Abstract
Anaplasma phagocytophilum is an obligately intracellular tick-transmitted bacterial pathogen of humans and other animals. During the course of infection, A. phagocytophilum utilizes gene conversion to shuffle ∼100 functional pseudogenes into a single expression cassette of the msp2(p44) gene, which codes for the major surface antigen and major surface protein 2 (MSP2). The role and extent of msp2(p44) recombination, particularly in hosts that only experience acute infections, is not clear. In the present study, we explored patterns of recombination and expression of the msp2(p44) gene of A. phagocytophilum in a serially infected mouse model. Even though the bacterium was passed rapidly among mice, minimizing the opportunities for the host to develop adaptive immunity, we detected the emergence of 34 unique msp2(p44) expression cassette variants. The expression of msp2(p44) pseudogenes did not follow a consistent pattern among different groups of mice, although some pseudogenes were expressed more frequently than others. In addition, among 263 expressed pseudogenes, 3 mosaic sequences each consisting of 2 different pseudogenes were identified. Population genetic analysis showed that genetic diversity and subpopulation differentiation tended to increase over time until stationarity was reached but that the variance that was observed in allele (expressed pseudogene) frequency could occur by drift alone only if a high variance in bacterial reproduction could be assumed. These findings suggest that evolutionary forces influencing antigen variation in A. phagocytophilum may comprise random genetic drift as well as some innate but apparently nonpurifying selection prior to the strong frequency-dependent selection that occurs cyclically after hosts develop strong adaptive immunity.
Keywords: Anaplasma phagocytophilum, msp2(p44), antigen variation, recombination, drift, selection
Introduction
Anaplasma phagocytophilum is an obligately intraleukocytic tick-transmitted rickettsial parasite of humans and other animals (Bakken, Krueth, Wilson-Nordskog, et al. 1996; Foley 2000; Dumler et al. 2005). Cases of human anaplasmosis are reported with increasing frequency (http://www.cdc.gov/anaplasmosis/) and the causative agents are widely distributed geographically infecting a large diversity of host species including humans, wildlife (rodents, carnivores, and deer), and domestic animals including dogs, cattle, sheep, and horses (Bakken, Krueth, Tilden, et al. 1996; Dumler and Brouqui 1997; Arnez et al. 2001; Foley et al. 2004; Nicholson et al. 2010; Zhan et al. 2010). This bacterium is transmitted by ticks in the Ixodes ricinus group and reservoirs include dusky-footed woodrats (Neotoma fuscipes), western gray squirrels (Sciurus griseus), and redwood chipmunks (Tamias ochrogenys) in the western United States, white-footed mice (Peromyscus leucopus) in the eastern United States, and voles and wood mice (Myodes spp. and Apodemus spp.) in the Old World (Telford et al. 1996; Nicholson et al. 1999; Foley et al. 2002, 2004; Nieto and Foley 2008, 2009). Infections are acute (self-limiting) or persistent depending on host species. Persistent infection occurs in some rodents, sheep, and possibly some dogs (Telford et al. 1996; Stuen et al. 1998; Stuen and Bergstrom 2001; Nieto et al. 2010; Scorpio et al. 2010).
In humans, horses, and some mouse models, acute infections self-limit concurrently with the development of adaptive immunity and activation of macrophages (Dumler et al. 2005). In contrast, the strategy for infection in chronically infected species appears to be a form of immune evasion. Anaplasma phagocytophilum, which resides in neutrophilic vacuoles, has several strategies for intracellular survival including inhibition of phagosome–lysosome fusion to prevent cell-mediated destruction as well as evasion of host adaptive immunity by serially presenting genetically variant major antigens (Dumler et al. 2005). Recombination to produce variant antigens and evade immunity occurs via different mechanisms in a diverse group of pathogens, including some rickettsiae, Borrelia burgdorferi and relapsing fever borrelias, and eukaryotes such as trypanosomes (Kitten et al. 1993; Zhang and Norris 1998; Taylor and Rudenko 2006). Anaplasma phagocytophilum uses the strategy of gene conversion to sequentially shuffle ∼100 different antigen “functional pseudogenes” of the pfam01617 family (common within Anaplasmataceae) sequentially into a hypervariable region of a single expression site of the msp2(p44) gene with conserved 5′ and 3′ ends (Barbet et al. 2006; Lin et al. 2006). The MSP2 protein is a major surface antigen of A. phagocytophilum and is homologous with the MSP2 and MSP3 surface proteins of Anaplasma marginale, a related pathogen that infects only ungulates (Eid et al. 1996; French et al. 1998; Brayton et al. 2003; Dunning Hotopp et al. 2006). As for A. phagocytophilum, diversity in msp2 of A. marginale is generated from a syntenic expression site by recombination, but A. marginale generates protein diversity using only seven pseudogenes and segmental recombination (Barbet et al. 2000).
Antigen variation is a common and effective strategy that allows some parasite to overcome host defense. It is also an example of rapid evolution at a single locus, for example, the msp2 locus. Mutation (in the form of gene conversion) creates variation in the msp2 expression site by sampling from the pseudogene library of about 100 alleles. In order for this strategy to be effective at host immune evasion, the mutation rate must be fairly high, and some alleles could convert into the msp2 site at higher rates than others. Random genetic drift operates to produce random changes in the allele frequencies, with the amount of change due to drift inversely proportional to the effective population size of the parasite. Despite the small size of these changes, the long-term effect of random genetic drift is to remove rare alleles. Mutation and drift together would lead to a stable equilibrium level of antigen variability (Kimura 1983).
However, natural selection affects the success of individual alleles for several reasons. The host adaptive immune system targets common alleles leading to frequency-dependent selection. In fact, as each of the approximately 100 available msp2 alleles is shuffled into the expression cassette, the library, although unchanged (since gene conversion is asymmetric recombination), becomes less effective as a source of novel variants. How quickly this library is exhausted depends on the mutation rate and the strength of selection and drift in removing old variants. In addition, dynamics in alleles could appear to be random or drift like if selection were applied by the innate immune system favoring certain antigen alleles but variably among host individuals or cells or if selection were applied to alleles that are linked with the msp2 locus. This second process is especially important in haploid asexual organisms such as bacteria where linkage is hard to break. The more random-appearing modes of selection may be difficult to model without a great deal of information but may also be responsible for much of apparently neutral molecular evolution (Gillespie 1991).
In the present study, we evaluate the patterns and causes of msp2 allelic dynamics in a model system of A. phagocytophilum strain HZ infection in serially infected mice. Specifically, we report allelic dynamics over time in this experimental model, identify spatial and temporal trends in msp2 alleles, and predict the rate of utilization or exhaustion of the pseudogene repertoire. Analysis of these data is used to understand the contributions of different forms of selection, random genetic drift, and mutation to the observed dynamics. Our analysis complements traditional molecular pathogenesis studies that seek to understand pathogen antigen interactions with host immunity, and we provide insight into the overall evolutionary forces that shape the available and expressed bacterial genetic variability in this and other systems of antigen variation.
