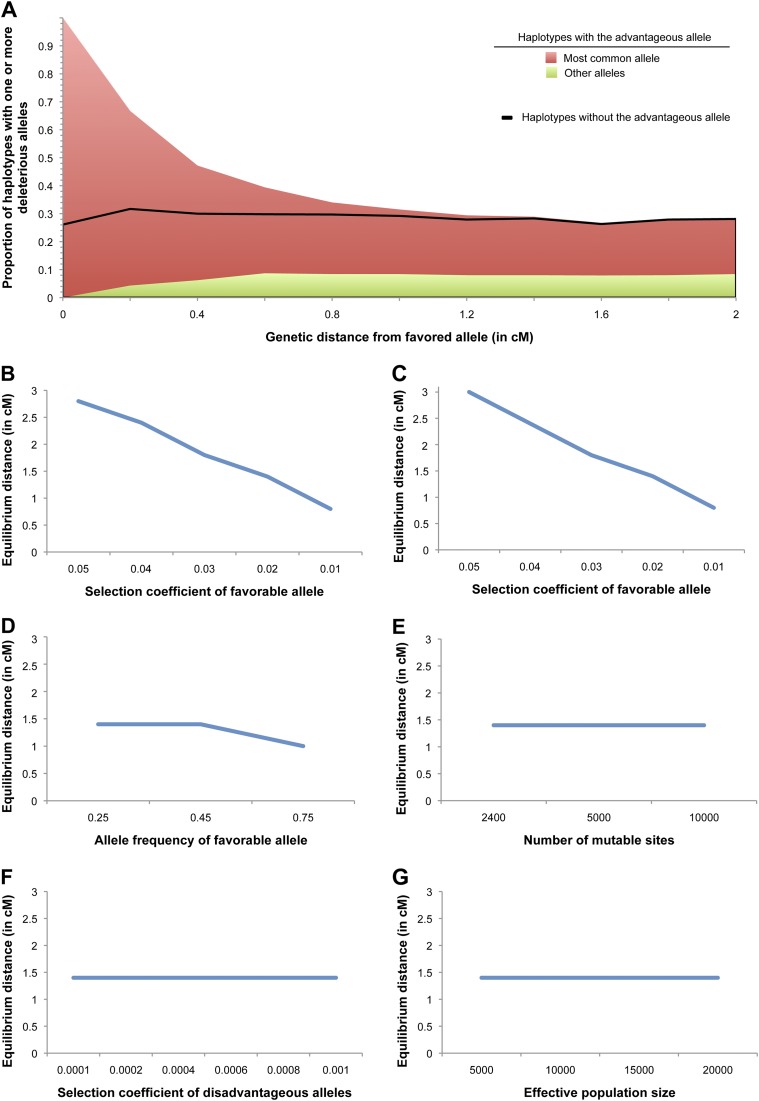
Expected increase in frequency of deleterious alleles on the sweeping haplotype after an incomplete selective sweep. Parameter values for the forward-in-time simulations in (A) loosely model the pattern at OCTN1, with a selective advantage of the favorable allele of 0.02, an initial effective population size 10,000, a final allele frequency of the favorable allele of 0.45, and 2,400 potential mutational targets (modeled after the number of nonsynonymous sites in IRF1). (A): The stacked red and green graph represents the expected proportion of haplotypes that contain one or more deleterious alleles among haplotypes with the favorable allele, and the black line indicates the proportion of haplotypes that contain one or more deleterious alleles among haplotypes without the favorable allele. “Most common allele” designates the proportion of haplotypes with the most common deleterious variant, and “All other alleles” indicates the proportion of haplotypes that contain one or more deleterious variants but do not contain the most common variant. Unless otherwise specified, parameter values in panels (B–G) are the same as in (A). (B–G): The relationship between equilibrium distance and (B) the selection coefficient of the favorable allele in a population of constant size; (C) the selection pressure of the favorable allele in a population that begins expanding at a rate of 0.5% per generation after the introduction of the advantageous allele; (D) the final allele frequency of the favorable allele; (E) the number of mutable sites constrained by purifying selection (equivalent to the mutation rate at the locus); (F) the selection pressure against deleterious alleles; and (G) the effective population size.
iHS Test
To test for recent positive selection on 503F, we calculated iHS scores from phased haplotype data across the genome in a Utah population (HapMap CEU) and in European populations from the HGDP (Consortium 2005; Li et al. 2008). For HapMap data, we used phased data from HapMap phase 2 (http://hapmap.ncbi.nlm.nih.gov/downloads/phasing/2006-07_phaseII/phased/; Frazer et al. 2007); for HGDP data, we phased the Illumina 550K SNP microarray data from a Crohn’s disease cohort reported previously (Imielinski et al. 2009) using fastPHASE (Scheet and Stephens 2006). We evaluated the statistical significance of the iHS test from the empirical distribution of iHS scores across the genome for each population and for all sites with a derived allele frequency between 20% and 80%. The iHS statistic is defined as the log of the ratio of integrated Extended Haplotype Homozygosity (iHH) scores at each site for the derived and ancestral alleles, standardized by the derived allele frequency (Voight et al. 2006). To calculate iHH for each allele at each site, we integrated the expected Extended Haplotype Homozygosity (EHH) in both directions from the core SNP until either expected EHH reached 0.05 or all haplotypes were unique (Voight et al. 2006; Huff et al. 2010). Because iHS has limited statistical power for sample sizes smaller than 20 (Pickrell et al. 2009), we restricted our analysis to population samples with at least 20 individuals.
Allele Frequency of the 503F Variant
The population frequencies of 503F were determined by genotyping SNP L503F (rs1050152) in 85 populations across the Old World, including 954 individuals from 48 HGDP populations (Li et al. 2008) and 772 individuals from 37 additional populations described in (Jorde et al. 1995; Bamshad et al. 1998; Watkins et al. 1999; Bulayeva et al. 2003; Xing et al. 2009, 2010). The SNP was genotyped by fluorescent primer extension using SNaPshot chemistry (Applied Biosystems) and analyzed on ABI 3100 genetic analyzer (Applied Biosystems). The populations genotyped and their population frequencies of the 503F allele are shown in supplementary table S2, Supplementary Material online.
