Key Points
The ADP-ribosylation factor (ARF) family of guanine-nucleotide-binding (G) proteins, including the ARF proteins, ARF-like (ARL) proteins and SAR1, regulates membrane traffic and organelle structure, and each family member is regulated through a cycle of GTP binding and GTP hydrolysis, which activate and inactivate, respectively, the G protein.
Traditionally, ARFs have been characterized for their immediate effects in the recruitment of coat proteins to drive cargo sorting, the recruitment of enzymes that can alter membrane lipid composition and the regulation of cytoskeletal factors. Now, new roles for ARFs have been discovered at the Golgi complex, for example in driving lipid transport. ARL proteins are also being increasingly linked to coordination of trafficking with cytoskeletal processes, for example during ciliogenesis.
There is particular interest in the mechanisms that control recruitment of the ARF guanine nucleotide exchange factors (GEFs) that mediate GTP binding to ARFs and, in the case of the cytohesin (also known as ARNO) GEF, membrane recruitment is coupled to relief of autoinhibition. GEFs such as cytohesin may also participate in a cascade of activation between particular pairs of ARFs.
Traditionally, G protein signalling has been viewed as a linear pathway, with the GDP-bound form of an ARF protein being inactive; however, more recent studies have highlighted novel roles for these GDP-bound forms and have also shown that GEFs and GTPase-activating proteins (GAPs) themselves can engage in distinct signalling responses through scaffolding functions.
Subject terms: Cytoskeleton, GTP-binding protein regulators, Membrane trafficking, Cell signalling, Diseases
The ADP-ribosylation factor (ARF) and ARF-like (ARL) family of G proteins, which are known to regulate membrane traffic and organelle structure, are emerging as regulators of diverse processes, including lipid and cytoskeletal transport. Although traditionally viewed as part of a linear signalling pathway, ARFs and their regulators must now be considered to exist within functional networks, in which both the 'inactive' ARF and the regulators themselves can mediate distinct effects.
Abstract
Members of the ADP-ribosylation factor (ARF) family of guanine-nucleotide-binding (G) proteins, including the ARF-like (ARL) proteins and SAR1, regulate membrane traffic and organelle structure by recruiting cargo-sorting coat proteins, modulating membrane lipid composition, and interacting with regulators of other G proteins. New roles of ARF and ARL proteins are emerging, including novel functions at the Golgi complex and in cilia formation. Their function is under tight spatial control, which is mediated by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs) that catalyse GTP exchange and hydrolysis, respectively. Important advances are being gained in our understanding of the functional networks that are formed not only by the GEFs and GAPs themselves but also by the inactive forms of the ARF proteins.
Main
The ADP-ribosylation factor (ARF) family of low molecular weight guanine-nucleotide-binding (G) proteins controls membrane traffic and organelle structure, and each member is regulated through a cycle of GTP binding and GTP hydrolysis, which activate and inactivate the G protein, respectively1,2. ARFs have several important functions, including the recruitment of coat proteins that promote sorting of cargo into vesicles, the recruitment and activation of enzymes, such as the phosphatidylinositol (PtdIns) kinases, that alter membrane lipid composition, and interaction with cytoskeletal factors (Table 1). There are six mammalian ARF proteins that can be divided into three classes based on sequence homology: Class I (ARF1, ARF2 and ARF3), Class II (ARF4 and ARF5) and Class III (ARF6) (Fig. 1). The Class II ARFs arose late in animal cell evolution, possibly in metazoans, but Class I ARFs are highly conserved and are present in all eukaryotes examined to date. Hence, in Drosophila melanogaster and Caenorhabditis elegans, each class has a single ARF orthologue, but yeast lacks the Class II ARFs. Plants have Class I ARFs, and potentially also Class III ARFs (Box 1).
Table 1.
ARF and ARL effectors
| Effector | ARF | Location | ARF-interacting region |
|---|---|---|---|
| Coat complexes | |||
| COPII | SAR1 | ER exit sites | SEC23 |
| COPI | ARF1,3 | Golgi, ERGIC | γ-COP, β-COP |
| AP1–clathrin | ARF1,3 | TGN, endosomes | γ-AP1, β-AP1 |
| GGA1,2,3–clathrin | ARF1,3 | TGN, endosomes | GAT domain |
| AP3 | ARF1,3 | Endosomes, TGN | – |
| AP4 | ARF1,3 | TGN | ɛ-AP4, μ-AP4 (also binds GDP-bound form) |
| BBSome | ARL6 | PM | – |
| Lipid-modifying enzymes | |||
| FAPP1,2 | ARF1 | Golgi | PH domain |
| CERT | ARF1 | Golgi | PH domain |
| PtdIns4K | ARF1 | Golgi | – |
| PtdIns4P5K | ARF1–6 | PM (ARF6) | – |
| PLD | ARF1–6, ARL1 | PM (ARF6) | – |
| Tethers | |||
| GMAP210 | ARF1 | cis-Golgi | C-terminal GRAB domain |
| CC golgins* | ARL1 | TGN | C-terminal GRIP domain |
| Exocyst | ARF6 | PM | SEC10 |
| GARP (VFT) | ARL1 | TGN, endosome | – |
| G protein regulators | |||
| ARHGAP21 | ARF1,6 | Golgi, PM | PH domain, C-terminal helix |
| Cytohesin (ARNO) | ARF6, ARL4 | PM | PH domain |
| Scaffolding proteins | |||
| JIP3,4 | ARF6 | Endosomes, intercellular bridge | LZII |
| FIP3,4 | ARF5,6 | Recycling endosomes, midbody | CC domain |
| Tubulin folding chaperone | |||
| Cofactor D | ARL2 | Cytosol | – |
| Cargo | |||
| Rhodopsin | ARF4 | TGN | VXPX targeting motif |
| Other | |||
| NM23-H1 | ARF6 | PM, cell junctions | – |
| PDEδ | ARL2,3 | Recruitment of prenylated proteins | β-sheet region |
| HRG4 | ARL2,3 | – | – |
| ARFAPTIN1,2 | ARF1, ARL1 | Golgi, TGN | BAR domain |
| SCOCO | ARL1 | Golgi | CC |
| BART2 | ARL2 | Mitochondria, nucleus | α-helices 3, 4 and 5 of BART2 |
| AP, adaptor protein; ARF, ADP-ribosylation factor; ARFAPTIN, ARF-interacting protein; ARHGAP21, Rho GTPase-activating protein 21; ARL, ARF-like; BAR, Bin–amphiphysin–Rvs; CC, coiled-coil; CERT, ceramide transfer; COP, coatomer protein; ER, endoplasmic reticulum; ERGIC, ER–Golgi intermediate compartment; GGA, Golgi-localized, γ-ear-containing, ADP-ribosylation factor-binding protein; GMAP210, Golgi-associated microtubule-binding protein 210; GRAB, GRIP-related ARF-binding; JIP, JNK-interacting protein; LZII, Leu zipper domain II; PDEδ, phosphodiesterase-δ; PLD, phospholipase D; PH, pleckstrin homology; PM, plasma membrane; PtdIns4K, phosphatidylinositol 4-kinase; PtdIns4P5K, phosphatidylinositol-4-phosphate 5-kinase; TGN, trans-Golgi network. | |||
| *Including golgin 245, golgin 97, GCC88, GCC185 (mammalian cells) and Imh1 (yeast). | |||
Figure 1. The domain structure and regulation of ARF and ARLs.
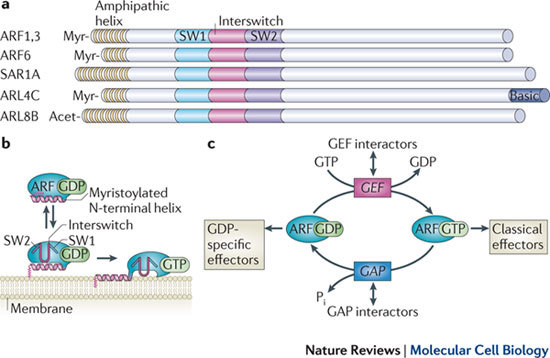
a | A schematic of representative ADP-ribosylation factor (ARF), SAR1 and ARF-like (ARL) proteins, indicating the conserved amino-terminal amphipathic helix and the protein-specific lipid modifications at the N terminus. These include myristoylation (Myr) and acetylation (Acet), both of which ensure tight membrane association. The effector regions of the guanine-nucleotide-binding (G) protein, switch 1 (SW1) and SW2, and the interswitch region between them, are depicted. These regions change conformation upon exchange of GDP for GTP and are involved in interactions with effectors. b | ARF•GDP reversibly associates with the membrane surface, and the myristoylated N-terminal helix ensures tight membrane association of ARF•GTP. The switch and interswitch regions are also shown, and these undergo a conformational change upon GTP binding to enter the hydrophobic pocket that the N-terminal amphipathic helix occupies in the GDP-bound form. c | ARF family G proteins undergo a cycle of GTP binding and hydrolysis, mediated by guanine nucleotide exchange factors (GEFs) and GTPase-activating proteins (GAPs), respectively. The GTP-bound form is thought to carry out G protein functions through interaction with 'classical effectors', including vesicle coat proteins and enzymes that can modify membrane lipid composition; however, increased attention has focused on networks of effectors that are targeted by proteins that interact with GEFs and GAPs themselves and unique effectors that associate specifically with the GDP-bound form of ARF proteins.