Materials and Methods
Mouse Inoculation and Sampling
Twenty-five 10-week-old male C3H/HeJ mice (Jackson Laboratories, Bar Harbor, ME) were maintained in a barrier facility and received food and water ad libitum. All animal experiments were conducted with approval of the Institutional Animal Care and Use Committee at the University of California, Davis, which is accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care International. The first cohort of mice (individuals designated HZA-1, HZC-1, HZE-1, HZF-1, and HZG-1) was inoculated intraperitoneally (i.p.) with 0.3 ml each purified horse leukocytes containing A. phagocytophilum HZ (real-time polymerase chain reaction [PCR] CT value = 28.5). Strain HZ is a fully sequenced human-origin strain from New York State (Dunning Hotopp et al. 2006). Starting on days 5 or 6 postinoculation, mice were anesthetized with ketamine (20 mg/kg) and xylazine (4 mg/kg), and 50 μl of blood were obtained via the retroorbital sinus. Mice were then bled once in every 48 h until A. phagocytophilum DNA was detected in their blood with CT values ≤ 35. Once infection was detected, mice were euthanized by ketamine/xylazine overdose followed by cervical dislocation. Mice were then exsanguinated, and 0.3 ml of blood in ethylenediaminetetraacetic acid were immediately needle-passed i.p. into the next naïve mouse in the serial passage group (SPG). This process was repeated for five serial passages within each SPG.
DNA Extraction and PCR
DNA from all blood samples including the inoculum was extracted using a Qiagen blood and tissue kit (Qiagen, Valencia, CA) per manufacturer's instructions. All DNA samples were initially screened for the presence of A. phagocytophilum DNA using a real-time PCR assay targeting the msp2(p44) gene (Drazenovich et al. 2006). Based on previous findings (Rejmanek D, unpublished data), it was determined that CT values ≤ 35 obtained by real-time PCR were necessary to successfully PCR amplify the entire msp2(p44) expression site by conventional PCR methods. The msp2(p44) expression site was amplified from all DNA samples using a nested PCR assay. In the first round of PCR, primers AB 1058 (GAACCATCCCCTTAAAATACTTTC) and AB 1207 (GGGAGTGCTCTGGTTAGATTTAGG), which generate a fragment of approximately 3 kb containing P44Sup1/omp-1n, msp2(p44), and truncated recA, were used (Barbet et al. 2006). In the second round of PCR, primer MSP2iF (GCTGAAGTGAGGAGACGAAG), which anneals in the 5′ region flanking the msp2(p44) gene, and MSP2iR (AATGGTAGCAGAACCAGAAG), which anneals just 3′ to the truncated recA, gene were used to generate a fragment of approximately 1.5 kb. The PCR conditions were an initial denaturation cycle of 3 min at 94 °C, followed by 35 cycles of 30 s at 94 °C, 1 min at 55 °C, and 4 min at 72 °C, and a final extension of 10 min at 72 °C. Products were prepared for cloning using the Qiagen gel extraction kit.
Cloning and Sequencing Expression Site Variants
PCR-amplified fragments were cloned into the pGEM-T Easy vector (Promega, Madison, WI) followed by transformation into Escherichia coli DH5α cells and plated onto LB agar containing 100 μg/ml ampicillin. Individual colonies were grown overnight in LB broth containing 100 μg/ml ampicillin and plasmids were purified using a Quantum Prep plasmid miniprep kit (BioRad, Hercules, CA). Plasmids were assessed for appropriate insert size following EcoRI digestion. In order to evaluate the diversity of expressed msp2(p44) pseudogenes from each individual, ten clones were randomly chosen for sequencing. In addition, 20 msp2(p44) expression site clones from the initial inoculum were also sequenced. Sequencing was performed using an ABI 3730 sequencer (Davis Sequencing, Davis, CA). Expression site sequences were manually trimmed to the nucleotides coding for the LAKT amino acid residues present on both sides of the hypervariable region. The appropriate msp2(p44) pseudogene designation for each trimmed sequence was determined by searching the A. phagocytophilum HZ genome using the Comprehensive Microbial Resource (CMR) website (http://cmr.jcvi.org/cgi-bin/CMR/GenomePage.cgi?org=gaph).
Statistical and Population Genetic Analysis
Data were maintained in Excel (Microsoft, Redmond, WA) and analyzed with the statistical package “R” (R-Development Core Team, http://www.r-project.org). For all tests, a value of P ≤ 0.05 was considered evidence of statistical significance. Chi-square analysis was conducted to compare the frequency of individual expressed msp2(p44) pseudogenes and to assess potential differences in pseudogene expression among individuals and cohorts. Differences in CT values among mouse SPGs were assessed using analysis of variance. Normality of total numbers of clones for each pseudogene was confirmed with a Kolmogorov–Smirnov test and then linear regression was performed to determine whether total numbers of each pseudogene were significantly associated with pseudogene length. A disproportionately large number of pseudogenes are found near the expression cassette in the HZ strain: In order to detect whether expressed pseudogenes tended to cluster near the expression cassette even more than was accounted for by their position in the genome, we divided the genome into four equal quadrants, from A (origin to 90°), B (90°–180°), C (180°–270°), and D (270°–360°, which contains the expression cassette). A chi-square goodness-of-fit test was performed to test whether the number of expressed pseudogenes observed in any of the mice from each quadrant differed from what was expected based on their underlying position in the genome.
For population genetic evaluation, we define a bacterial “subpopulation” as the group of bacteria within a particular mouse passage group. Then, the gene diversity (also referred to in the literature as “expected heterozygosity”) of each subpopulation at a given time was measured as H = 1 − Σpi2 (Weir 1990; Nei and Kumar 2000; Templeton 2006). We used the sample H as an estimator for the population H since it is a maximum likelihood estimator, and its bias is consistent with consistent sample size (Weir 1990). In order to evaluate the divergence that the five subpopulations experienced over five cohorts of hosts, these gene diversities were used to estimate the population subdivision fixation value Fst = (HT − HS)/HT, where HT gives the overall population heterozygosity expected if each subpopulation had the same allele (expressed pseudogene) frequency and HS gives the mean subpopulation heterozygosity (Templeton 2006). Fst = 0 indicates a homogeneous population and Fst = 1 indicates completely independent populations with no overlap in alleles.
Since the classic Fst statistics are known to be misleading when gene diversity is high, we also used alternative measures of diversity and differentiation (Jost 2008). For each subpopulation, we calculated the effective number of alleles Ae, using the formula Ae = 1/Spi2 defined by Crow and Kimura (1970) (equivalent in ecology to Simpson's inverse index of diversity). For a given time point, we calculated the total population effective number of alleles Aet by merging all five subpopulations. Aes was calculated as the mean Ae over subpopulations. The effective number of subpopulations Δst is defined as Aet/Aes, a measure of population differentiation. (These are equivalent to the ecological measure alpha diversity for Aes, gamma diversity for Aet, and beta diversity for Δst.) A simple measure of population differentiation Dst = 1 − 1/Δst was calculated for each time point. Dst is similar to the widely used Fst which also ranges from 0 to 1, increasing with greater subpopulation differentiation.
Analysis of covariance (ANCOVA) (Crawley 2002) was performed to evaluate whether SPG, time, or interaction had an effect on gene diversity.
The library of functional msp2 pseudogenes includes approximately 100 loci (Foley et al. 2009). As new pseudogenes duplicate and move into the expression site over time, then the number of pseudogenes remaining unexpressed must decline over time from 100 to 0. The rate at which this library was exhausted over time was estimated assuming that gene conversion events occur as a Poisson random process. We then calculated a decay rate, 95% confidence interval (CI), and half-life assuming an exponential decay of the library. We use the term “exhaustion” in this model system, although in vivo pseudogenes may be capable of reexpression on decay of the primary immune responses below a threshold bactericidal level.