Haplotype Bifurcation Diagram
The genotypes of the IBD5 region in 1,868 Crohn’s disease cases and 5,540 controls (Imielinski et al. 2009) were determined by genotyping individuals using the Illumina 550K SNP microarray at the Center for Applied Genomics at the Children’s Hospital of Philadelphia. All patients met the standard diagnostic criteria for Crohn’s disease (Silverberg et al. 2007). We used genotypes from 639 SNPs spanning 1 MB upstream and downstream of OCTN1 and selected only subjects of European ancestry as determined both by self-reported ancestry and by STRUCTURE (Falush et al. 2003) runs using ancestry informative markers as in Imielinski et al. (2009). We phased the genotype data using BEAGLE (Browning SR and Browning BL 2007). After phasing, we constructed haplotype bifurcation diagrams using the program SWEEP (Sabeti et al. 2002) from the phased genotype data of the cases and a subset of controls (the 1,262 samples collected in Utah and Atlanta), with the core haplotypes defined by the 503F and 503L alleles of OCTN1.
mRNA Expression
We examined colonic expression of selected candidate genes located in the IBD5 LD block. Gene expression was assayed in individual colonic biopsy specimens from subjects with early-onset Crohn’s disease (n = 30) and from healthy controls (n = 11). Inflammation was quantified in colon biopsies by using the Crohn’s Disease Histological Index of Severity. After informed consent, colonic biopsies were obtained from subjects with Crohn’s disease and healthy controls. All of the biopsies for Crohn cases and healthy controls were obtained from the ascending colon. Colon biopsies were immediately placed in RNAlater stabilization reagent (Qiagen, Germany) at 4 °C. Total RNA was isolated by an RNeasy Plus Mini Kit (Qiagen) and stored at −80 °C. Samples were then submitted to the Cincinnati Children's Hospital Medical Center Digestive Health Center Microarray Core where the quality and concentration of RNA were measured by the Agilent Bioanalyser 2100 (Hewlett Packard) using an RNA 6000 Nano Assay to confirm a 28S/18S ratio of 1.6:2.0. We amplified 100 ng of total RNA by using a Target 1-round Aminoallyl-aRNA Amplification Kit 101 (Epicentre, WI). The biotinylated complementary RNA was hybridized to Affymetrix GeneChip Human Genome HG-U133 Plus 2.0 arrays, containing probes for 22,634 genes. The images were captured by an Affymetrix Genechip Scanner 3000. The complete data set is available at the NCBI Gene Expression Omnibus (GEO): colonic gene expression data set, GSE10616. Data were normalized to an internal control within each batch and to the healthy control samples to allow for array-to-array comparisons.
Results and Discussion
Analysis of Deleterious Hitchhiking
To analyze the properties of deleterious hitchhiking alleles in large recombining genomes during an incomplete selective sweep, we employed forward-in-time simulations of a Wright–Fisher model (see Materials and Methods). The simulations model a newly arising advantageous mutation and a linked locus at which variation is constrained by purifying selection (see fig. 2). We are interested in the conditions that produce LD between the advantageous allele and one or more deleterious alleles in the linked locus. These conditions produce statistical association between disease risk and alleles in a sweeping haplotype block.
We restrict the simulations such that the new advantageous mutation originates on a chromosome with one or more deleterious alleles. In the absence of this restriction, the advantageous allele is expected to be at or near linkage equilibrium with deleterious alleles at the end of the modeled sweep in all of the scenarios we evaluated. Therefore, the most important factor controlling the behavior of deleterious hitchhiking is the probability that the advantageous mutation originates on a chromosome with a deleterious allele. This probability is equal to the population frequency of chromosomes with one or more deleterious alleles. At mutation–selection equilibrium, this probability is equal to μ/s, where μ is the deleterious mutation rate of the locus under purifying selection and s is the strength of selection against new mutants at this locus. Therefore, the scenario most likely to give rise to deleterious hitchhiking is one in which many weakly constrained mutational targets are in close proximity to the favorable allele. This scenario may be particularly relevant to complex genetic diseases for two reasons. First, because complex genetic diseases involve more genes than Mendelian diseases, there may be more mutational targets that can potentially contribute to disease susceptibility (higher μ). Second, the genes involved in complex diseases may be substantially less constrained by purifying selection than those involved in Mendelian diseases (lower s; Blekhman et al. 2008).
Our simulation results show that the two other major factors controlling the behavior of deleterious hitchhiking are the selective advantage of the advantageous allele and the genetic distance between the advantageous and deleterious alleles (fig. 2). As genetic distance increases, LD between advantageous and deleterious alleles decreases due to recombination between them (fig. 2A). The expected distance at which approximate linkage equilibrium will be reached (the equilibrium distance) depends almost exclusively on the selective advantage of the favorable allele (fig. 2). This result holds for the following reasons. First, recombination is the primary force for reestablishing equilibrium. Second, controlling for genetic distance, the expected amount of recombination since the origin of the favorable allele depends on the span of time since the start of the selective sweep. Finally, this time span depends primarily on the selective advantage of the favorable allele. Thus, selective advantage controls equilibrium distance.
The time since the start of the selective sweep and therefore equilibrium distance are relatively insensitive to both the initial population size and changes in population size (Hawks et al. 2007; fig. 2C and G). Recombination can introduce additional deleterious alleles into the sweeping haplotype block, as shown in the green area of figure 2A, but the rate at which additional deleterious alleles are introduced is lower than the rate at which the initial deleterious allele is lost, for example, at a distance of 0.4 cM, the deleterious allele frequency has dropped from 100% to 47%, whereas the frequency of other deleterious alleles has increased to only 6% (fig. 2A). Although mutation can introduce new deleterious alleles in the selective sweep, the mutation rate has no appreciable effect on equilibrium distance (fig. 2E).
When deleterious hitchhiking occurs in a disease susceptibility gene, common SNPs in LD with the advantageous allele should consistently show signals of disease association resulting from LD with multiple disease-causing variants. However, these disease-causing variants may be individually too rare to produce association signals. This is a type of synthetic association: Observed disease association at a common SNP resulting from variants separated from the common SNP by large genetic distances (Dickson et al. 2010). In the absence of positive selection, if rare mutations make a large contribution to disease risk, synthetic association at common SNPs can occasionally occur at distances greater than 2.5 cM from the disease-causing variants (Dickson et al. 2010). In contrast, for a genomic region influenced by a strong selective sweep, synthetic association is more predictable, with an equilibrium distance of less than 3 cM from the advantageous allele for most selective sweeps (fig. 2A).