In addition, there are over 20 ARF-like (ARL) proteins, which seem to have broader roles than ARFs. Some ARL proteins, including ARL1, ARL2 and ARF-related protein 1 (ARFRP1), are ancient and have homologues in plants, yeast and metazoans, whereas others, such as ARL11, arose later in evolution and are present only in vertebrates2. SAR1, which is present in all eukaryotes examined to date, is also considered to be a member of the ARF family, owing to the presence of an amino-terminal amphipathic helix and the functional similarity of this protein to ARF1 in recruiting a coat complex during vesicle budding.
The study of ARF protein function was aided greatly by the early discovery of the regulators of ARF GTP binding and GTP hydrolysis. The ARF guanine nucleotide exchange factors (GEFs) contain a conserved SEC7 domain that catalyses GDP release from, and GTP binding to, their substrate ARFs. The GTPase-activating proteins (GAPs) catalyse the hydrolysis of GTP-bound ARFs and are critically important because ARFs have negligible intrinsic GTP-hydrolysis activity. ARF GAPs contain a conserved zinc-finger GAP catalytic domain. The conserved, signature catalytic domains in the GEFs and GAPs are what facilitated the identification of these ARF regulators in all organisms from yeast to man. However, the ARF substrate specificity for these GEFs and GAPs remains unclear, and whether they also work on ARL proteins has yet to be determined.
In the past, G protein activity was viewed as a linear signalling pathway, with the GDP-bound form being inactive and the GTP-bound form initiating effector functions until it returned to the inactive, GDP-bound state. The GEFs and GAPs were thought of as 'activators' and 'inactivators', respectively, that controlled this on–off switch (Fig. 1c). However, work on ARF and ARL proteins over the past decade has revealed that their signalling is more complex and that GEFs and GAPs can initiate their own physiological responses. We see evidence of ARF proteins acting in pairs or in series in the endoplasmic reticulum (ER)–Golgi system and at the plasma membrane. In this Review, we emphasize how ARF proteins function as a network in which the ARF regulators participate. These regulators also integrate ARF activities with other G protein signalling networks, as well as initiating their own distinct signalling pathways. We highlight new ARF and ARL activities, discuss how GEFs and GAPs can act as scaffolds, both as effectors and in initiating signalling responses, and describe how they participate in development and disease. The reader is referred to two excellent prior reviews: one deals comprehensively with ARF1 and ARF6 function1 and the other more broadly with ARF, ARL, GEF and GAP proteins2.
Localization and activity of ARFs and ARLs
ARFs localize to membranes throughout the cell, including the plasma membrane and the membranes of the secretory, endosomal and lysosomal pathways. A distinguishing feature of ARF family G proteins is the presence of an N-terminal amphipathic helix that is critical for membrane binding (Fig. 1a,b). In addition, all ARF proteins are modified by myristoylation at the N terminus and this co-translational modification is required for membrane recruitment and biological activity. The myristoyl group and associated N-terminal amphipathic helix are inserted into the membrane upon GTP binding3. Thus, in addition to changes in the effector-binding regions upon exchange of GDP for GTP, ARF proteins undergo a second change in conformation that brings them into very close contact with the membrane4 (Fig. 1). This distinguishes them from other small G proteins of the Ras superfamily, including the Ras, Rho and Rab families, which have a long carboxy-terminal linker to which their lipid membrane anchor is attached2. ARF effectors are thus constrained to a position close to the membrane surface, in contrast to those of Rab and Rho, which can be located some distance from the membrane2. Some ARL proteins are myristoylated but most seem to lack this modification. In ARL8B, loss of hydrophobic residues in the amphipathic helix abolishes lysosomal localization5. Interestingly, ARFRP1 (Arl3 in Saccharomyces cerevisiae), ARL8A and ARL8B are acetylated rather than myristoylated at their N terminus2. In SAR1, the N-terminal amphipathic helix binds directly to membranes and induces membrane curvature6.
Unlike for Rab and Rho G proteins, no GDP dissociation inhibitor (GDI) proteins have been identified for ARFs or ARLs. ARF1 and ARF3 appear to be released from membranes on GTP hydrolysis in cells. ARF6, however, remains bound to membranes in its GDP-bound conformation, and there is evidence that ARF4 and ARF5 remain bound to ER–Golgi intermediate compartment (ERGIC) membranes in their GDP-bound form7,8. This raises the possibility that ARF proteins that are bound to membranes in their GDP-bound form might interact with membrane-localized partners and mediate signalling. Evidence for this idea is emerging for ARF6 (see below), and suggests that distinct signalling pathways might be coordinated through the nucleotide state of these constitutively membrane-bound ARF family proteins. SAR1 and some of the ARLs, such as ARL1, ARL4 and ARL8, are cytosolic when GDP-bound, similarly to ARF1 (Refs 2,9), and it remains to be determined whether this is true for other ARL proteins.
In humans, there are 15 ARF GEFs, which are divided into six subfamilies, as well as the SAR1 GEF SEC12 (Table 2). No specific ARL GEFs have yet been identified, although the ARF1 GEF Syt1 (suppressor of Ypt3 1) in yeast apparently also has activity towards ARL1 (Ref. 10). The 31 identified mammalian ARF GAPs fall into nine major subgroups based on their domain structure (Table 2). Two ARL GAPs have been identified (see below). GEFs and GAPs are recruited to very specific sites within cells to not only catalyse GTP exchange and hydrolysis, respectively, but also to assemble protein complexes at these sites independently of their catalytic activity (Fig. 1c). In this way, versatile signalling networks can be assembled that can respond dynamically to extracellular and intracellular signals.
Table 2.