Results
Mouse Infections
The initial five mice inoculated with A. phagocytophilum-infected horse leukocytes as well as the remaining four mice in each of the five SPGs (i.e., those that were serially inoculated with A. phagocytophilum-infected mouse blood), all developed PCR-detectable infections within 5–11 days (median = 5 days). Real-time PCR CT values for all mice ranged from 25.1 to 34.9 (mean = 30.8 ± 2.8 standard deviation [SD]). Although the mean CT values between successive cohorts increased from 27.0 (±2.8 SD) in passage 1 to 32.8 (±1.2 SD) in passage 5, this trend was not significant (P = 0.96).
Expression of msp2(p44) Pseudogenes
The entire A. phagocytophilum msp2(p44) expression site from all of the infected mice was successfully amplified and cloned. At least 10 clones were available for sequencing from 22 of the mice. Cloning of the expression site from the other three mice (HZC-2, HZC-5, and HZG-4) resulted in 6, 9, and 8 clones, respectively. In total, 20 clones from the initial inoculum and 243 clones from individual mice were sequenced and identified based on their sequence similarity to known pseudogenes in the A. phagocytophilum HZ genome.
Thirty-eight unique pseudogenes were detected during the course of the study. Within the original inoculum, the expression of four different pseudogenes was detected. Among the 20 clones sampled, 15 (75%) were P44-51, 3 (15%) were P44-23, 1 (5%) was P44-1, and 1 (5%) was P44-2b. The pseudogene expression profiles from the first passage mice were nearly identical to that of the inoculum, with the exception of two additional pseudogenes (P44-19 and P44-35) being expressed. However, by the second passage, eight new pseudogenes were expressed, and one of the initial inoculum pseudogenes (P44-1) was no longer detected. This trend continued through the third and fourth passages with 12 and 10 new pseudogenes detected, respectively. By the fifth passage, four new pseudogenes were detected and only two of the original inoculum pseudogenes (P44-51 and P44-1) remained. These results are summarized in table 1.
Table 1.
Number of Expressed msp2(p44) Pseudogenes during Each Mouse Passage.a
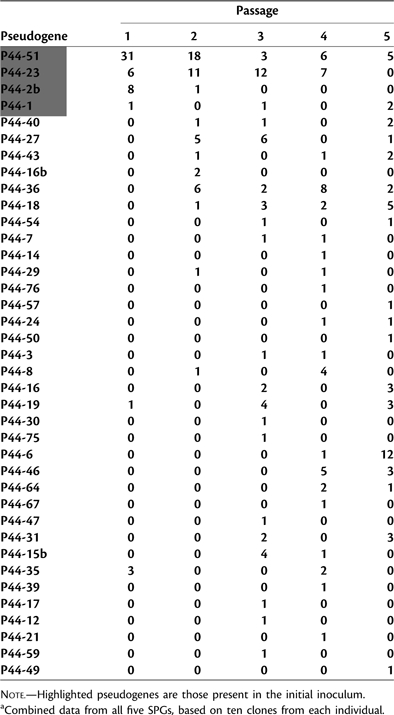 |
Among individual mice, the number of expressed pseudogenes ranged from 2 to 8 (mean = 4.7 ± 1.9 SD, median = 5). The differences in numbers of pseudogenes expressed in individual mice were not significant (P = 0.99). The total number of unique pseudogenes ranged from 11 to 19 per SPG (mean = 15.4 ± 3.1 SD, median = 16). This difference was also not significant (P = 0.86). Although mice from all five SPGs initially expressed the same four pseudogenes that were present in the inoculum, the detection of particular pseudogenes during ensuing serial passages varied considerably among the different SPGs resulting in no discernable temporal pattern of pseudogene expression (see Supplementary Material online).
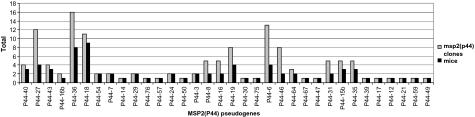
A high proportion of all the msp2(p44) pseudogenes present in the HZ genome (∼40%) was expressed at some point during the study. Ignoring the four inoculum pseudogenes (P44-51, P44-23, P44-2b, and P44-1) because of inherent bias, several of the genes including P44-27, P44-36, P44-18, and P44-6 were more commonly expressed than others (fig. 1). A significant difference (P = 0.01) in the frequency of expression among the noninoculum pseudogenes was detected, although this difference became nonsignificant (P = 0.14) if P44-36, the most frequently expressed noninoculum pseudogene, was excluded from the analysis. Two of the pseudogenes (P44-18 and P44-36) were not only more often expressed but were each detected in at least eight individual mice from several different SPGs. On visual inspection, pseudogenes expressed most frequently appeared to be somewhat closer to the expression cassette than those with no or less frequent expression (data not shown). However, this relationship was not significant statistically (P = 0.21). There was also no support for a relationship between pseudogene expression frequency and pseudogene length (P = 0.81).
FIG. 1.
The number of expression site clones of each msp2(p44) pseudogene excluding the 4 inoculum pseudogenes (P44-51, P44-23, P44-2b, and P44-1) detected throughout the study (total clones = 130) and the number of individual mice in which each pseudogene was expressed.
Population Genetic Analysis
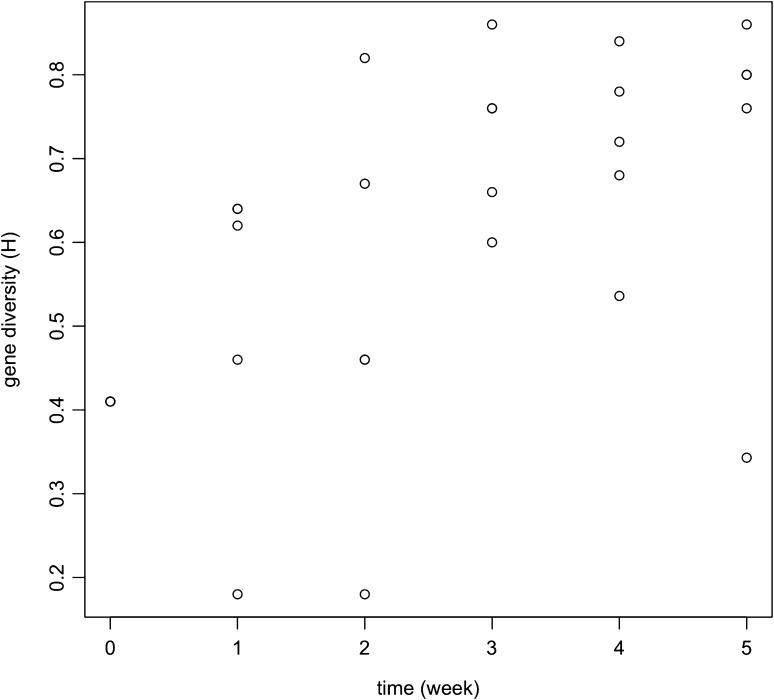
Table 2 gives the calculated gene diversities (H), number of observed pseudogenes (A, for allele), and unused pseudogene library size for each subpopulation. Gene diversity tended to increase over time as shown in figure 2. Time but not SPG was a significant predictor for gene diversity in the ANCOVA analysis, with an unadjusted R2 value of 0.50 (F = 13.9, P = 0.0013). Stationarity may have been reached by week 3 after its early increase as seen in table 3 in gene diversity, population subdivision, effective number of alleles, and effective number of subpopulations. The mean gene diversity over weeks 3, 4, and 5 is 0.717. The expected gene diversity for mutation-drift equilibrium depends on the effective population size Ne and the mutation rate μ. At equilibrium,
Table 2.