Positive Selection at IBD5
The 503F variant (rs1050152) of OCTN1, a gene located near the center of the IBD5 haplotype, has been associated with Crohn’s disease in several studies (Rioux et al. 2001; Mirza et al. 2003; Peltekova et al. 2004; Fisher et al. 2006; Silverberg et al. 2007; Imielinski et al. 2009; Franke et al. 2010). However, no convincing causal link between this variant and Crohn’s disease has been established. The key substrate of the transporter encoded by OCTN1 is ergothioneine (ET), an antioxidant synthesized by fungi and present in most plants and animals (Grundemann et al. 2005; Ey et al. 2007). 503F is a gain-of-function mutation that increases ET transport efficiency by 50% and ET substrate affinity by 3-fold (Taubert et al. 2005). This variant is common in European and Middle Eastern populations but is rare throughout the rest of the world (fig. 3B). Thus, 503F is characterized by several unusual properties: It is absent in Africa and East Asia but common in Europe; it is associated with a specific haplotype background characterized by extensive LD in Europeans; it confers a gain-of-function on the protein; and it is associated with disease, although there is no direct evidence for causation. Collectively, these observations are consistent with the hypothesis that 503F was influenced by positive selection and that the association of this variant with disease is the result of genetic hitchhiking. This hypothesis can account for the extensive LD in the IBD5 haplotype, the geographic distribution of the 503F allele, and disease association with 503F in the absence of direct causation (Wagener and Cavalli-Sforza 1975; Rice 1987).
FIG. 3.
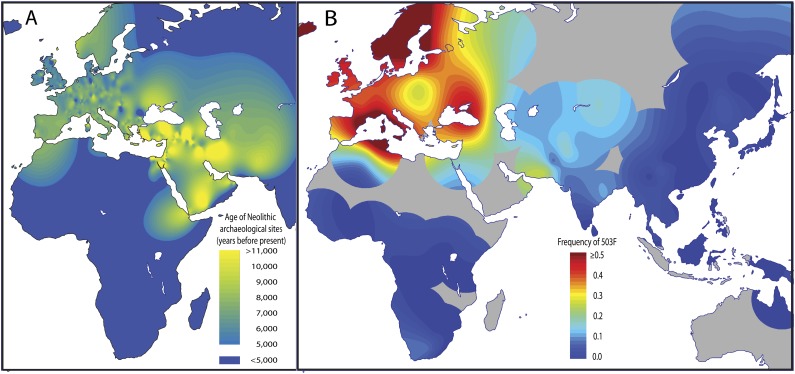
Geographic distribution of (A) the age of early Neolithic archaeological sites and (B) the 503F allele of OCTN1. The contour map in (A) is similar to figure 1A in Balaresque et al. (2010) and is based on the dates of 774 archaeological sites provided in Hassan (1985) and Pinhasi et al. (2005). The contour map in (B) is based on the population frequency of 503F in 85 populations across the Old World (supplementary table S2, Supplementary Material online). The intensity surfaces for the contour maps were generated using bicubic spline interpolation (Matlab r2009a griddata, 'v4' option), followed by truncation of values to fit the ranges indicated in the legends (r2009a). In (B), allele frequencies were averaged for populations located within 1°, and values were not interpolated for regions further than 10° from the nearest frequency data point (gray areas). The allele frequency of 503F in a population and the distance of that population to the nearest early Neolithic site are highly correlated (r2 = 0.44, P = 0.0067, t-test of Spearman’s ρ).
We propose that 503F is an adaptation to low dietary levels of ET among early Neolithic farmers in the Fertile Crescent. Although ET content is relatively high in meat and a variety of plant foods, it is conspicuously low in many of the plants first domesticated in the Fertile Crescent, including wheat, barley, lentils, and peas (Ey et al. 2007; table 1). Because the function of ET is not well understood, it is difficult to identify the specific fitness consequences that Neolithic farmers would have faced as a result of low levels of ET in their diet. Despite the lack of direct evidence, there are several lines of evidence supporting the importance of ET. ET functions both as an antioxidant and a neuroprotective agent (Moncaster et al. 2002) and OCTN1 functions exclusively as an ET transporter (Grundemann et al. 2005). OCTN1 is highly conserved among vertebrates, with >90% protein identity between human and other primates and 75% protein identity between human and chicken (Altschul et al. 1997). In addition, despite being exclusively synthesized by fungi and mycobacteria, ET is found in a wide variety of plants and animals (table 1; Ey et al. 2007). Further, depletion of ET has been shown to increase susceptibility to oxidative stress in mammalian cells, resulting in increased mitochondrial DNA damage, protein oxidation, and lipid peroxidation (Paul and Snyder 2010). Finally, ET has been shown to play a specific role in response to UV-induced oxidative stress (Markova et al. 2009), which may have been particularly relevant to Fertile Crescent farmers if they were as light skinned as many of their descendants are. If the transition from hunting and gathering to early agriculture in the Fertile Crescent resulted in a dietary ET deficiency, a genetic variant increasing the absorption of ET might have been favored by positive selection, allowing it to spread rapidly through the population.
Table 1.
Ergothioneine Content of Various Foods (Data from Ey et al. (2007)).
| Food | Ergothioneine (mg/kg wet weight) |
| Oyster mushrooms | 118.91 |
| Garlic | 3.11 |
| Pork | 1.68 |
| Beef | 1.33 |
| Chicken | 1.15 |
| Portabella mushrooms | 0.93 |
| Wheat bran | 0.84 |
| Broccoli | 0.24 |
| Onion | 0.23 |
| Spinach | 0.11 |
| Milk | <0.01 |
| Lentils | <0.01 |
| Green peas | <0.01 |
| Wheat flour (refined) | <0.01 |
| Barley flour (refined) | <0.01 |
Previous studies have suggested that a recent selective sweep may have occurred in one or more Eurasian populations near the IBD5 haplotype in the region containing IL13, which is approximately 350 kb downstream of OCTN1 (Sakagami et al. 2004; Zhou et al. 2004; Tarazona-Santos and Tishkoff 2005). Because signals of positive selection can extend for long genomic distances (Grossman et al. 2010), the patterns observed at IL13 could potentially be explained by positive selection on the 503F variant of OCTN1. To test for evidence of positive selection on the 503F variant, we employed the iHS statistic, which is the most powerful method for detecting recent positive selection when the favorable allele is still polymorphic in the population (Voight et al. 2006; Huff et al. 2010). This test measures the decay of LD around a polymorphic site and is designed to detect extended haplotype blocks that are produced by a recent selective sweep. When the test is applied to the advantageous allele, the statistical power is greater than 80% at the 0.01 significance level in a sample size of 50 individuals (Voight et al. 2006; Huff et al. 2010). The empirical one-tailed test for 503F in the HapMap CEU sample resulted in a P value of 0.007 (table 2). Of the four Human Genome Diversity Panel (HGDP) European populations we tested, three (Russia, Sardinia, and France) were significant at the 0.01 level, with P = 0.012 in the Basque sample. This result provides strong evidence that the 503F variant has been influenced by recent positive selection in European populations (table 2). By itself, the iHS test is rarely able to conclusively identify the variant that has been targeted by selection due to significant iHS signals from nearby hitchhiking alleles (see supplementary table S5, Supplementary Material online). Therefore, although these results support the hypothesis of positive selection acting on 503F, we cannot rule out the possibility that the target of selection was a variant other than 503F on the IBD5 haplotype.