ARF family GEF and GAP proteins
| Name | Aliases and orthologues | Substrate | Location | Motifs and domains | Interactors |
|---|---|---|---|---|---|
| ARF GEFs | |||||
| GBF1 | Gea1,2 (Sc), GARZ (Dm), GNL1 (At) | ARF1,3,5 | ERGIC, Golgi | DCB | p115 tether, Rab1, γCOP, Drs2 (with Gea2) |
| BIG1 | p200 ARF GEP, Sec7 (Sc) | ARF1,3 | TGN, endosome, nucleus | DCB, AKAP | Myosin IXb, Exo70 |
| BIG2 | BIG5 (BEN1, AtMIN7) (At) | ARF1,3 | TGN, endosome | DCB, AKAP | GABA receptor |
| Cytohesin 1 | PSCD1, Steppke (Dm) | ARF1,6 | PM, endosome | CC, PH, Polybasic | ARFRP1, CASP, ARL4, CNK1 |
| Cytohesin 2 | ARNO, PSCD2, Steppke (Dm) | ARF1,3,6 | PM, endosome | CC, PH, Polybasic | CASP, GRASP (tamalin), IPCEF, A2AR, β-arrestin, V-ATPase, ARL4, ARF6, CNK1, ERBB receptor |
| Cytohesin 3 | GRP1, ARNO3, PSCD3, Steppke (Dm) | ARF1,6 | PM, endosome | CC, PH, Polybasic | CASP, GRASP (tamalin), THR, ARL4, ARF6, CNK1 |
| Cytohesin 4 | PSCD4 | ARF1,5 | – | CC, PH, Polybasic | – |
| EFA6A–D | PSD1–4, Yel1 (Sc), Syt1 (Sc), EFA6 (Dm) | ARF6 | PM, endosome | PH, CC, pro | TWIK1 K+ channel |
| BRAG1 | IQSEC2, IQARFGEF, Loner (Dm), Schizo (Dm) | ARF6 | PSD | PH, CC, IQ | IRSp53 |
| BRAG2 | GEP100, IQSEC1, Loner (Dm), Schizo (Dm) | ARF6 | PM, endosome, nucleus | PH, CC, IQ | AMPA receptor |
| BRAG3 | IQSEC3, SYNARFGEF, Loner (Dm), Schizo (Dm) | ARF6 | PSD | PH, CC, IQ | PSD95, Homer, utrophin (dystrophin), S-SCAM |
| ARF GAPs * | |||||
| ARFGAP1 | Gcs1 (Sc) | ARF1–5 | Golgi | ALPS | – |
| ARFGAP2,3 | ZNF289, Glo3 (Sc) | ARF1–5 | Golgi | Polybasic, Glo3 (ISS repeat) | COPI coat |
| ADAP1,2 | Centaurin α1,β | – | – | Two PH | – |
| SMAP1,2 | – | ARF1,6 | – | Clathrin box, CALM | – |
| AGFG1,2 | HRB1,2 | – | – | FG repeats | – |
| GIT1,2 | CAT1,2, p95APP1,2, GIT (Dm) | ARF6 | PM | ANK, SHD, CC, PBS | PIX, PLCγ, MEK1, FAK, GPCR kinase |
| ASAP1–3 | AMAP1,2, DEF1, PAG2,3, PAP, ASAP (Dm) | ARF1,5,6‡ | PM, FA | BAR, PH, ANK, Pro, SH3 | CIN85 (SH3KBP1), cortactin, CRK, SRC, FAK, PYK2 |
| ACAP1–3 | Centaurin β1,β2,β5 | ARF6 | PM, endosome | BAR, PH, ANK | β1 integrin, cellubrevin, transferrin R |
| AGAP1–11 | Centaurin-γ | – | – | GLD, PH, ANK | – |
| ARAP1–3 | Centaurin δ1,δ2,δ3 | – | – | SAM, five PH, ANK, RhoGAP, RA | RhoA, CIN85, Rap1 |
| ARL GAPs | |||||
| RP2 | Cin2 (Sc), XRP2 | ARL3 | PM, periciliary ridge, cilial basal body | – | – |
| ELMOD2 | – | ARL2,3 | – | – | – |
| A2AR, adenosine A2A receptor; ADAP, ARF GAP with dual PH domain-containing; AGFG, ARF GAP domain and FG repeats-containing; AKAP, A kinase-anchoring protein; ALPS, amphipathic lipid packing sensor; ANK, ankyrin repeat; ARF, ADP-ribosylation factor; ARL, ARF-like; ASAP, ARF GAP containing SH3, ankyrin repeat and PH domains; At, Arabidopsis thaliana; BAR, Bin–amphiphysin–Rvs; CALM, clathrin assembly lymphoid myeloid; CASP, cytohesin-associated scaffolding protein; CC, coiled-coil; Cin2, chromosome instability protein 2; CIN85, CBL-interacting protein 85; COPI, coatomer complex I; DCB, dimerization and cyclophilin-binding domain; Dm, Drosophila melanogaster; ERGIC, ER–Golgi intermediate compartment; FA, focal adhesion; FAK, focal adhesion kinase; FG, phenylalanine, glycine repeats; GABA, γ-aminobutyric acid; GAP, GTPase-activating protein; Gea, ARF guanine nucleotide exchange factor; GEF, guanine nucleotide exchange factor; GEP, guanine nucleotide exchange protein; GLD, GTP-binding protein like domain; GNL1, guanine nucleotide-binding protein-like 1; GPCR, G protein-coupled receptor; GRASP, GRP1-associated scaffold protein; IPCEF, interaction protein for cytohesin exchange factors; IQ, IQ motif; MEK, MAPK/ERK kinase; PAG, Paxillin-associated protein with ARF GAP activity; PBS, paxillin binding site; PH, pleckstrin homology; PLC, phospholipase C; PM, plasma membrane; Pro, proline-rich; PSD, post-synaptic density; PYK, proline-rich tyrosine kinase; RA, Ras association motif; RP2, retinitis pigmentosa 2; SAM, sterile motif; Sc, Saccharomyces cerevisiae; SH3, SRC homology 3; SHD, SRC homology domain; S-SCAM, synaptic scaffolding molecule; TGN, trans-Golgi network; THR, thyroid hormone receptor; TWIK1, tandem of P domains in a weak inward-rectifying K+ channel 1; ZNF289, zinc-finger 289. | |||||
| *Consensus name used from Ref. 140. | |||||
| ‡ASAPs work better on ARF1 and ARF5 than on ARF6. | |||||
Expanding the roles of ARFs and ARLs
Following activation on membranes, GTP-bound ARFs recruit coat proteins, lipid-modifying enzymes, tethers and other effector molecules that influence membrane trafficking and organelle structure1,2 (Table 1). For example, ARF1 recruits the cytosolic coatomer complex I (COPI) to Golgi membranes, allowing sorting of cargo proteins into COPI-coated vesicles11. ARF proteins at the trans-Golgi network (TGN) also recruit heterotetrameric clathrin adaptor protein 1 (AP1), AP3 and AP4, as well as the three monomeric Golgi-localized, γ-ear-containing, ADP-ribosylation factor-binding proteins (GGAs), GGA1, GGA2 and GGA312. These various coat proteins specifically bind cargo proteins and incorporate them into newly forming vesicles for sorting and transport to their correct destinations. ARFs can also recruit and activate enzymes that alter membrane lipid composition. Phospholipase D (PLD), which hydrolyses phosphatidylcholine to generate phosphatidic acid, is activated by all ARF proteins and also by ARL1 (Ref. 13). All ARF proteins can both recruit and activate PtdIns-4-phosphate 5-kinase (PtdIns4P5K), an enzyme that phosphorylates PtdIns4P at the 5-position to generate PtdIns-4,5-bisphosphate (PtdIns(4,5)P2)1. For example, ARF6 at the cell periphery directly affects the activity of PtdIns4P5K at the plasma membrane, and thus regulates PtdIns(4,5)P2 levels there1. At the Golgi, ARF1 recruits and stimulates the activity of PtdIns 4-kinase (PtdIns4K), forming PtdIns4P, which is an important membrane lipid for Golgi function14. ARF1 also binds to PtdIns4P-specific pleckstrin homology (PH) domains contained in a family of oxysterol-binding proteins that are believed to function in lipid homeostasis at the Golgi14.
New functions for Golgi-associated ARFs. The five ARF proteins in humans, ARF1, ARF3, ARF4, ARF5, and ARF6, are ubiquitously expressed. Studies to date have focused mainly on ARF1 at the Golgi and ARF6 at the plasma membrane, but ARF3, ARF4 and ARF5 are also present on Golgi membranes (Fig. 2a). Surprisingly, depletion experiments using RNA interference (RNAi) show that no single ARF, including ARF1, is required for Golgi function; instead, ARFs function in pairs at particular steps in Golgi transport15. For example, ARF1 and ARF4 act redundantly during transport in the early secretory pathway15. Consistent with this observation, ARF4 localizes to the ERGIC and cis-Golgi8 and, together with ARF1 at the cis-Golgi, it organizes trafficking between these compartments16.
Figure 2. ARF and ARL functions in the secretory pathway and in specialized transport.
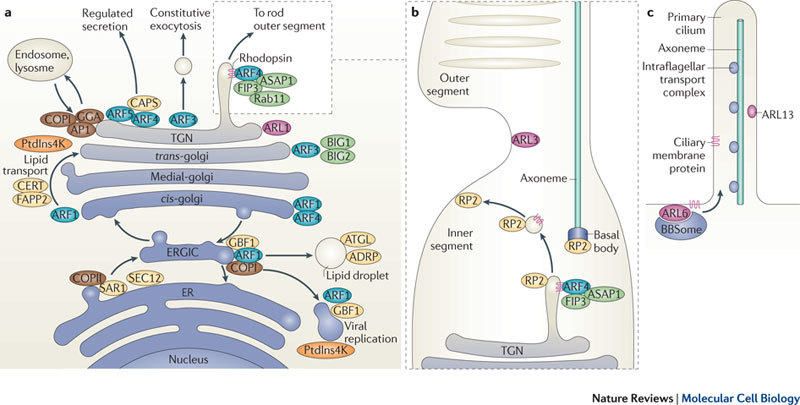
a | ADP-ribosylation factor (ARF) proteins have distinct localizations and functions in the endoplasmic reticulum (ER)–Golgi system. ARF1 and ARF4 localize to the early cis-Golgi and ARF3 specifically localizes to the trans-Golgi network (TGN). In addition to the recruitment of coat proteins (coatomer complex I (COPI), GGA (Golgi-localized, γ-ear-containing, ADP-ribosylation factor-binding protein) and adaptor protein 1 (AP1)) to the Golgi, ARF1 binds to ceramide transfer (CERT) and FAPP2 to mediate the transport of ceramide and glucosylceramide lipids from the cis-Golgi to the trans-Golgi. At the ER–Golgi intermediate compartment (ERGIC), ARF1 and its guanine nucleotide exchange factor (GEF) GBF1 act with COPII to regulate the formation of lipid droplets and for the replication of several viruses. CAPS (Calcium-dependent activator protein for secretion), which is involved in regulated secretion, is recruited to the TGN by ARF4 and ARF5. At the ER, SAR1, activated by SEC12, recruits COPII to allow vesicle transport to the Golgi. b | In retinal cells, ARF4 binds specifically to rhodopsin in the TGN membrane and, together with FIP3, ASAP (ARF GAP containing SH3, ankyrin repeat and PH domains) and Rab11, it facilitates the transport of rhodopsin in transport vesicles from the inner segment to the outer segment, which is a specialized cilium. ARF-like 3 (ARL3) has been found to be localized to the connecting cilium, and retinitis pigmentosa 2 (RP2; also known as XRP2), an ARL3 GAP, localizes to the TGN, the basal body and the membrane adjacent to the connecting cilium. c | In primary cilia, ARL6 recruits the BBSome coat complex that facilitates the transport of membrane proteins into the cilium. ARL13 is localized to the cilium and has been implicated in intraflagellar transport. ADRP, adipose differentiation-related protein (also known as adipophilin); ATGL, adipose triglyceride lipase; PtdIns4K, phosphatidylinositol 4-kinase.