Genetic Summary Statistics for five SPGs of Anaplasma phagocytophilum in Mice.
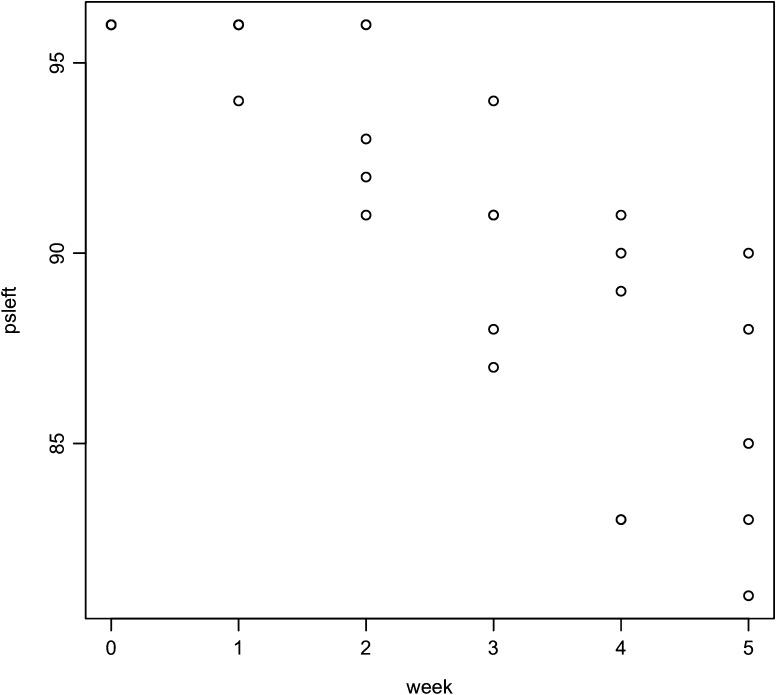
| SPG | Time | H | A | Ae | psleft |
| HZA | 0 | 0.410 | 4 | 1.69 | 96 |
| HZA | 1 | 0.620 | 3 | 2.63 | 96 |
| HZA | 2 | 0.820 | 7 | 5.56 | 91 |
| HZA | 3 | 0.600 | 5 | 2.50 | 88 |
| HZA | 4 | 0.840 | 8 | 6.25 | 83 |
| HZA | 5 | 0.860 | 8 | 7.14 | 81 |
| HZC | 0 | 0.410 | 4 | 1.69 | 96 |
| HZC | 1 | 0.460 | 3 | 1.85 | 96 |
| HZC | 2 | 0.670 | 4 | 3.03 | 92 |
| HZC | 3 | 0.760 | 6 | 4.17 | 87 |
| HZC | 4 | 0.680 | 5 | 3.12 | 83 |
| HZC | 5 | 0.343 | 2 | 1.52 | 83 |
| HZE | 0 | 0.410 | 4 | 1.69 | 96 |
| HZE | 1 | 0.180 | 2 | 1.22 | 96 |
| HZE | 2 | 0.460 | 3 | 1.85 | 96 |
| HZE | 3 | 0.660 | 4 | 2.94 | 94 |
| HZE | 4 | 0.780 | 6 | 4.55 | 91 |
| HZE | 5 | 0.760 | 5 | 4.16 | 90 |
| HZF | 0 | 0.410 | 4 | 1.69 | 96 |
| HZF | 1 | 0.640 | 4 | 2.78 | 94 |
| HZF | 2 | 0.460 | 3 | 1.85 | 93 |
| HZF | 3 | 0.760 | 6 | 4.17 | 91 |
| HZF | 4 | 0.720 | 5 | 3.57 | 90 |
| HZF | 5 | 0.800 | 6 | 5.00 | 88 |
| HZG | 0 | 0.410 | 4 | 1.69 | 96 |
| HZG | 1 | 0.640 | 4 | 2.78 | 96 |
| HZG | 2 | 0.180 | 2 | 1.22 | 96 |
| HZG | 3 | 0.860 | 8 | 7.14 | 91 |
| HZG | 4 | 0.536 | 3 | 2.16 | 89 |
| HZG | 5 | 0.800 | 6 | 5.00 | 85 |
Note.—Time zero corresponds to the initial inoculum population which was used for each line. The time is in weeks, approximately. H is the population heterozygosity (a measure of genetic diversity). A gives the numbers of distinct pseudogenes sampled. Ae gives the effective number of alleles and psleft gives the size of the remaining unused msp2(p44) pseudogene library.
FIG. 2.
Gene diversity at the msp2(p44) expression cassette of each subpopulation of Anaplasma phagocytophilum through serial passages over time in mice.
Table 3.
msp2(p44) Pseudogene Population Diversity and Differentiation Statistics Over Time.
| Week | Ht | Hs | Fst | Aet | Aes | Dst | Dst = 1 − 1/Dst |
| 0 | 0.41 | 0.41 | 0 | 1.69 | 1.69 | 1 | 0 |
| 1 | 0.571 | 0.51 | 0.107 | 2.332 | 2.252 | 1.04 | 0.04 |
| 2 | 0.792 | 0.518 | 0.346 | 4.81 | 2.702 | 1.78 | 0.44 |
| 3 | 0.898 | 0.728 | 0.189 | 9.77 | 4.184 | 2.34 | 0.57 |
| 4 | 0.903 | 0.712 | 0.219 | 10.35 | 3.93 | 2.63 | 0.62 |
| 5 | 0.899 | 0.712 | 0.208 | 9.92 | 4.564 | 2.17 | 0.54 |
Note.—Ht gives the gene diversity in the population as a whole. Hs gives the mean subpopulation gene diversity. Fst (sometimes called the fixation index) measures the amount of population subdivision or differentiation among the five mouse SPGs. Aet, Aes, and Dst are the total effective number of alleles, the subpopulation effective number of alleles, and the effective number of subpopulations (Aet/Aes), respectively. Dst is an index of population subdivision analogous to Fst.
a result that can be derived for haploids similarly to the standard diploid result (e.g., Charlesworth B and Charlesworth D 2010). In our system, this would indicate that Neμ ∼ 1.26, indicating that about one A. phagocytophilum individual undergoes a gene conversion event per generation. Either a low effective population size with a high gene conversion rate or a high effective population size with a low gene conversion rate could produce this result.
The effective population size is difficult to determine since the population of pathogens grows approximately exponentially from day 0 to day 6 within each week, each bacterial cell undergoing binary fission about once a day (Branger et al. 2004). If these new cells circulated freely in the bloodstream (an unrealistic assumption), then Ne would be approximately the harmonic mean population size (Slatkin and Hudson 1991). However, A. phagocytophilum population growth occurs within host cells and, in the course of a week in the mouse host, one A. phagocytophilum cell may divide about six times, with its ∼26 = 64 progeny ready to invade a new neutrophil one to two times before we harvested blood for passage.