Table 2.
iHS Test Results at 503F for European Populations.
| Sample | iHS | Empirical P value |
| HapMap CEU | −3.10 | 0.0007 |
| HGDP Russian | −2.75 | 0.0044 |
| HGDP Sardinian | −2.76 | 0.0075 |
| HGDP French | −2.64 | 0.0076 |
| HGDP Basque | −2.37 | 0.0128 |
To estimate the age of 503F, we measured the decay of LD in the HapMap CEU sample with the method described in Reich and Goldstein (1999) using the implementation details from Sabeti et al. (2005). Our estimate incorporated all SNPs in HapMap Phase 2 that are within 0.04 cM of 503F (Consortium 2005) (see supplementary table S4, Supplementary Material online). The estimated age of origin of 503F was 12,550 years ago (95% confidence interval = 7,750–19,025, 25-year generation time), which is consistent with the earliest archaeological evidence for the domestication of wheat (10,600 years ago) and barley (9,500 years ago) (Hillman 1975; Zeist and Bakker-Heeres 1982; Badr et al. 2000; Ozkan et al. 2002). The origin age of 503F is also consistent with the domestication of the earliest pulses, which are particularly low in ET content (see peas and lentils in table 1) and appear in the archaeological record soon after wheat and barley (Ladizinsky 1979).
To better assess the geographic distribution of 503F, we genotyped this variant in 954 individuals from 48 HGDP populations and 772 individuals from 37 additional populations. Figure 3 compares the frequency of 503F across the Old World with the ages of early Neolithic archaeological sites (see supplementary table S3, Supplementary Material online), suggesting a close relationship between the geographic distribution of 503F and the origin of agriculture in the Fertile Crescent. With an allele frequency of 0.4 and an age of origin of 12,550 (7,750–19,025) years ago, we estimate that the selective advantage was approximately 1.9% (1.3–3.2%) for early Neolithic farmers with one copy of 503F (estimated assuming additive deterministic selection with the best-fitting model of European population history from Schaffner et al. (2005)).
Evidence for Deleterious Hitchhiking at IBD5
Modeling disease variants as genetic hitchhikers provides a framework for constructing haplotype association tests to assist in disease-gene mapping efforts. The haplotype bifurcation diagram in figure 4A depicts recombination events that have occurred on haplotypes with the 503F allele. Each bifurcation point represents an SNP that defines one or more recombinant haplotypes, which were created by recombination events upstream of that SNP. The disease-hitchhiking model predicts that haplotypes created by recombination events between the favorable allele and the disease locus will not be associated with disease and that the signal of disease association should be concentrated in haplotypes created by more distant recombination events (fig. 1). Our simulation results predict that hitchhiking disease variants should be located within 1.4 cM of 503F (fig. 2A).
FIG. 4.
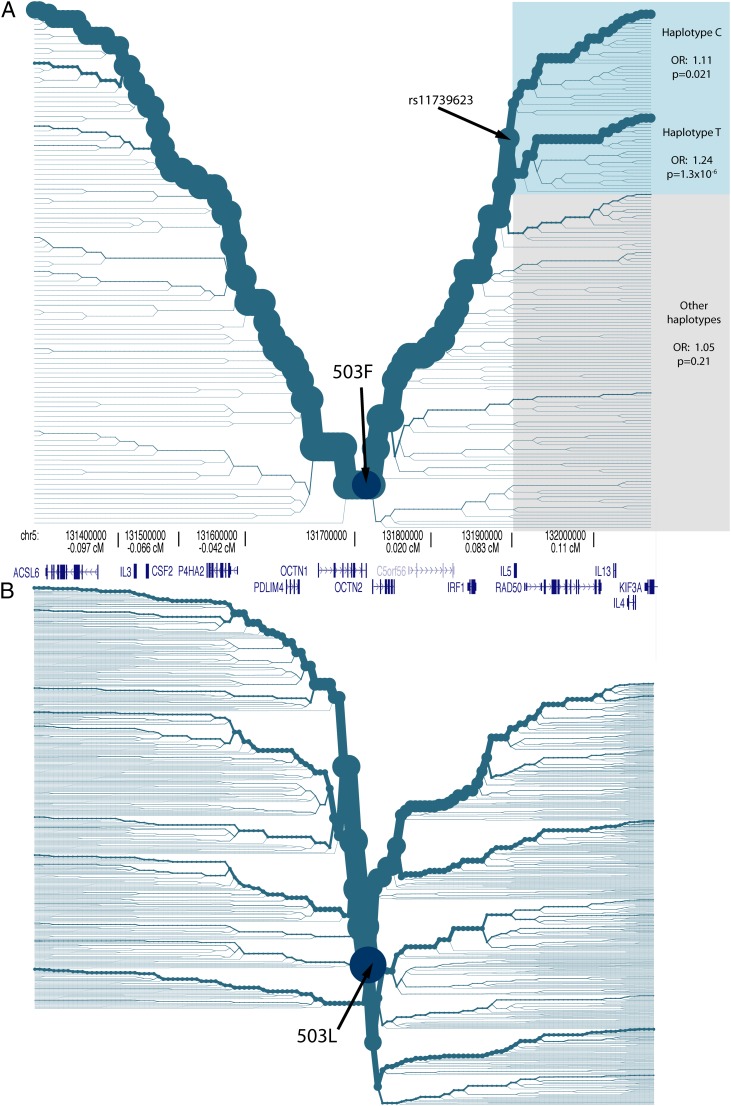
Haplotype bifurcation and disease association at (A) 503F and (B) 503L in Europeans. A haplotype bifurcation diagram is a visual display of the breakdown of LD on the core haplotype, with the thickness of the lines corresponding to the number of samples with the indicated haplotype. We constructed haplotype bifurcation diagrams from the phased genotype data of 1,262 controls (see Materials and Methods). Haplotypes C and T indicate the allelic state at rs11739623. “Other haplotypes” indicate the combined group of haplotypes with an inferred recombination event between 503F and rs11739623. The shaded regions in A designate the haplotypes tested for disease association. The structure of genes in this region is shown. The physical positions along chromosome 5 are listed in hg18 coordinates. The genetic distances from L503F (chr5:131704219) are listed in centiMorgan.