ARF1 and ARF3 are identical except for seven amino acid differences in their N-terminal and C-terminal regions, and previously they were thought to function and localize identically. However, a Golgi-targeting sequence contained within the α3 helix of ARF1 and ARF3 targets a chimaera of ARF6 and ARF1 to the early Golgi17. Furthermore, ARF3 localizes specifically to the TGN (Fig. 2a), and this localization depends on four ARF3-specific amino acids contained in the N-terminal amphipathic helix, which are conserved among ARF3 homologues18. ARF3, but not ARF1, becomes cytosolic at 20oC, the temperature at which exit from the TGN is blocked18. Thus, ARF3 might have an additional crucial role during exit from the Golgi.
Several important functions for Class II ARFs at the TGN have now been defined (Fig. 2a). In an elegant series of studies, ARF4 was found to specifically recognize the VXPX cytosolic targeting motif in retinal rhodopsin to facilitate its transport into the rod outer segment, which is a specialized cilium19 (Fig. 2b). This ciliary targeting complex includes, in addition to ARF4, Rab11, FIP3 (a shared ARF and Rab11 effector) and ASAP1 (ARF GAP containing SH3, ankyrin repeat and PH domains 1)20. Exactly how this complex facilitates the packaging of rhodopsin into post-Golgi carriers has yet to be determined but, interestingly, rhodopsin itself initiates complex formation by recruiting ARF4. The rhodopsin-binding site on ARF4 is in the α3 helix19, the same region that in ARF1 binds the SNARE protein membrin (also known as GOSR2) to mediate targeting to the early Golgi17; thus, this region might generally allow ARF protein binding to membrane receptors. ARF4 and ARF5 can also directly bind to CAPS (calcium-dependent activator protein for secretion), which regulates exocytosis of dense core vesicles from nerve terminals21. It is the GDP-bound form of the ARF that binds to the PH domains of CAPS proteins, and knockdown of CAPS, ARF4 or ARF5 causes retention of chromagrannin (a marker for dense core vesicles) in the Golgi, suggesting that ARF4 and ARF5, together with CAPS, regulate the release of dense core vesicles from the Golgi (Fig. 2a). How these roles of ARF4 and ARF5 at the TGN in specialized cells can be reconciled with findings of ARF4 localization to, and ARF4 and ARF5 functioning at, the early Golgi in other cells8,15,16 is not known.
Recent discoveries show that ARF1 regulates lipid transfer proteins within the Golgi and promotes the formation of lipid droplets at the ERGIC (Fig. 2a). At the Golgi, ARF1 recruits the lipid transfer proteins ceramide transfer (CERT) and FAPP2 (Ref. 14) through interaction with their PH domains, which can also bind PtdIns4P. CERT mediates the non-vesicular transport of ceramide from the ER to the Golgi and FAPP2 mediates the transfer of glucosylceramide from the cytosolic side of the early Golgi to the trans-Golgi22. Exactly how the directionality of this transfer occurs, and the role that ARF1 has, is not yet clear. The finding that ARF1 associates with GBF1 and COPI during lipid droplet formation was unexpected. These proteins were identified in an RNAi screen of lipid droplet formation in D. melanogaster23 and also appeared in proteomic analyses of lipid droplets along with other trafficking proteins, which led to the idea that lipid droplets interface with multiple membrane trafficking pathways24. In particular, the delivery of two proteins, adipose triglyceride lipase (ATGL) and adipose differentiation-related protein (ADRP; also known as adipophilin), to the surface of lipid droplets requires ARF1, GBF1 and COPI, and possibly the COPII machinery, in mammalian cells25; similar results were obtained in D. melanogaster S2 cells26. Another ARF family member, ARFRP1, is highly expressed in adipocytes, and mice that lack ARFRP1 in adipose tissue show severe defects in lipid storage and enhanced lipolysis27. Finally, in some cell types ARF1 at the plasma membrane affects endocytosis of proteins anchored to the membrane by a glycosyl PtdIns (GPI) linkage28. This may also require the ARF GEF GBF1 (Ref. 29) and could be related to the other lipid-regulating functions of ARF1.
New understanding of ARF6 function. A great deal of work on ARF6 function has been summarized in a previous review1, so here we focus on more recent advances. In mammals, ARF6 is not required for early embryonic development, but ARF6-knockout mice die at mid-gestation or shortly after birth and exhibit impaired liver development30. This phenotype suggests that the critical physiological roles of ARF6 take place after birth and is consistent with reported effects of ARF6 on cell adhesion, cell migration, wound healing and metastasis.
ARF6 is present at the plasma membrane and influences both the cortical actin cytoskeleton and endosomal membrane recycling1 (Fig. 3). At the plasma membrane, ARF6 changes the membrane lipid composition through activation of PtdIns4P5K and PLD, resulting in the generation of PtdIns(4,5)P2 and phosphatidic acid. These lipids are important for sorting proteins within the membrane, for the formation of clathrin-coated pits during endocytosis, and for the recruitment and activation of Rho family G proteins, such as Rac, to alter actin polymerization. There is some evidence that ARF6 can interact with AP2 (Ref. 31) and clathrin during G protein-coupled receptor (GPCR) cell signalling32. A recent study has found that ARF6 enters cells in clathrin-coated vesicles to facilitate the rapid recycling of the transferrin receptor back to the plasma membrane through interaction with the microtubule motor adaptor protein JNK-interacting protein 4 (JIP4) after clathrin uncoating33. In some cells, ARF6 associates with endosomal membranes derived from clathrin-independent forms of endocytosis and mediates recycling of this membrane back to the plasma membrane34. Recycling endosomes return membrane proteins that are important for cell adhesion and migration back to the plasma membrane34,35. ARF6 regulation of such endosomal membrane trafficking is required for the polarized delivery of CDC42, Rac and the PAR6 complex to the leading edge of migrating cells36, which can alter adhesion to the extracellular matrix through focal adhesions and actin-based protrusions. Hence, regardless of the mode of endocytosis, ARF6 is important for membrane recycling.
Figure 3. The localization and function of ARF and ARL proteins in endosomal–lysosomal trafficking.