An approximation of the population size at the time of inoculation can be obtained by noting that approximately 1% of neutrophils were seen to be occupied by A. phagocytophilum. A young mouse of 20 g has about 2 ml blood with about 4 × 106 neutrophils/ml. So at the end of the week, about 8 × 104 host cells contain A. phagocytophilum. At the start of the week, a 0.3-ml inoculation would contain about 1.2 × 104 parasitized cells. The exact number of bacterial individuals may not be critical to obtaining an effective population size, since, within each host cell the bacteria are mostly clonal. A gene conversion frequency of approximately 1 per 10−4 organisms is similar to that calculated for Neisseria pilin variation that proceeds by an analogous mechanism (Criss et al. 2005; Helm and Seifert 2010).
However, random genetic drift does not easily explain the large changes in H from week to week. In the case of drift in a haploid population, the allele frequency pit+1 has an expected value pi and variance pi(1 − pi)/N (Crow and Kimura 1970). If N is on the order of 104, as we calculated above, and pi = 0.1, then the change in pi due to drift has a SD of about 0.003. This is too small to explain the changes we observed in H. However, if there is a large variance in the reproductive success of bacterial ancestral cells with differing expression cassette genotypes, then the Ne is much lower than it would be otherwise; in this case, drift could account for the observed genetic diversity.
Exhaustion of the Pseudogene Library
The msp2(p44) pseudogene library was exhausted at a rate of about 2 (out of approximately 100) per week (fig. 3). Not only is this seen to be approximately true by examining table 2, but calculation assuming an exponential decay of the library gives a decay rate of a = 0.0235 with a 95% CI of 0.013–0.034. The library's useful half-life is thus T1/2 = 29 weeks in the mouse, in this serial passage model. This would also be true in longer term infections if there is immune selection against reexpression of pseudogenes.
FIG. 3.
msp2(p44) pseudogenes in the Anaplasma phagocytophilum genome library that have not already been expressed in serially passaged mice over time.
msp2(p44) Sequence Variation (Mosaics)
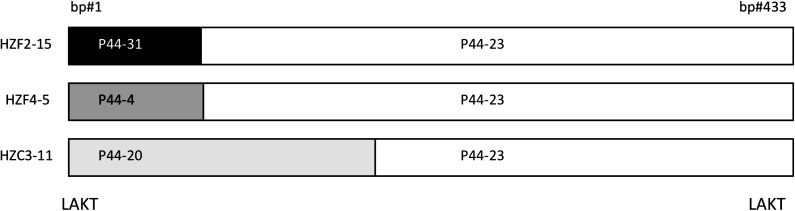
Although most msp2(p44) expression site clones matched with high similarity (99–100%) across the hypervariable (hv) region to a particular pseudogene within the HZ genome, several clones were less similar (93–98%). Two of these clones (HZF2-15 and HZF4-5) were both 95% identical to P44-23 across the entire hv region. However, across the first 66 bp of the hv region, HZF2-15 was only 83% (50/66 bp) identical to P44-23 but 100% identical to another pseudogene (P44-31). Similarly, across the first 60 bp of the hv region, HZF4-5 was 78% (47/60 bp) identical to P44-23 but 100% identical to P44-4. A more extreme example was evident in HZC3-11. For the first 170 bp of the hv region HZC3-11 was 84% (142/170) identical to P44-23 but 100% identical to P44-20. These differences, which are consistent with segmental recombination, are illustrated in figure 4. The sequences reported here have been assigned GenBank accession numbers JN248717 (HZF2-15), JN248718 (HZF4-5), and JN248716 (HZC3-11).
FIG. 4.
Graphical representation of expressed mosaic pseudogenes HZF2-15, HZF4-5, and HZC3-11 across the msp2(p44) hypervariable region. LAKT denotes the conserved amino acids (leucine, alanine, lysine, and tryptophan) located on either end of the hypervariable region.
Discussion
Utilization of a hyperrecombination phenotype to generate antigen variability is hypothesized to represent an evolutionary adaptation to facilitate evasion of host immunity. Such a phenomenon is well supported for A. marginale, which induces chronic infection in its ungulate hosts and sequentially and cyclically expresses variant MSP2 antigens in synchrony with changing host adaptive immunity (French et al. 1998; Brayton et al. 2003). In contrast, infection with A. phagocytophilum in many hosts is not chronic and the role of antigen recombination in host–pathogen interactions is not obvious. In the current study, we detected the emergence of a large number of A. phagocytophilum msp2 expression cassette variants during acute serial infections of naïve mice and found that several pseudogenes did appear to be utilized more commonly than expected by chance alone. There was little evidence for utilization of particular pseudogenes in any temporal order, as a function of spatial location on the chromosome, or associated with donor pseudogene size. Population genetic analysis showed that genetic diversity and subpopulation differentiation tended to increase over time until stationarity was reached but that the variance that was observed in allele (expressed pseudogene) frequency could occur by drift alone only if a high variance in bacterial reproduction could be assumed. This could be due to fitness differences that vary in a random drift-like way or they could be due extremely low Ne. These results suggest that evolutionary forces influencing antigen variation in A. phagocytophilum may comprise drift as well as some innate but apparently nonpurifying selection prior to the strong frequency-dependent selection that occurs cyclically after hosts develop strong adaptive immunity.
Mutation, in this case gene conversion, occurred extensively over the course of this study with an evolution from 6 expressed pseudogene alleles in passage 1 (where four originated from the horse inoculum) to 18 in passage 5. Similar findings have been reported from mice and horses during early time points of infection (Wang et al. 2004; Scorpio et al. 2008). Not only does recombination occur frequently in Anasplasma species but also mosaics in the expression cassette can occur as we saw here. However, among the five SPGs of mice in the present study, there was little discernable pattern or explanation for the particular pseudogenes used with two exceptions: in passage 1, most expression cassettes contained pseudogenes that remained from the horse inoculum and a few particular pseudogenes, that is, numbers P44-18 and P44-36, occurred several times during the study. Pseudogene P44-18 also has been found commonly in A. phagocytophilum HZ growing in HL-60 cells (Sarkar et al. 2008) and during early stages of infection in mice and horses but interestingly not in ticks (Zhi et al. 2002). Also, the P44-18 variant (encoded by pseudogene APH_1194 in HZ) is expressed in different US strains of A. phagocytophilum: the human Webster (Wisconsin) and HGE2 (Minnesota) strains and a California wood rat strain (Barbet et al. 2006). It may be that these pseudogenes have some biochemical or other inherent feature that facilitates their recombination into the expression cassette or that their expressed proteins are particularly good at host attachment and intracellularization. Our data did not support genome position or length (which varied from 473 bp, e.g., P44-76, to 1302 bp [P44-14]) of the pseudogene as an explanation for likelihood of recombination into the expression cassette in contrast with results from a previous study of A. phagocytophilum strains in domestic and wild animals in the United States and Europe in which proximity of an A. phagocytophilum pseudogene to the expression site was significantly associated with frequency of pseudogene expression (Foley et al. 2009). This could be a Type II statistical error in the current study, that is, the distance between pseudogene and expression cassette is in fact biologically relevant, but the present study lacked power to detect it. Thus the underlying features that contribute to more frequent expression of some pseudogenes remain unknown.