Among the genes in the IBD5 region, there are two strong a priori biological candidates for Crohn’s disease causation: IRF1 (0.057 cM from 503F) and IL5 (0.1 cM from 503F). Both genes are involved in mechanisms that may contribute to Crohn’s disease pathogenesis. IL5 encodes the cytokine IL5, whose expression is increased in lymphocytes isolated from lamina propria cells in ulcerative colitis patients, though not from Crohn’s disease patients (Fuss et al. 1996). Overexpression of IL5 is associated with intestinal inflammation, though distinct in character from murine colitis (Lee et al. 1997). IL5 knockout mice demonstrate an apparently normal adaptive immune response but demonstrate significantly attenuated eosinophilia in response to parasitic challenge (Kopf et al. 1996). IRF1’s potential role in Crohn’s disease causation is supported by several lines of biological evidence: IRF1 deficient mice develop severe infection from the intracellular bacteria Mycobacterium bovis (Kamijo et al. 1994; Yamada et al. 2002). The absence of IRF1 results in the decoupling of the MyD88toll-like receptor signaling pathway (Negishi et al. 2006), a pathway critical for host defense against intracellular pathogens. Thus, a deficiency in IRF1 appears to result in defects in innate immunity, particularly in pathways important in the clearing of intracellular bacteria. Defects in intracellular bacterial clearance are an emerging theme in the pathogenesis of Crohn’s disease, highlighted by the identification of the Crohn’s disease susceptibility loci ATG16L1, NOD2, and IRGM (Singh et al. 2006; Cooney et al. 2010; Travassos et al. 2010).
To test for evidence of disease hitchhiking in IRF1 and IL5, we performed a haplotype association analysis using Illumina 550k SNP microarray data from 1,868 Crohn’s disease cases and 5,540 controls (Imielinski et al. 2009; fig. 4). The odds ratio (OR) for the 503F allele itself was 1.24 (P = 5.5 × 10−9, allele frequency 48.2% in cases vs. 42.7% in controls). At about 200 kb downstream of 503F, past IRF1 and IL5, the core haplotype splits into two common haplotypes at rs11739623 (see fig. 4). These two haplotypes are defined by 31 SNPs (supplementary table S1, Supplementary Material online); we refer them as haplotypes C and T, designating the alleles that differentiate them at rs11739623:C/T (see fig. 4). For the remaining group of individually rare 503F haplotypes, where a recombination event has occurred between 503F and IL5 (gray shading in fig. 4), the difference in frequency between cases and controls was not significant, with an OR of 1.05 (P = 0.21, allele frequency 10.7% in cases vs. 10.2% in controls, see fig. 4). In contrast, the two haplotypes with no detectable recombination between 503F and IRF1 or IL5 (blue shading in fig. 4) were both associated with Crohn’s disease (haplotype C: OR = 1.11, P = 0.021; haplotype T: OR = 1.26, P = 1.0 × 10−6; fig. 4). The OR for haplotypes C and T combined was 1.24 (P = 2.6 × 10−8, allele frequency 37.4% in cases vs. 32.4% in controls). The pattern of haplotype association matches the prediction under the hypothesis that the ancestral sweeping haplotype harbored one or more disease-causing mutations in IRF1 or IL5; 503F haplotypes with no recombination between 503F and IRF1 or IL5 are strongly associated with disease, whereas the remaining 503F recombinant haplotypes are not associated with disease (fig. 4). From the simulation results in figure 2A, we can obtain a rough approximation of how severe a disease-causing mutation would need to be in IBD5. An OR of 1.4 for disease-causing mutations in IRF1 was required to produce the observed allele frequency difference (5.5%) at 503F.
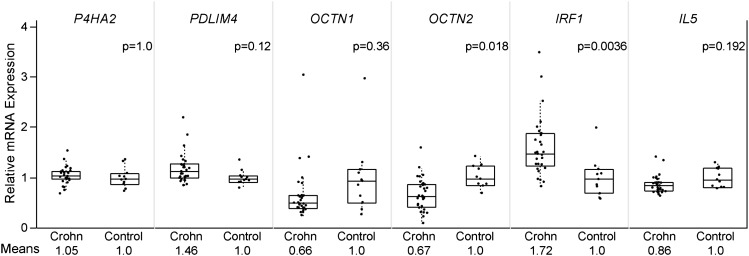
To further evaluate the potential functional importance of genes in the IBD5 region, we measured mRNA expression levels in six genes (P4HA2, PDLIM4, OCTN1, OCTN2, IRF1, and IL5) from colonic biopsies of 30 childhood-onset Crohn’s disease cases and 11 healthy controls. Notably, we observed no significant difference in expression of OCTN1 between cases and controls (fig. 5). After correction for multiple comparisons, significant mRNA expression level differences were observed only in IRF1 and OCTN2. For OCTN2, we observed lower mRNA mean expression levels among Crohn’s disease cases compared with controls (0.67 vs. 1.0; uncorrected P = 0.003). OCTN2 is a paralogue of OCTN1 and encodes a membrane transporter for carnitine. As with many genetic variants on the IBD5 haplotype, genetic variants in OCTN2 are associated with Crohn’s disease (Peltekova et al. 2004). However, a role for OCTN2 in Crohn’s disease causation is not supported by our haplotype association results and has limited biological plausibility. We observed a striking difference in expression of IRF1, with a 72% increase in Crohn’s disease cases relative to controls (mean expression levels in Crohn 1.72 vs. controls 1.0; uncorrected P = 0.0006, fig. 5). This result is consistent with a previous study of IRF1 expression levels from colonic biopsies, which reported a 90% increase in Crohn’s disease cases relative to controls (Clavell et al. 2000). The increased expression in Crohn’s patients provides additional support for a potential role in disease causation for IRF1, although the IBD5 disease-causing variants may not be directly responsible for the difference in expression. In the absence of genotype data on these subjects, we are unable to determine whether IRF1 mRNA expression differences are associated with the IBD5 haplotype, but such studies are justified by our observations.