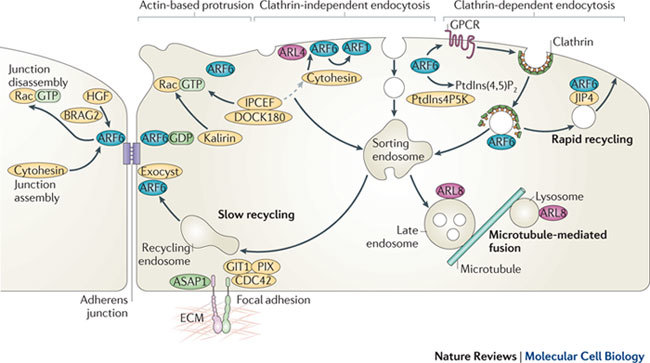
At the plasma membrane, ADP-ribosylation factor 6 (ARF6) activates phosphatidylinositol-4-phosphate 5-kinase (PtdIns4P5K) to generate PtdIns-4,5-bisphosphate (PtdIns(4,5)P2) and, together with ARF-like 4 (ARL4), recruits cytohesin (also known as ARNO) guanine nucleotide exchange factors (GEFs) that can lead to further activation of ARF6 or ARF1. Cytohesins associate with the IPCEF (interactor protein for cytohesin exchange factors)–DOCK180 complex, which activates Rac, but another Rac GEF, Kalirin, can be recruited to membranes by ARF6•GDP. ARF6 at the plasma membrane can regulate the membrane lipid composition, alterations in cortical actin to drive protrusions (for example, during cell migration), and endocytosis of ligand-activated guanine-nucleotide-binding (G) protein-coupled receptors (GPCR) via clathrin-dependent endocytosis. ARF6 and the microtubule motor adaptor protein JNK-interacting protein 4 (JIP4) promote rapid recycling of endosomal membrane back to the cell surface, and ARF6, together with the exocyst complex, also affects slow recycling from sorting endosomes. ARF1 has been implicated in clathrin-independent endocytosis of glycosyl PtdIns (GPI)-anchored proteins in some cells. ARF6 and the ARF6 GEFs cytohesin and BRAG2 have been implicated in both assembly and disassembly of adherens junctions. Two ARF GTPase-activating proteins (GAPs), ASAP1 (ARF GAP containing SH3, ankyrin repeat and PH domains 1) and GIT1, localize to focal adhesions that mediate adhesion to the extracellular matrix (ECM), and GIT1 interacts with PIX, a GEF for CDC42. ARL8 is required for fusion of multivesicular late endosomes with lysosomes and is involved in transport along microtubules. HGF, hepatocyte growth factor.
The crucial functions of ARF6 in membrane lipid modification, establishment of cell polarity and promotion of endocytic recycling are conserved in yeast and D. melanogaster1. Arf3, the yeast ARF6 homologue, contributes to PtdIns(4,5)P2 levels at the plasma membrane37 and also affects polarization events, such as bud site selection38 in S. cerevisiae and new end take-off growth in Schizosaccharomyces pombe39. ArfB, the ARF6 homologue in the filamentous fungi Aspergillus nidulans, localizes to both the plasma membrane and endomembranes, and regulates endocytosis and polarity establishment during hyphal growth40. In D. melanogaster, deletion of the ARF6 homologue blocks the rapid endocytic recycling required for cytokinesis in spermatocytes, resulting in male sterility, but no other phenotypes were reported41. Interestingly, in mammalian cells ARF6 interacts with JIP4 to control a motor switch mechanism regulating endosomal trafficking in cytokinesis42. The crystal structure of ARF6 in complex with JIP4 shows that residues adjacent to the switch regions are structural determinants for the specific binding of JIP4 to ARF6 (Ref. 43).
ARF6 has been implicated in both the assembly and disassembly of adherens junctions in polarized epithelial cells1 (Fig. 3). During adherens junction formation, PAR3 recruits a scaffolding protein, FRMD4A, that binds to cytohesin GEFs, which leads to activation of ARF6 (Ref. 44). Treatment of fully polarized epithelial cells with hepatocyte growth factor leads to activation of ARF6, most likely through the ARF GEF BRAG2 (Ref. 45), and activation of Rac, which causes disassembly of adherens junctions by stimulating endocytosis of epithelial cadherin (E-cadherin)1. Hence, depending on the signalling complex assembled, either formation or disassembly of adherens junctions can be achieved through activation of ARF6. There is also some evidence that the ARF6 GEF EFA6 affects tight-junction assembly46. ARF6 activation has also been reported at the onset of tubulogenesis (a developmental progression from polarized epithelia to tubular structures), and perturbation of the ARF6 GTP–GDP cycle inhibits tubule formation47.
Importance of turning off ARF6. ARF proteins carry out their actions through a regulated cycle of GTP binding and hydrolysis. This allows ARFs to engage and disengage with their effectors with spatial and temporal specificity, and in some cases may allow ARF•GDP to bind other classes of effector. ARF6•GDP binds several TBC (Tre2–Bub2–Cdc16) domain-containing proteins, which often have Rab GAP activity48. ARF6•GDP binds both TBC1 domain family member 24 (TBC1D24; a protein mutated in familial infantile myoclonic epilepsy49) and the TRE17 oncogene50. TRE17 binding to ARF6 increases its activation50; although TRE17 does not itself have GEF activity towards ARF6, it may facilitate interaction of ARF6 with another GEF. ARF6•GDP also binds to the Kalirin family of Rho GEFs, through their spectrin-like repeat domain51, and recruits Kalirin to the membrane, where it subsequently activates Rac and RhoG to regulate actin dynamics51 (Fig. 3). Hence, ARF6•GDP and ARF6•GTP both interact with regulatory proteins of other small G proteins, allowing alternative signalling pathways to be activated depending on which nucleotide is bound (Fig. 1c). This raises the intriguing possibility that other GDP-bound ARF or ARL proteins might also bind unique effector proteins.
Turning off ARF6 is important for its biological function. In some cells, expression of the constitutively active mutant of ARF6, Q67L, leads to the accumulation of early endosomes containing plasma membrane proteins that enter cells independently of clathrin; failure to inactivate ARF6 blocks further trafficking of this membrane towards recycling or to other destinations52. Immediately upon platelet activation, ARF6•GTP levels fall, and this inactivation precedes, and is required for, the subsequent activation of Rac53. ARF6 is important for the disassembly of adherens junctions1 and, more recently, active ARF6 was shown to disrupt the formation of epithelial cysts54. The Slit2–ROBO4 signalling pathway is important for maintaining barrier function in the vascular network, and ROBO4 interacts with paxillin to recruit ARF GAP proteins, such as GIT1, to inactivate ARF6 (Ref. 55); this ARF6 inactivation suppresses protrusive activity of the endothelial cells and neovascularization. GIT2 and ARF6 inactivation are also important for maintaining the podosome, an actin-rich sealing zone in osteoclasts56. Finally, non-canonical ubiquitylation of ARF6, catalysed by FBX8 (an F-box and SEC7 domain-containing protein) seems to be another, unusual, way to turn off ARF6 (Ref. 57). FBX8 is diminished or lacking in several cancer cell lines, which is consistent with roles for ARF6 in cancer cell metastasis58.
Insights into ARL function. Similarly to ARF1, ARL1 and ARL2 arose early in evolution and share common effectors in plants, yeast and mammals. ARL1 recruits GRIP-domain golgins to the TGN2. It also mediates TGN-localization of ARF-interacting proteins (ARFAPTINs), which contain Bin–amphiphysin–Rvs (BAR) domains that induce the formation of tubules and vesicles at the TGN59. Whereas ARL1 functions in vesicle trafficking similarly to ARFs, ARL2 has a highly conserved function in regulating microtubule-based processes2. ARL3 is closely related to ARL2, but is found only in cells with cilia, where it regulates microtubule-based processes at the cilial basal body2,60 (Fig. 2b). ELMOD2 has been reported to be a GAP for ARL2, but also has activity against ARF1 and ARF6, which is surprising given that it has no homology to ARF GAPs61; the physiological relevance of this activity remains to be determined. Retinitis pigmentosa 2 (RP2; also known as XRP2) acts as a GAP for ARL3 during intraflagellar transport and ciliogenesis.
ARL3, ARL6 and ARL13 affect intraflagellar transport and ciliogenesis (Fig. 2b,c). Cilia are vital for cell signalling and differentiation, and their impaired formation is responsible for many genetic disorders62. Bardet–Biedl syndrome is a complex genetic disease that can be caused by mutation in any one of 14 genes associated with ciliogenesis. Transport of membrane proteins into the cilium is driven by a complex of proteins, called the BBSome. BBSome subunits have 'coat-like' attributes and similar structural folds to those found in COPI and adaptor protein complexes, suggesting that the BBSome can sort specific cargo for transport (Fig. 2c). ARL6 is a BBS subunit (BBS3) and is required in its GTP-bound form to recruit the BBSome onto the plasma membrane to drive cargo sorting into cilia63. Structural and biochemical analyses have shown that one of the mutations in ARL6 that causes Bardet–Biedl syndrome, T31R, leads to a non-functional ARL6 that cannot bind GTP64. This supports the idea that ARL6 recruits the BBSome complex to membranes for formation of BBSome-coated vesicles. ARL13 is mutated in patients with Joubert syndrome, which is a rare, complex cerebral disorder that is characterized by developmental delays and cognitive disability. It is also involved in intraflagellar transport (Fig. 2c) and, in C. elegans, ARL-13 associates with the doublet segment of the cilium and its loss results in shortened cilia65,66.