The lack of pattern of overrepresentation by most pseudogenes or order in the use of particular pseudogenes is in agreement with previous studies in which mice were sampled on multiple occasions during the course of a single infection with no apparent patterns of pseudogene recombination between individuals (Choi et al. 2007; Scorpio et al. 2008). This is not the case for some other pathogens, however: for example, the emergence of antigen variants of relapsing fever borrelias reportedly follows a “loose order” during the course of infection (Barbour and Stoenner 1985).
Although gene conversion was required to generate the diversity that was observed, the numbers and dynamics of alleles (expressed pseudogenes) observed appear to reflect a balance between gene conversion and a random drift-like process. For high diversity populations, population differentiation is meaningfully measured by comparing the total effective number of alleles Aet, and the subpopulation effective number of alleles Aes, to obtain the effective number of subpopulations Δst = Aet/Aes·Δst, which is a measure of population subdivision, does not suffer the anomalies of Fst, since Δst is independent of Aes (Jost 2008). For all of these estimators, we detected an increase over the first three passages, although the Fst results show the methodological problems discussed by Jost. Since all five mouse lines were inoculated with aliquots from the same Anaplasma culture, Fst was zero at week 0. In the following weeks, Fst increased initially and then settled at a steady level, and more importantly, so did Δst.
One possible explanation to account for the observed allele frequency changes is sample size. The null hypothesis here is that there were no population allele frequency changes, merely sample frequency changes. This hypothesis is not consistent with the almost complete turnover in alleles from week 1 to week 3 and from week 3 to week 6. Nor is it consistent with the increase in gene diversity over time.
Random genetic drift typically does account for patterns of genetic diversity such as those in this study but would not suffice in the case of the effective population sizes we estimated above. However, “nonadaptive” variation (i.e., not due to fitness differences) in bacterial reproduction could produce a much lower Ne, and in this case, drift could be the main driver. During infection by A. phagocytophilum, only one of many competing bacterial clones is likely to enter a particular leucocyte. And perhaps only one clone is ready to invade at a given moment when appropriately susceptible leucocytes become available. This could cause rapid changes in pseudogene frequencies resulting in a lower Ne.
Host adaptive immunity, inducing frequency-dependent balancing selection, was minimized in this study by the experimental design. But could there have been some nondirectional natural selection? A molecular mechanism for a variable selection regime that produces patterns similar to those induced by drift is not known. Little natural selection is likely during bacterial replication within a host cell and drift is minimal at this point as well since progeny are produced clonally. But at the time of invasion of host cells, receptor affinity and other factors that influence successful invasion could contribute to changes in frequency of expressed pseudogenes. If certain pseudogenes were consistently preferentially converted or selected, this could lead to more genetic uniformity across the lines, but we see little evidence of this. However, if each mouse has a slightly different innate immune response (or provides other environmental differences), then pseudogenes may be selected for in a way that mimics random genetic drift. No consistent selection would occur over the five mice in an SPG. But over time and SPG, apparently random shifts in pseudogene frequency would occur. Interestingly, in A. phagocytophilum grown in cell culture (where drift would seem to be much more important than natural selection), only a limited number of different msp2(p44) pseudogenes were expressed over time (Scorpio et al. 2004; Sarkar et al. 2008), although Ne and environmental factors were very different in that study. It has been shown, however, that the ability of A. phagocytophilum to invade cells expressing the sialyl Lewis-modified P-selectin ligand is associated with specific msp2(p44) expression (Troese et al. 2009).
Data in this study and evolutionary models that are consistent with these data also lend insight into what accounts for the overall size of the pseudogene “library,” that is, the resource the pathogen requires for the production of variant antigens. In A. marginale, which contains seven different pseudogenes distributed across the genome (Barbet et al. 2000; Brayton et al. 2005; Futse et al. 2009), additional antigen diversity is generated over time through segmental recombination of fragments of several different pseudogenes into the expression cassette, resulting in a large number of potential mosaic sequences (French et al. 1999; Barbet et al. 2000; Brayton et al. 2002; Palmer et al. 2007). It has been suggested that segmental recombination does not occur in A. phagocytophilum possibly because there is little selection advantage to do so given the large number of available donor pseudogenes (Felek et al. 2004). We showed that the library's decay half-life was 29 weeks in this serial passage model: It would be expected to decay faster in a host with chronic infection imposing strong selection on the bacterium. In fact, results from the current and several other studies (Lin and Rikihisa 2005; Barbet et al. 2006) suggest that expression cassette mosaic formation does occur. Short mosaic sequences were detected at the beginning of the hypervariable region in two of our samples (HZF2-15 and HZF4-5), which were very similar to the short mosaics reported, by Lin and Rikihisa (2005). One interesting mosaic sequence was present in sample HZC3-11 in which one pseudogene (P44-20) accounted for over 40% of the hypervariable region, whereas another pseudogene (P44-23) made up the rest. Importantly, of the >100 genomic pseudogenes, P44-20 (APH_1390) is the most closely related to P44-23 (APH_1256) having 80% nucleotide identity and 82% amino acid identity over the hypervariable region (for an alignment of all peptides encoded by HZ strain genomic pseudogenes, see supplementary fig. 1 of Barbet et al. 2006). Since P44-23 was present in the inoculum, the most plausible mechanistic explanation for the HZC3-11 expressed variant is recombination at sequences shared between the incoming donor pseudogene and the recipient expression site copy. These observations of recombination within either the shared hypervariable region end sequences or between pseudogenes that share greater homology suggest a mechanism for expression of mosaics dependent on pseudogene archive substructure.
So if expression cassette mosaics can develop and genetic diversity increases in the absence of strong adaptive host immunity, what accounts for the maintenance of the large pseudogene library in A. phagocytophilum? One ecological explanation is host niche polymorphism: Various alleles are necessary to optimize fitness across different host species. Another is natural selection that might occur at the time of host infection and cell invasion. The present study does show that high rates of recombination of different pseudogenes into the msp2(p44) expression site over the course of acute A. phagocytophilum infection in a mouse model. The expression of pseudogenes does not follow a consistent pattern and is unrelated to the adaptive immune response of the host, although some pseudogenes are expressed more frequently than others. Overall gene diversity, population subdivision, and variance in allele frequency support a mutation-drift-selection balance. Anaplasma phagocytophilum evolution, which shapes the large number of pseudogenes present in the HZ genome, is a complex process comprising interactions of drift, host adaptive immunity with frequency-dependent selection, host niche polymorphism and the early forms of selection implicated in the present study. Further work that could help understand this evolution could include evaluation of natural selection early in infection, changes in populations of A. phagocytophilum of other strains and in chronically infected hosts, and the strength of drift and possible founder effects that occur during tick infection and transmission.
Supplementary Material
Supplementary material is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Nathan Nieto for help with the study design and animal infections, members of the Foley laboratory for helpful discussion, and Nat Lim, Joy Worth, Susan Wang, Regina Dingler, Katie Azervand, Kat Dang, and Erica Sparacino for laboratory assistance. This work was supported by the National Institutes of Health Allergy and Infectious Disease and Evolution of Infectious Disease Program (RO1 GM081714).