FIG. 5.
Colonic mRNA expression levels for genes within IBD5. Colon biopsy specimens were obtained from Crohn patients (n = 30) and healthy controls (n = 11). mRNA expression was measured using the Affymetrix GeneChip Human Genome HG-U133 Plus 2.0 array. Bonferroni corrected P values are reported from a Wilcoxon rank sum test.
The pattern of LD surrounding the 503F allele of OCTN1 provides strong evidence for recent positive selection in populations ancestral to Europeans starting approximately 12,500 years ago. We propose that 503F was an adaptation to early agriculture in the Fertile Crescent, increasing the absorption of ET to compensate for the lack of ET in the diet. The association between 503F and Crohn’s disease is probably the result of one or more disease-causing variants that increased in frequency via genetic hitchhiking after becoming linked to 503F. Several lines of evidence collectively implicate IRF1 as a strong candidate for the disease-susceptibility locus at IBD5: our detailed haplotype association analysis, our mRNA expression results, which are consistent with previous reports (Clavell et al. 2000), and previous studies demonstrating that the IRF1 null mouse is susceptible to intracellular bacteria (Kamijo et al. 1994; Yamada et al. 2002; Negishi et al. 2006). Furthermore, our results demonstrate that genetic hitchhiking of multiple deleterious variants can be a common occurrence during a strong selective sweep at distances of up to 3 cM from the advantageous allele. A greater understanding of the phenomenon of deleterious hitchhiking should advance efforts to identify causal variants in genomic regions with overlapping signals of disease association and recent positive selection.
Supplementary Material
Supplementary tables S1–S5 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Alan Rogers and Jon Seger for their helpful comments and suggestions. An allocation of computer time from the Center for High Performance Computing at the University of Utah is gratefully acknowledged. This work was supported by the Primary Children’s Medical Center Foundation and National Institute of Diabetes and Digestive and Kidney Diseases grant DK069513 (to S.L.G.), The University of Luxembourg—Institute for Systems Biology Program, the Gene Expression and Sequencing Core of the National Institutes of Health (NIH)-supported Cincinnati Children’s Hospital Research Foundation Digestive Health Center (1P30DK078392-01), and NIH grant 1T32HL105321-01. L.A.D. is supported by NIH grants R01 DK078683 and R01 DK068164. J.X. is supported by NIH/National Human Genome Research Institute K99HG005846. D.W. and L.B.J. are supported by NIH grant GM59290. This investigation was also supported by Public Health Service research grant UL1-RR025764 and CO6-RR11234 from the National Center for Research Resources.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badr A, Muller K, Schafer-Pregl R, El Rabey H, Effgen S, Ibrahim HH, Pozzi C, Rohde W, Salamini F. On the origin and domestication history of Barley (Hordeum vulgare) Mol Biol Evol. 2000;17:499–510. doi: 10.1093/oxfordjournals.molbev.a026330. [DOI] [PubMed] [Google Scholar]
- Balaresque P, Bowden GR, Adams SM, et al. (16 co-authors) A predominantly neolithic origin for European paternal lineages. PLoS Biol. 2010;8:e1000285. doi: 10.1371/journal.pbio.1000285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Watkins WS, Dixon ME, Jorde LB, Rao BB, Naidu JM, Prasad BV, Rasanayagam A, Hammer MF. Female gene flow stratifies Hindu castes. Nature. 1998;395:651–652. doi: 10.1038/27103. [DOI] [PubMed] [Google Scholar]
- Blekhman R, Man O, Herrmann L, Boyko AR, Indap A, Kosiol C, Bustamante CD, Teshima KM, Przeworski M. Natural selection on genes that underlie human disease susceptibility. Curr Biol. 2008;18:883–889. doi: 10.1016/j.cub.2008.04.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulayeva K, Jorde LB, Ostler C, Watkins S, Bulayev O, Harpending H. Genetics and population history of Caucasus populations. Hum Biol. 2003;75:837–853. doi: 10.1353/hub.2004.0003. [DOI] [PubMed] [Google Scholar]
- Burton PR, Clayton DG, Cardon LR (258 co-authors) Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clavell M, Correa-Gracian H, Liu Z, Craver R, Brown R, Schmidt-Sommerfeld E, Udall J, Jr, Delgado A, Mannick E. Detection of interferon regulatory factor-1 in lamina propria mononuclear cells in Crohn's disease. J Pediatr Gastroenterol Nutr. 2000;30:43–47. doi: 10.1097/00005176-200001000-00016. [DOI] [PubMed] [Google Scholar]
- Consortium TIH. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooney R, Baker J, Brain O, Danis B, Pichulik T, Allan P, Ferguson DJ, Campbell BJ, Jewell D, Simmons A. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- Dickson SP, Wang K, Krantz I, Hakonarson H, Goldstein DB. Rare variants create synthetic genome-wide associations. PLoS Biol. 2010;8:e1000294. doi: 10.1371/journal.pbio.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ey J, Schomig E, Taubert D. Dietary sources and antioxidant effects of ergothioneine. J Agric Food Chem. 2007;55:6466–6474. doi: 10.1021/jf071328f. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SA, Hampe J, Onnie CM, et al. (14 co-authors) Direct or indirect association in a complex disease: the role of SLC22A4 and SLC22A5 functional variants in Crohn disease. Hum Mutat. 2006;27:778–785. doi: 10.1002/humu.20358. [DOI] [PubMed] [Google Scholar]
- Franke A, McGovern DP, Barrett JC, et al. (96 co-authors) Genome-wide meta-analysis increases to 71 the number of confirmed Crohn's disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazer KA, Ballinger DG, Cox DR, et al. (233 co-authors) A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuss IJ, Neurath M, Boirivant M, Klein JS, de la Motte C, Strong SA, Fiocchi C, Strober W. Disparate CD4+ lamina propria (LP) lymphokine secretion profiles in inflammatory bowel disease. Crohn's disease LP cells manifest increased secretion of IFN-gamma, whereas ulcerative colitis LP cells manifest increased secretion of IL-5. J Immunol. 1996;157:1261–1270. [PubMed] [Google Scholar]