Retinitis pigmentosa is a retinal degeneration disease, and mutations in the RP2 gene are responsible for a large fraction of the most severe X-linked form. RP2 was identified as a GAP for ARL3, and mutations associated with retinitis pigmentosa compromise ARL3 GAP activity67. ARL3 localizes to the photoreceptor segment connecting to the cilium (Fig. 2b), and ARL3−/− mice have abnormal kidney and photoreceptor development, indicating the importance of this protein in primary cilia68. RP2 localizes to the basal body and centriole at the base of the photoreceptor cilium, but also to the adjacent Golgi and apical plasma membrane69. Furthermore, RP2 promotes vesicle trafficking from the Golgi to the base of the cilium in mammalian cells69, presumably acting together with ARF4, ASAP1 and FIP3. Intriguingly, D. melanogaster ARL3 (also called Dead end) regulates actin polymerization and vesicular trafficking to the plasma membrane, which are important for tracheal morphogenesis70. Hence, ARL3 appears to link microtubule-based processes and vesicular trafficking during development.
ARL8 might also coordinate microtubule and vesicular trafficking. ARL8 localizes to late endosomes and lysosomes (Fig. 3) in both humans and worms, and mediates transport of endocytic proteins between these two compartments71. ARL8 also facilitates the axonal transport of presynaptic cargo proteins in vesicles, preventing their premature aggregation9. Exactly how these two functions of ARL8 are related is not clear but they might both involve transport along microtubules2.
ARF GEFs in physiology and disease
A great deal of progress has been made in identifying ARF GEFs, and an unexpectedly broad range of roles has been revealed for these regulators, including both the coordination of membrane trafficking with lipid homeostasis and signalling at the plasma membrane (Table 2). Because GEFs ensure the precise temporal and spatial activation of ARFs, their own localization mechanisms are crucial for understanding their cellular roles. These mechanisms are turning out to be quite complex, even for the simplest of the ARF GEFs, the members of the cytohesin (also known as ARNO) family. Membrane trafficking is crucial to numerous developmental and physiological processes, and the specific functions of different ARF GEFs in these pathways and their links to disease are now being revealed.
Mechanisms of ARF GEF recruitment. There is particular interest in understanding how ARF GEFs are recruited to membranes to regulate ARF activation. BIG1 and BIG2 localize to the TGN and endosomes, where they have both distinct and overlapping functions72,73. By contrast, GBF1 localizes predominantly to the cis-Golgi74 (Fig. 2a), where it controls transport of membrane proteins through the secretory pathway75. The activity of phosphodiesterase 3A is important for recruitment of BIG1 and BIG2 to the trans-Golgi76. However, Rab1 (Ref. 77) and PtdIns4P generated by PtdIns4KIIIα78 are involved in recruitment of GBF1 to membranes. Other close connections between Golgi ARFs and PtdIns4P have emerged recently. In yeast there is an interesting synergy observed between the ARF1 GEF Gea2 and PtdIns4P produced by Pik1 (the yeast homologue of PtdIns4KIIIβ). Both are simultaneously required to activate the aminophospholipid translocase (flippase) Drs2 at the TGN during formation of AP1–clathrin vesicles79.
PtdIns4Ks are essential for viral replication, and notably produce the PtdIns4P-enriched membrane environment that recruits the enteroviral RNA polymerases80. GBF1 is required for the replication of numerous viruses, including enteroviruses, hepatitis C virus and coronaviruses81,82,83,84. In enteroviral systems, GBF1 and PtdIns4KIIIβ are recruited coordinately to membranes by the viral 3A protein to promote formation of functional viral replication complexes80 near ER exit sites (Fig. 2a).
Yel1 is an EFA6-like GEF for the ARF6 orthologue Arf3 in yeast, and localizes to the plasma membrane of the emerging bud85. Similarly to its mammalian orthologues, the PH domain of Yel1 is required for membrane targeting but, interestingly, multiple regions of the protein are important for precise spatial localization of this GEF85.
BRAG2, an ARF6 GEF, also has a PH domain that is critical for membrane targeting and in breast cancer cells is specifically recruited to the EGF receptor upon EGF stimulation, through direct interaction of its PH domain with the EGF receptor86. This interaction requires phosphorylation on specific Tyr residues and thus the recruitment of BRAG2 couples receptor activation to ARF6 activation86. BRAG2 is overexpressed in many breast cancer cell lines and depletion of BRAG2 by small interfering RNA blocks cell invasion in vitro and in animal tumour models86. These observations add to others that have implicated ARF6 and its activation in a number of models of cancer cell invasion and metastasis1,58.
Autoinhibition of cytohesin GEFs. At the cell periphery, the cytohesin GEFs function in plasma membrane–endosomal membrane trafficking pathways, and in signal transduction pathways that are important for cell proliferation, immune response and growth control87,88. Members of this GEF family can catalyse exchange on both ARF1 and ARF6 in vitro and in cells, although in vitro they are more efficient GEFs for ARF1 (Ref. 87). Recent insights have been gained into how cytohesin activation is spatially regulated, and how its autoinhibition is relieved (Fig. 4). In addition to phosphoinositide binding at the membrane, the PH domains of cytohesin family members interact with the GTP-bound forms of ARF6 (Ref. 89) and ARL4 (Refs 90, 91), leading to cytohesin recruitment and further activation of ARF6 or ARF1 at the membrane. A crystal structure of the SEC7 domain in tandem with the PH domain of cytohesin 3 (also known as GRP1) revealed that it adopts an autoinhibited conformation. The C-terminal helix that follows the PH domain and the linker between the SEC7 and PH domains block the catalytic site92. Interaction of the PH domain with ARF6•GTP and phosphoinositides (either PtdIns(4,5)P2 or PtdIns-3,4,5-trisphosphate (PtdIns(3,4,5)P3)), as well as the interaction of the polybasic C terminus of cytohesin with acidic phospholipids, all contribute to relieving this autoinhibition92 (Fig. 4). Reconstitution of the cytohesin-exchange assay on liposomes, in the presence of both activating ARF6•GTP and substrate ARF1, revealed that mutations in the PH domain of cytohesin that abolished interaction with ARF6•GTP were completely inactive93. Together, these studies demonstrate how precise spatial regulation of cytohesin activation is achieved. A specific phosphoinositide (PtdIns(4,5)P2 and/or PtdIns(3,4,5)P3), additional acidic phospholipids and an active ARF localized in the plasma membrane must all coincide to relieve autoinhibition, thus restricting the membrane domain at which these GEFs can become active.
Figure 4. The recruitment of an ARF GEF to the membrane is coupled to relief of autoinhibition.
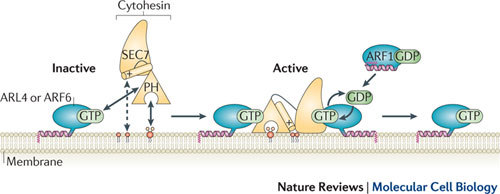
An active GTP-bound ADP-ribosylation factor (ARF) family member (either ARF-like 4 (ARL4) or ARF6), phosphoinositides (phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P2) or PtdIns-3,4,5-trisphosphate (PtdIns(3,4,5)P3)), and additional acidic phospholipids such as phosphatidylserine, are all required for membrane recruitment of the cytohesin (also known as ARNO) guanine nucleotide exchange factor (GEF), to convert it from its cytosolic inactive form to its fully active membrane-bound form. Before recruitment, the SEC7 catalytic GEF domain, the pleckstrin homology (PH) domain and the carboxy-terminal α-helix of cytohesin are in an autoinhibited conformation (left), with the C-terminal α-helix (charged residues within this are shown as '+') and linker situated between the catalytic SEC7 domain and the PH domain, which blocks the ARF-binding site. Upon binding of the PH domain to the GTP-bound GEF at the membrane, the catalytic site is released from autoinhibition (right). This can in turn drive further activation of ARF proteins, such as ARF1, at the membrane, and may form the basis of an ARF protein activation cascade. Figure is modified, with permission, from Ref. 93 © (2011) American Society for Biochemistry and Molecular Biology.
We do not know whether ARF6, ARF1 or both are the primary substrates for the cytohesins. However, ARF6•GTP is more efficient in relieving autoinhibition of cytohesins than ARF1•GTP, both in vitro and in cells89,92. The activation of cytohesins by a GTP-bound ARF family member raises the question of whether they can engage in a positive feedback loop, whereby the substrate itself can stimulate further exchange. Indeed, such a loop has been demonstrated in vitro for ARF1 (Ref. 93).