References
- Arnez M, Petrovec M, Lotric-Furlan S, Zupanc TA, Strle F. First European pediatric case of human granulocytic ehrlichiosis. J Clin Microbiol. 2001;39:4591–4592. doi: 10.1128/JCM.39.12.4591-4592.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakken JS, Krueth J, Tilden RL, Dumler JS, Kristiansen BE. Serological evidence of human granulocytic ehrlichiosis in Norway. Eur J Clin Microbiol Infect Dis. 1996;15:829–832. doi: 10.1007/BF01701530. [DOI] [PubMed] [Google Scholar]
- Bakken JS, Krueth J, Wilson-Nordskog C, Tilden RL, Asanovich K, Dumler JS. Clinical and laboratory characteristics of human granulocytic ehrlichiosis. JAMA. 1996;275:199–205. [PubMed] [Google Scholar]
- Barbet AF, Lundgren A, Yi J, Rurangirwa FR, Palmer GH. Antigenic variation of Anaplasma marginale by expression of MSP2 mosaics. Infect Immun. 2000;68:6133–6138. doi: 10.1128/iai.68.11.6133-6138.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbet AF, Lundgren AM, Alleman AR, Stuen S, Bjoersdorff A, Brown RN, Drazenovich NL, Foley JE. Structure of the expression site reveals global diversity in MSP2 (P44) variants in Anaplasma phagocytophilum. Infect Immun. 2006;74:6429–6437. doi: 10.1128/IAI.00809-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour AG, Stoenner HG. Antigenic variation of Borrelia hermsii. In: Herkowitz I, Simon M, editors. Genome rearrangement. New York: Alan R. Liss; 1985. p. 123–135. [Google Scholar]
- Branger S, Rolain JM, Raoult D. Evaluation of antibiotic susceptibilities of Ehrlichia canis, Ehrlichia chaffeensis, and Anaplasma phagocytophilum by real-time PCR. Antimicrob Agents Chemother. 2004;48:4822–4828. doi: 10.1128/AAC.48.12.4822-4828.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brayton KA, Kappmeyer LS, Herndon DR, Dark MJ, Tibbals DL, Palmer GH, McGuire TC, Knowles DP., Jr Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc Natl Acad Sci U S A. 2005;102:844–849. doi: 10.1073/pnas.0406656102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brayton KA, Meeus PF, Barbet AF, Palmer GH. Simultaneous variation of the immunodominant outer membrane proteins, MSP2 and MSP3, during Anaplasma marginale persistence in vivo. Infect Immun. 2003;71:6627–6632. doi: 10.1128/IAI.71.11.6627-6632.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brayton KA, Palmer GH, Lundgren A, Yi J, Barbet AF. Antigenic variation of Anaplasma marginale msp2 occurs by combinatorial gene conversion. Mol Microbiol. 2002;43:1151–1159. doi: 10.1046/j.1365-2958.2002.02792.x. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. Elements of evolutionary genetics. Greenwood Village (CO): Roberts & Company; 2010. [Google Scholar]
- Choi KS, Scorpio DG, Barat NC, Stephen Dumler J. Msp2 variation in Anaplasma phagocytophilum in vivo does not stimulate T cell immune responses or interferon-gamma production. FEMS Immunol Med Microbiol. 2007;49:374–386. doi: 10.1111/j.1574-695X.2007.00214.x. [DOI] [PubMed] [Google Scholar]
- Crawley M. Statistical computing. An introduction to data analysis using S-Plus. New York: Wiley; 2002. [Google Scholar]
- Criss AK, Kline KA, Seifert HS. The frequency and rate of pilin antigenic variation in Neisseria gonorrhoeae. Mol Microbiol. 2005;58:510–519. doi: 10.1111/j.1365-2958.2005.04838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crow JF, Kimura M. An introduction to population genetics theory. New York: Burgess Publishing Company; 1970. [Google Scholar]
- Drazenovich N, Foley J, Brown RN. Use of real-time quantitative PCR targeting the msp2 protein gene to identify cryptic Anaplasma phagocytophilum infections in wildlife and domestic animals. Vector Borne Zoonotic Dis. 2006;6:83–90. doi: 10.1089/vbz.2006.6.83. [DOI] [PubMed] [Google Scholar]
- Dumler J, Brouqui P. Human granulocytic ehrlichiosis. In: Anderson B, Friedman H, Bendinelli M, editors. Rickettsial infection and immunity. New York: Plenum Press; 1997. pp. 149–161. [Google Scholar]
- Dumler JS, Choi KS, Garcia-Garcia JC, Barat NS, Scorpio DG, Garyu JW, Grab DJ, Bakken JS. Human granulocytic anaplasmosis and Anaplasma phagocytophilum. Emerg Infect Dis. 2005;11:1828–1834. doi: 10.3201/eid1112.050898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunning Hotopp JC, Lin M, Madupu R, et al. (40 co-authors) Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2006;2:e21. doi: 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid G, French DM, Lundgren AM, Barbet AF, McElwain TF, Palmer GH. Expression of major surface protein 2 antigenic variants during acute Anaplasma marginale rickettsemia. Infect Immun. 1996;64:836–841. doi: 10.1128/iai.64.3.836-841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felek S, Telford S, 3rd, Falco RC, Rikihisa Y. Sequence analysis of p44 homologs expressed by Anaplasma phagocytophilum in infected ticks feeding on naive hosts and in mice infected by tick attachment. Infect Immun. 2004;72:659–666. doi: 10.1128/IAI.72.2.659-666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley J. Human ehrlichiosis: a review of clinical disease and epidemiology for the physician. Infect Dis Clin Pract. 2000;9:93–98. [Google Scholar]
- Foley JE, Foley P, Brown RN, Lane RS, Dumler JS, Madigan JE. Ecology of granulocytic ehrlichiosis and Lyme disease in the western United States. J Vector Ecol. 2004;29:41–50. [PubMed] [Google Scholar]
- Foley JE, Kramer VL, Weber D. Experimental ehrlichiosis in dusky footed woodrats (Neotoma fuscipes) J Wildl Dis. 2002;38:194–198. doi: 10.7589/0090-3558-38.1.194. [DOI] [PubMed] [Google Scholar]
- Foley JE, Nieto NC, Barbet A, Foley P. Antigen diversity in the parasitic bacterium Anaplasma phagocytophilum arises from selectively-represented, spatially clustered functional pseudogenes. PLoS One. 2009;4:38265. doi: 10.1371/journal.pone.0008265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French D, Brown W, Palmer G. Emergence of Anaplasma marginale antigenic variants during persistent rickettsemia. Infect Immun. 1999;67:5834–5840. doi: 10.1128/iai.67.11.5834-5840.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French DM, McElwain TF, McGuire TC, Palmer GH. Expression of Anaplasma marginale major surface protein 2 variants during persistent cyclic rickettsemia. Infect Immun. 1998;66:1200–1207. doi: 10.1128/iai.66.3.1200-1207.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futse JE, Brayton KA, Nydam S, Palmer GH. Generation of antigenic variants via gene conversion: evidence for recombination fitness selection at the locus level in Anaplasma marginale. Infect Immun. 2009;77:3181–3187. doi: 10.1128/IAI.00348-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillespie JH. The causes of molecular evolution. New York: Oxford University Press; 1991. [Google Scholar]