- Grossman SR, Shylakhter I, Karlsson EK, et al. (13 co-authors) A composite of multiple signals distinguishes causal variants in regions of positive selection. Science. 2010;327:883–886. doi: 10.1126/science.1183863. [DOI] [PubMed] [Google Scholar]
- Grundemann D, Harlfinger S, Golz S, Geerts A, Lazar A, Berkels R, Jung N, Rubbert A, Schomig E. Discovery of the ergothioneine transporter. Proc Natl Acad Sci U S A. 2005;102:5256–5261. doi: 10.1073/pnas.0408624102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan FA. Radiocarbon chronology of Neolithic and Predynastic sites in upper Egypt and the Delta. Afr Archaeol Rev. 1985;3:95–115. [Google Scholar]
- Hawks J, Wang ET, Cochran GM, Harpending HC, Moyzis RK. Recent acceleration of human adaptive evolution. Proc Natl Acad Sci U S A. 2007;104:20753–20758. doi: 10.1073/pnas.0707650104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- Hillman G. The plant remains from Tell Abu Hureyra: a preliminary report. Proc Prehistoric Soc. 1975;41:70–73. [Google Scholar]
- Huff CD, Harpending HC, Rogers AR. Detecting positive selection from genome scans of linkage disequilibrium. BMC Genomics. 2010;11:8. doi: 10.1186/1471-2164-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imielinski M, Baldassano RN, Griffiths A, et al. (111 co-authors) Common variants at five new loci associated with early-onset inflammatory bowel disease. Nat Genet. 2009;41:1335–1340. doi: 10.1038/ng.489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorde LB, Bamshad MJ, Watkins WS, Zenger R, Fraley AE, Krakowiak PA, Carpenter KD, Soodyall H, Jenkins T, Rogers AR. Origins and affinities of modern humans: a comparison of mitochondrial and nuclear genetic data. Am J Hum Genet. 1995;57:523–538. doi: 10.1002/ajmg.1320570340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamijo R, Harada H, Matsuyama T, et al. (11 co-authors) Requirement for transcription factor IRF-1 in NO synthase induction in macrophages. Science. 1994;263:1612–1615. doi: 10.1126/science.7510419. [DOI] [PubMed] [Google Scholar]
- Kopf M, Brombacher F, Hodgkin PD, et al. (11 co-authors) IL-5-deficient mice have a developmental defect in CD5+ B-1 cells and lack eosinophilia but have normal antibody and cytotoxic T cell responses. Immunity. 1996;4:15–24. doi: 10.1016/s1074-7613(00)80294-0. [DOI] [PubMed] [Google Scholar]
- Ladizinsky G. Seed dispersal in relation to domestication of middle East legumes. Econ Bot. 1979;33:284–289. [Google Scholar]
- Lee NA, McGarry MP, Larson KA, Horton MA, Kristensen AB, Lee JJ. Expression of IL-5 in thymocytes/T cells leads to the development of a massive eosinophilia, extramedullary eosinophilopoiesis, and unique histopathologies. J Immunol. 1997;158:1332–1344. [PubMed] [Google Scholar]
- Li JZ, Absher DM, Tang H, et al. (11 co-authors) Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- Ma Y, Ohmen JD, Li Z, Bentley LG, McElree C, Pressman S, Targan SR, Fischel-Ghodsian N, Rotter JI, Yang H. A genome-wide search identifies potential new susceptibility loci for Crohn's disease. Inflamm Bowel Dis. 1999;5:271–278. doi: 10.1097/00054725-199911000-00005. [DOI] [PubMed] [Google Scholar]
- Markova NG, Karaman-Jurukovska N, Dong KK, Damaghi N, Smiles KA, Yarosh DB. Skin cells and tissue are capable of using L-ergothioneine as an integral component of their antioxidant defense system. Free Radic Biol Med. 2009;46:1168–1176. doi: 10.1016/j.freeradbiomed.2009.01.021. [DOI] [PubMed] [Google Scholar]
- Mirza MM, Fisher SA, King K, et al. (13 co-authors) Genetic evidence for interaction of the 5q31 cytokine locus and the CARD15 gene in Crohn disease. Am J Hum Genet. 2003;72:1018–1022. doi: 10.1086/373880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moncaster JA, Walsh DT, Gentleman SM, Jen LS, Aruoma OI. Ergothioneine treatment protects neurons against N-methyl-D-aspartate excitotoxicity in an in vivo rat retinal model. Neurosci Lett. 2002;328:55–59. doi: 10.1016/s0304-3940(02)00427-5. [DOI] [PubMed] [Google Scholar]
- Negishi H, Fujita Y, Yanai H, Sakaguchi S, Ouyang X, Shinohara M, Takayanagi H, Ohba Y, Taniguchi T, Honda K. 2006. Evidence for licensing of IFN-gamma-induced IFN regulatory factor 1 transcription factor by MyD88 in toll-like receptor-dependent gene induction program. Proc Natl Acad Sci USA. 103:15136–15141. [DOI] [PMC free article] [PubMed]
- Onnie C, Fisher SA, King K, Mirza M, Roberts R, Forbes A, Sanderson J, Lewis CM, Mathew CG. Sequence variation, linkage disequilibrium and association with Crohn's disease on chromosome 5q31. Genes Immun. 2006;7:359–365. doi: 10.1038/sj.gene.6364307. [DOI] [PubMed] [Google Scholar]
- Ozkan H, Brandolini A, Schafer-Pregl R, Salamini F. AFLP analysis of a collection of tetraploid wheats indicates the origin of emmer and hard wheat domestication in southeast Turkey. Mol Biol Evol. 2002;19:1797–1801. doi: 10.1093/oxfordjournals.molbev.a004002. [DOI] [PubMed] [Google Scholar]
- Paul BD, Snyder SH. The unusual amino acid L-ergothioneine is a physiologic cytoprotectant. Cell Death Differ. 2010;17:1134–1140. doi: 10.1038/cdd.2009.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltekova VD, Wintle RF, Rubin LA, et al. Functional variants of OCTN cation transporter genes are associated with Crohn disease. Nat Genet. 2004;36:471–475. doi: 10.1038/ng1339. [DOI] [PubMed] [Google Scholar]