GEF-mediated cascades. There is also evidence that cytohesins might mediate a cascade of activation from ARF6 to ARF1. Cells expressing constitutively active ARF6Q67L have increased levels of ARF1•GTP89. ARF1 affects several processes at the plasma membrane, including recruitment of proteins to focal adhesions and during phagocytosis. In the forming phagocytic cup, ARF6•GTP is recruited earlier than ARF1•GTP, at a stage that requires rapid insertion of new membrane94. Hence, the ARF6–cytohesin–ARF1 cascade might ensure a high level of activated ARF protein here. ARF6 is less abundant than ARF1 in cells, and as both ARF1 and ARF6 can recruit effectors such as PtdIns4P5K and PLD, processes requiring acute activation of such effectors may rely on the more abundant ARF1 to provide an adequate supply. In support of this idea, both ARF1 and ARF6, through cytohesins, contribute to activation of PtdIns4P5K and PLD in the insulin signalling pathway95. In addition to ARF6–cytohesin–ARF1 or possible ARL4–cytohesin–ARF6 cascades, there is a conserved ARL cascade, in which yeast ARL3•GTP recruits ARL1 to TGN membranes2. In this case, it is not known whether an ARL GEF is involved. Hence, ARF family cascades could be common and could explain the Golgi ARFs that act in pairs.
ARF GEFs in scaffolding complexes. Use of the specific cytohesin inhibitor SecinH3 has revealed roles for this family of GEFs in the insulin and ERBB receptor Tyr kinase signalling pathways96,97,98. Cytohesins are positive activators of insulin signalling in both D. melanogaster and mammalian cells, and they are important for cell growth and for insulin sensitivity in human liver cells97,98. They regulate insulin signalling by binding CNK1, a scaffolding molecule that is important for Ras, phosphoinositide 3-kinase (PI3K) and AKT signalling95. CNK1 recruits cytohesins in an insulin-dependent manner to the plasma membrane, where they generate a PtdIns(4,5)P2-enriched microdomain that is essential for PI3K–AKT activation. Other scaffolding proteins interact with the coiled-coil domain of cytohesin; these proteins include Golgi reassembly-stacking protein (GRASP) and IPCEF (interactor protein for cytohesin exchange factors), which mediate the interaction of DOCK180 with cytohesin99. Interestingly, assembly of this scaffolding complex promotes Rac activation and cell migration, indicating that these scaffolds assemble a signalling complex that determines a specific downstream output upon ARF activation99. Cytohesins also affect integrin signalling in the immune system, and cytohesin 1 can activate β2 integrins in dendritic cells100, possibly through a scaffolding role of cytohesins.
GEFs in neuronal development and disease. Levels of ARF6 and the EFA6 and cytohesin family GEFs markedly increase in the mammalian brain after birth, suggesting important roles in postnatal nervous system development101. Experiments in isolated hippocampal neurons indicate that ARF6, EFA6 and the cytohesins might affect neurite and dendritic spine development102,103.
In humans, mutations in the ARF1 GEF BIG2 are linked to autosomal recessive periventricular heterotopia (ARPH), a disease in which the cerebral cortex is severely underdeveloped owing to failure of neurons in the lateral ventricular proliferative zone to migrate to the cortex104. This impaired migration arises from a defect in vesicular trafficking that alters the adhesive properties of these neurons105. Disease alleles include an early frameshift mutation that deletes most of the BIG2 protein104.
Members of the BRAG (or IQSEC) family of ARF GEFs are extremely abundant in neuronal postsynaptic densities, and can serve as GEFs for ARF6 (Ref. 87). BRAG1 (also known as IQSEC2) and BRAG2 are vital for neuronal development. BRAG1 is mutated in X-linked nonsyndromic intellectual disability (also referred to as mental retardation). Three point mutations isolated from patients map to the SEC7 domain and result in proteins that cannot activate ARF6 normally106. BRAG2 has been linked to alterations in synaptic content during long-term depression (LTD). Signalling through AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid)-type glutamate receptors facilitates LTD, and downregulation of activated AMPA receptors is normally regulated by AMPA receptor-mediated recruitment of BRAG2, which in turn activates ARF6 and endocytosis107. Thus, BRAG GEFs and ARF6 are vital for neuronal development and learning.
ARF-independent GEF functions. Cytohesin GEFs may affect signalling through epidermal growth factor (EGF) or ERBB receptor Tyr kinase receptors independently of their GEF activity. EGF receptors (EGFRs) undergo ligand-induced dimerization and subsequent trans-phosphorylation, mediated by conformational changes in their cytoplasmic portion. Cytohesins bind directly to these cytoplasmic domains and promote conformational changes that increase phosphorylation96. Furthermore, treatment of an EGF receptor-dependent lung cancer cell line with the cytohesin inhibitor SecinH3 reduced proliferation96. Surprisingly, this function of the cytohesins does not require their GEF activity. Similarly, in C. elegans, the GEF EFA-6 regulates microtubule dynamics at the cell cortex independently of its substrate ARF6 (Ref. 108). Furthermore, essential functions of GBF1 in poliovirus replication are independent of ARF1 activation109. The extent to which other ARF GEFs may have broader roles beyond ARF activation warrants further investigation. There are also suggestions that some multidomain ARF GAP proteins have functions that are independent of their GAP activity.
ARF GAPs as scaffolding effectors
All ARF GAPs contain the conserved zinc-finger ARF GAP catalytic domain in addition to other domains responsible for membrane recruitment, regulation of GAP activity and other scaffolding functions (Table 2). ARFGAP1, the first ARF GAP to be cloned110, is Golgi-localized and, together with ARFGAP2 and ARFGAP3, mediates most ARF-bound GTP hydrolysis at the Golgi. The complex, multidomain structure of the other ARF GAP families has stimulated much research. Here, we highlight a few examples of how these multidomain ARF GAPs, by recognizing the GTP-bound form of their substrate ARF, act as downstream effectors in addition to signal terminators. Information about other ARF GAPs can be found in an excellent review article111.
The ASAP proteins are the prototypical multidomain GAPs that interact with many signalling molecules, including SRC and focal adhesion kinase111 (Table 2). ASAP1 resides in focal adhesions but, in response to SRC activation, it facilitates formation of podosomes112, which are discrete actin-based structures that are formed at the cell substratum to degrade matrix. The crystal structure of ARF6 in complex with the catalytic domain of ASAP3 revealed that a catalytic Arg-finger of ASAP3 is responsible for GTP hydrolysis113, similarly to many other GAPs, a finding that is consistent with an earlier structure of the GAP domain of ASAP2 (Ref. 114). There is also some evidence that calcium might bind to the complex and regulate GAP activity113, although this needs to be confirmed with full-length ASAP3 and ARF6 and in cells. The ASAPs all have N-terminal BAR domains that can induce membrane curvature and tubule formation in transfected cells and in cell-free systems. The BAR domain in ASAP1 negatively regulates its GAP activity towards ARF1 (Ref. 115), and binding of the Rab11 effector FIP3 to the BAR domain of ASAP1 stimulates its GAP activity116. As mentioned earlier, ASAP1 also promotes ciliary targeting together with ARF4 and FIP3 (Ref. 20) (Fig. 2b). ASAP1 is upregulated in breast, pancreatic and colorectal cancer58. CBL-interacting protein 85 (CIN85; also known as SH3KBP1) binds to ASAP1, recruiting the E3 ubiquitin ligase CBL, to trigger the monoubiquitylation of ASAP1; this modification is important for invasion of breast cancer cells117 but the role for ubiquitylation of ASAP in cell invasion is not known. One caveat to observations made when ASAP is expressed in cells is that a study designed to systematically look at ARF GAP function and ARF specificity failed to detect an effect of ASAP1 expression on either ARF1•GTP or ARF6•GTP levels in cells118. This raises the possibility that the GAP activity of ASAP1 might not always be critical for some of ASAP1's specific functions.
The ARF GAP GIT1, originally identified as a GPCR kinase-interacting protein, can coordinate signalling by acting as a scaffold. GIT1 and its substrate ARF6 affect ligand-stimulated endocytosis of several GPCRs through either clathrin-dependent or clathrin-independent endocytic pathways119. Among the proteins interacting with GIT1 are the CDC42 and Rac GEF PIX, focal adhesion kinase and paxillin. GIT1, similarly to ASAP1, is sometimes observed in focal adhesions and its influence on the activation of CDC42 and Rac suggests that ARF inactivation and Rac activation are coordinated (Fig. 3). D. melanogaster GIT is required for muscle morphogenesis120 and the GIT1-knockout mouse is defective in fear learning121 and dendritic spine formation122. Rac3 interacts with GIT1, disrupting GIT1 binding to paxillin; this in turn stimulates GIT1 GAP activity, presumably towards ARF6 (Ref. 123), and inhibits cell spreading and neuritogenesis. In endothelial cells, ROBO4 interacts with paxillin, which recruits GIT1 to inactivate ARF6, and this leads to vascular stability, blocking cellular protrusions and neovascular leak55. Thus, these examples provide insights into how modular ARF GAPs promote spatially and temporally restricted assembly of signalling complexes, and allow a precise physiological output in response to a signal.