- Helm RA, Seifert HS. Frequency and rate of pilin antigenic variation of Neisseria meningitidis. J Bacteriol. 2010;192:3822–3823. doi: 10.1128/JB.00280-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jost L. GST and its relatives do not measure differentiation. Mol Ecol. 2008;17:4015–4026. doi: 10.1111/j.1365-294x.2008.03887.x. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge: Cambridge University Press; 1983. [Google Scholar]
- Kitten T, Barrera AV, Barbour AG. Intragenic recombination and a chimeric outer membrane protein in the relapsing fever agent Borrelia hermsii. J Bacteriol. 1993;175:2516–2522. doi: 10.1128/jb.175.9.2516-2522.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Rikihisa Y. Establishment of cloned Anaplasma phagocytophilum and analysis of p44 gene conversion within an infected horse and infected SCID mice. Infect Immun. 2005;73:5106–5114. doi: 10.1128/IAI.73.8.5106-5114.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Q, Zhang C, Rikihisa Y. Analysis of involvement of the RecF pathway in p44 recombination in Anaplasma phagocytophilum and in Escherichia coli by using a plasmid carrying the p44 expression and p44 donor loci. Infect Immun. 2006;74:2052–2062. doi: 10.1128/IAI.74.4.2052-2062.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M, Kumar S. Molecular evolution and phylogenetics. Oxford: Oxford University Press; 2000. [Google Scholar]
- Nicholson WL, Allen KE, McQuiston JH, Breitschwerdt EB, Little SE. The increasing recognition of rickettsial pathogens in dogs and people. Trends Parasitol. 2010;26:205–212. doi: 10.1016/j.pt.2010.01.007. [DOI] [PubMed] [Google Scholar]
- Nicholson WL, Castro MB, Kramer VL, Sumner JW, Childs JE. Dusky-footed wood rats (Neotoma fuscipes) as reservoirs of granulocytic Ehrlichiae (Rickettsiales: Ehrlichieae) in northern California. J Clin Microbiol. 1999;37:3323–3327. doi: 10.1128/jcm.37.10.3323-3327.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto NC, Foley JE. Evaluation of squirrels (Rodentia: Sciuridae) as ecologically significant hosts for Anaplasma phagocytophilum in California. J Med Entomol. 2008;45:763–769. doi: 10.1603/0022-2585(2008)45[763:eosrsa]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Nieto NC, Foley JE. Reservoir competence of the redwood chipmunk, Tamias ochrogenys, for Anaplasma phagocytophilum. Vector Borne Zoonotic Dis. 2009;9:573–577. doi: 10.1089/vbz.2008.0142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto NC, Madigan JE, Foley JE. The dusky-footed woodrat (Neotoma fuscipes) is susceptible to infection by Anaplasma phagocytophilum originating from woodrats, horses, and dogs. J Wildl Dis. 2010;46:810–817. doi: 10.7589/0090-3558-46.3.810. [DOI] [PubMed] [Google Scholar]
- Palmer GH, Futse JE, Leverich CK, Knowles DP, Jr, Rurangirwa FR, Brayton KA. Selection for simple major surface protein 2 variants during Anaplasma marginale transmission to immunologically naive animals. Infect Immun. 2007;75:1502–1506. doi: 10.1128/IAI.01801-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar M, Troese MJ, Kearns SA, Yang T, Reneer DV, Carlyon JA. Anaplasma phagocytophilum MSP2(P44)-18 predominates and is modified into multiple isoforms in human myeloid cells. Infect Immun. 2008;76:2090–2098. doi: 10.1128/IAI.01594-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorpio DG, Caspersen K, Ogata H, Park J, Dumler JS. Restricted changes in major surface protein-2 (msp2) transcription after prolonged in vitro passage of Anaplasma phagocytophilum. BMC Microbiol. 2004;4:1. doi: 10.1186/1471-2180-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorpio DG, Dumler JS, Barat NC, Cook JA, Barat CE, Stillman BA, Debisceglie KC, Beall MJ, Chandrashekar R. Comparative strain analysis of Anaplasma phagocytophilum infection and clinical outcomes in a canine model of granulocytic anaplasmosis. Vector Borne Zoonotic Dis. 2010;3:223–229. doi: 10.1089/vbz.2009.0262. [DOI] [PubMed] [Google Scholar]
- Scorpio DG, Leutenegger C, Berger J, Barat N, Madigan JE, Dumler JS. Sequential analysis of Anaplasma phagocytophilum msp2 transcription in murine and equine models of human granulocytic anaplasmosis. Clin Vaccine Immunol. 2008;15:418–424. doi: 10.1128/CVI.00417-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M, Hudson RR. Pairwise comparisons of mitochondrial DNA sequences in stable and exponentially growing populations. Genetics. 1991;129:555–562. doi: 10.1093/genetics/129.2.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuen S, Bergstrom K. Persistence of Ehrlichia phagocytophila infection in two age groups of lambs. Acta Vet Scand. 2001;42:453–458. doi: 10.1186/1751-0147-42-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuen S, Engvall EO, Artursson K. Persistence of Ehrlichia phagocytophila infection in lambs in relation to clinical parameters and antibody responses. Vet Record. 1998;143:553–555. doi: 10.1136/vr.143.20.553. [DOI] [PubMed] [Google Scholar]
- Taylor JE, Rudenko G. Switching trypanosome coats: what's in the wardrobe? Trends Genet. 2006;22:614–620. doi: 10.1016/j.tig.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Telford SR, Dawson JE, Katavolos P, Warner CK, Kolbert CP, Persing DH. Perpetuation of the agent of human granulocytic ehrlichiosis in a deer tick-rodent cycle. Proc Natl Acad Sci U S A. 1996;93:6209–6214. doi: 10.1073/pnas.93.12.6209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Templeton AR. Population genetics and microevolutionary theory. Hoboken (NJ): Wiley; 2006. [Google Scholar]
- Troese MJ, Sarkar M, Galloway NL, Thomas RJ, Kearns SA, Reneer DV, Yang T, Carlyon JA. Differential expression and glycosylation of Anaplasma phagocytophilum major surface protein 2 paralogs during cultivation in sialyl Lewis x-deficient host cells. Infect Immun. 2009;77:1746–1756. doi: 10.1128/IAI.01530-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Rikihisa Y, Lai TH, Kumagai Y, Zhi N, Reed SM. Rapid sequential changeover of expressed p44 genes during the acute phase of Anaplasma phagocytophilum infection in horses. Infect Immun. 2004;72:6852–6859. doi: 10.1128/IAI.72.12.6852-6859.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weir BS. Genetic data analysis. Sunderland (MA): Sinauer; 1990. [Google Scholar]
- Zhan L, Cao WC, Jiang JF, et al. (12 co-authors) Anaplasma phagocytophilum in livestock and small rodents. Vet Microbiol. 2010;144:405–408. doi: 10.1016/j.vetmic.2010.02.018. [DOI] [PubMed] [Google Scholar]
- Zhang JR, Norris SJ. Genetic variation of the Borrelia burgdorferi gene vlsE involves cassette-specific, segmental gene conversion. Infect Immun. 1998;66:3698–3704. doi: 10.1128/iai.66.8.3698-3704.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhi N, Ohashi N, Tajima T, Mott J, Stich RW, Grover D, Telford SR, 3rd, Lin Q, Rikihisa Y. Transcript heterogeneity of the p44 multigene family in a human granulocytic ehrlichiosis agent transmitted by ticks. Infect Immun. 2002;70:1175–1184. doi: 10.1128/IAI.70.3.1175-1184.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.