- Pickrell JK, Coop G, Novembre J, et al. (11 co-authors) Signals of recent positive selection in a worldwide sample of human populations. Genome Res. 2009;19:826–837. doi: 10.1101/gr.087577.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinhasi R, Fort J, Ammerman AJ. Tracing the origin and spread of agriculture in Europe. PLoS Biol. 2005;3:e410. doi: 10.1371/journal.pbio.0030410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Goldstein DB. Estimating the age of mutations using variation at linked markers. In: Goldstein DB, Scholotter C, editors. Microsatellites: evolution and applications. Oxford: Oxford University Press; 1999. pp. 128–138. [Google Scholar]
- Rice WR. Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics. 1987;116:161–167. doi: 10.1093/genetics/116.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rioux JD, Daly MJ, Silverberg MS, et al. (31 co-authors) Genetic variation in the 5q31 cytokine gene cluster confers susceptibility to Crohn disease. Nat Genet. 2001;29:223–228. doi: 10.1038/ng1001-223. [DOI] [PubMed] [Google Scholar]
- Rioux JD, Silverberg MS, Daly MJ, et al. (17 co-authors) Genomewide search in Canadian families with inflammatory bowel disease reveals two novel susceptibility loci. Am J Hum Genet. 2000;66:1863–1870. doi: 10.1086/302913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach JC, Glusman G, Smit AF, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:626–629. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabeti PC, Reich DE, Higgins JM, et al. (17 co-authors) Detecting recent positive selection in the human genome from haplotype structure. Nature. 2002;419:832–837. doi: 10.1038/nature01140. [DOI] [PubMed] [Google Scholar]
- Sabeti PC, Walsh E, Schaffner SF, et al. (15 co-authors) The case for selection at CCR5-Delta32. PLoS Biol. 2005;3:e378. doi: 10.1371/journal.pbio.0030378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakagami T, Witherspoon DJ, Nakajima T, Jinnai N, Wooding S, Jorde LB, Hasegawa T, Suzuki E, Gejyo F, Inoue I. Local adaptation and population differentiation at the interleukin 13 and interleukin 4 loci. Genes Immun. 2004;5:389–397. doi: 10.1038/sj.gene.6364109. [DOI] [PubMed] [Google Scholar]
- Schaffner SF, Foo C, Gabriel S, Reich D, Daly MJ, Altshuler D. Calibrating a coalescent simulation of human genome sequence variation. Genome Res. 2005;15:1576–1583. doi: 10.1101/gr.3709305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P, Stephens M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am J Hum Genet. 2006;78:629–644. doi: 10.1086/502802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seger J, Smith WA, Perry JJ, Hunn J, Kaliszewska ZA, Sala LL, Pozzi L, Rowntree VJ, Adler FR. Gene genealogies strongly distorted by weakly interfering mutations in constant environments. Genetics. 2010;184:529–545. doi: 10.1534/genetics.109.103556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverberg MS, Duerr RH, Brant SR, et al. (15 co-authors) Refined genomic localization and ethnic differences observed for the IBD5 association with Crohn's disease. Eur J Hum Genet. 2007;15:328–335. doi: 10.1038/sj.ejhg.5201756. [DOI] [PubMed] [Google Scholar]
- Simonson TS, Yang Y, Huff CD, et al. (12 co-authors). 2010. Genetic evidence for high-altitude adaptation in tibet. Science 329:72–75. [DOI] [PubMed]
- Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagy to eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- Tarazona-Santos E, Tishkoff SA. Divergent patterns of linkage disequilibrium and haplotype structure across global populations at the interleukin-13 (IL13) locus. Genes Immun. 2005;6:53–65. doi: 10.1038/sj.gene.6364149. [DOI] [PubMed] [Google Scholar]
- Taubert D, Grimberg G, Jung N, Rubbert A, Schomig E. Functional role of the 503F variant of the organic cation transporter OCTN1 in Crohn's disease. Gut. 2005;54:1505–1506. doi: 10.1136/gut.2005.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosa M, Negoro K, Kinouchi Y, et al. (13 co-authors) Lack of association between IBD5 and Crohn's disease in Japanese patients demonstrates population-specific differences in inflammatory bowel disease. Scand J Gastroenterol. 2006;41:48–53. doi: 10.1080/00365520510023864. [DOI] [PubMed] [Google Scholar]
- Travassos LH, Carneiro LA, Ramjeet M, et al. (16 co-authors) Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- Voight BF, Kudaravalli S, Wen X, Pritchard JK. A map of recent positive selection in the human genome. PLoS Biol. 2006;4:e72. doi: 10.1371/journal.pbio.0040072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagener DK, Cavalli-Sforza LL. Ethnic variation in genetic disease: possible roles of hitchhiking and epistasis. Am J Hum Genet. 1975;27:348–364. [PMC free article] [PubMed] [Google Scholar]
- Watkins WS, Bamshad M, Dixon ME, et al. (15 co-authors). 1999. Multiple origins of the mtDNA 9-bp deletion in populations of South India. Am J Phys Anthropol. 109:147–158. [DOI] [PubMed]
- Xing J, Watkins WS, Shlien A, et al. (13 co-authors) Toward a more uniform sampling of human genetic diversity: a survey of worldwide populations by high-density genotyping. Genomics. 2010;96:199–210. doi: 10.1016/j.ygeno.2010.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Watkins WS, Witherspoon DJ, Zhang Y, Guthery SL, Thara R, Mowry BJ, Bulayeva K, Weiss RB, Jorde LB. Fine-scaled human genetic structure revealed by SNP microarrays. Genome Res. 2009;19:815–825. doi: 10.1101/gr.085589.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada H, Mizuno S, Sugawara I. Interferon regulatory factor 1 in mycobacterial infection. Microbiol Immunol. 2002;46:751–760. doi: 10.1111/j.1348-0421.2002.tb02760.x. [DOI] [PubMed] [Google Scholar]
- van Zeist W, Bakker-Heeres JAH. Archaeobotanical studies in the Levant. 1. Neolithic sites in the Damascus basin: Aswad, Ghoraife, Ramad. Palaeohistoria. 1982;24:165–256. [Google Scholar]
- Zhou G, Zhai Y, Dong X, et al. (12 co-authors) Haplotype structure and evidence for positive selection at the human IL13 locus. Mol Biol Evol. 2004;21:29–35. doi: 10.1093/molbev/msg231. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.