Intracellular pathogens can use a fascinating GAP-blocking mechanism to rewire the host cell's signalling network for their own purposes. Enterohaemorrhagic Escherichia coli produce the EspG protein, which binds to GTP-bound ARF1 and ARF6, blocking their access to GAPs and disrupting the function of both early Golgi and recycling endosomes124. Moreover, EspG simultaneously binds to p21-activated kinase (PAK), an effector of a distinct G protein family member, CDC42, and promotes PAK localization at Golgi membranes rather than at the plasma membrane. This raises the possibility that EspG assembles its own signalling complex on intracellular membranes to subvert membrane trafficking and polarity processes in host cells.
Conclusions & perspectives
ARF activity is regulated in a spatiotemporal manner by the GEFs and GAPs, underlining the importance of precise localization of these regulators. In the case of cytohesins, such specificity can be achieved through a coincidence-detection mechanism, requiring both an activating ARF or ARL protein and a specific lipid composition. This example also reveals the existence of ARF family activation cascades and how relief of autoinhibition can be coupled to precise spatial cues. It will be interesting to see how widespread these mechanisms are among ARF family members. ARF cascades, similarly to those demonstrated for Rab G proteins, could transform one membrane domain into another during highly dynamic membrane trafficking. These transformations involve coordinated changes in the lipid and protein composition of each membrane domain, a specialty of many ARF family members, which recruit both lipid-modifying enzymes and protein effectors such as coats and tethers. The signature feature of ARF family proteins, their N-terminal membrane-binding amphipathic helix, ensures that they are closely associated with the lipid bilayer in their GTP-bound form. Future studies on how ARF family proteins function will therefore require in vitro reconstitution on model membranes. There appears to be a particularly important link between ARF1 function and PtdIns4P, a lipid that has a central role in the function of the Golgi, which parallels the coordination of membrane trafficking and PtdIns(4,5)P2 signalling by ARF6 at the plasma membrane.
The GAPs and GEFs for the ARF family proteins are multidomain proteins that can assemble signalling complexes and so place the ARFs and ARLs into larger networks. These networks include cytoskeleton regulators, and it appears that some ARL proteins (ARL2, for example) have evolved exclusively to regulate the cytoskeleton. The role of ARF6 in networks linking membrane trafficking to the actin cytoskeleton also involves interaction of ARF6 with GEFs and GAPs of the Rac and Rho small G proteins, actin cytoskeleton regulators. Another emerging concept is that some ARF family members remain membrane-bound in their GDP-bound form so that they can interact with signalling complexes and promote alternative signalling pathways. Ultimately, these ARF family signalling networks will need to be studied through systems level analysis.
So far, no GEFs and only two GAPs that are specific for an ARL have been identified. Several ARL proteins affect ciliogenesis and, in some cases, ciliopathies; other ARLs function in neurons and have been associated with neurodegenerative disorders. Hence, increased understanding of ARLs and their regulators should inform both fundamental questions in cell biology and disease mechanisms.
Finally, the use of model organisms to complement studies in mammalian cells has already provided valuable insights into the physiological roles of ARF family proteins. This approach holds great promise for uncovering the unknown functions of most ARLs, as well as defining the full range of activities of all ARF and ARL proteins.
Box 1 | ARF function in plants and protists.
Plants have numerous ADP-ribosylation factors (ARFs) that are homologous to human ARF1 (Ref. 125) and were originally thought to lack Class III ARF6-like proteins. However, in Arabidopsis thaliana, ARFB (also called ARFB1A) localizes to the plasma membrane and lacks the Golgi-targeting motif (MXXE) that is found in other ARF1 homologues in plants126 and in mammals17. Nevertheless, only the GBF and BIG subfamilies of ARF guanine nucleotide exchange factors (GEFs) seem to be present in plants, and these function in both endocytic and Golgi trafficking pathways127. A. thaliana GNOM (also known as EMB30) is a homologue of mammalian GBF1 but acts at endosomes and the plasma membrane during the polar transport of the plant hormone auxin during development127,128. Another GBF-like protein in A. thaliana, GNOM-like 1 (GNL1), functions at the Golgi similarly to mammalian GBF1, but is also involved in endosomal trafficking129. BIG5 (also known as BEN1 and ATMIN7) was identified in a screen for A. thaliana mutants defective for internalization of the PIN auxin transporter from the plasma membrane. This ARF GEF is most closely related to BIG1 and BIG2 in mammalian cells, localizes predominantly to the trans-Golgi network (TGN) and early endosomes, and is involved in early endosomal trafficking130. Interestingly, BIG5 is targeted for degradation by a plant bacterial pathogen, Pseudomonas syringae, to protect the latter from host defence systems at the cell wall131.
A. thaliana ARF GAPs include four members of a family of mammalian ACAP homologues that are known as VASCULAR NETWORK 3 (VAN3)-like after the first member to be characterized125. VAN3 (also known as SCARFACE and AGD3) regulates formation of plant vascular networks132,133. In addition to its roles on endosomes, VAN3 cooperates with GNOM during clathrin-mediated endocytosis of the PIN auxin transporter128. Another ARF GAP in A. thaliana, ARF GAP DOMAIN 5 (AGD5; also known as NEVERSHED), is a homologue of yeast ARF GAP effector 2 (Age2) and mammalian SMAP family ARF GAPs that localizes to the TGN134. AGD5 is required for floral organ cell separation125 and regulates membrane trafficking though TGN–early endosomal compartments to trigger organ abscission135.
Interestingly, the protozoan parasite Trypanosoma brucei expresses a single ARF protein that has characteristics of both ARF1 and ARF6. T. brucei ARF1 is a basic protein with a calculated isoelectric point (pI) value of 9.1, which is similar to that of human ARF6, but T. brucei ARF1 contains the Golgi-targeting motif MXXE136 that is found in human ARF1 and ARF3 (Ref. 17). Depletion of T. brucei ARF1 by small interfering RNA causes a major decrease in endocytosis and the formation of intracellular flagella, but the Golgi remains intact136. Trypanosomes also express an ARF-like 2 (ARL2) homologue, which is involved in microtubule biogenesis and cytokinesis137, and an ARL1 homologue, which is important for Golgi structure and exocytosis of glycosyl phosphatidylinositol (GPI)-anchored proteins138. ARF and ARL proteins in trypanosomes are myristoylated, a modification that is required for their activity. Trypanosomes cause African sleeping sickness, a disease with no successful therapy. A selective inhibitor of trypanosomal N-myristoyl transferase has been shown to be effective in blocking trypanosome viability in a mouse model of this disease139.
Acknowledgements
We apologize to authors whose work we could not cite owing to space limitations. We thank C. Eyster, L. Maldonado-Baez, J. Ménétrey and C. Le Clainche for critical reading of the manuscript. Work in our laboratories is supported by the Division of Intramural Research in the National Heart, Lung, and Blood Institute, US National Institutes of Health (J.G.D.) and grants from the Agence Nationale de la Recherche and the Centre National de la Recherche Scientifique, France (C.L.J.).
Glossary
- Guanine nucleotide exchange factors
(GEFs). Proteins that promote the release of GDP from guanine-nucleotide-binding (G) proteins, which allows GTP to bind. These proteins often stabilize the nucleotide-free form and then are released upon GTP binding.
- GTPase-activating proteins
(GAPs). Proteins that promote GTP hydrolysis on GTP-bound guanine-nucleotide-binding (G) proteins. For ADP-ribosylation factor (ARF) proteins, GAPs are critical, as ARFs have negligible intrinsic GTPase activity. The catalytic regions of GAPs often include an Arg-finger motif that inserts into the GTP-binding pocket to stimulate hydrolysis of GTP.
- Myristoylation
A lipid modification, occurring co- or post-translationally, in which a myristoyl moiety is attached to a Gly residue at the second position from the amino terminus, after cleavage of the N-terminal Met residue.
- GDP dissociation inhibitor
(GDI). A protein that binds specifically to the GDP-bound form of a GTP-binding protein, preventing guanine nucleotide exchange.
- Cilium
A slender extension on the cell surface. A non-motile, primary cilium is present on nearly all epithelial cells in the body and serves as a sensory organ that is important for regulating cell differentiation and division.
- Lipid droplets
Lipid storage organelles that are surrounded by a phospholipid monolayer.
- New end take-off
A switch in cellular growth of fission yeast, from monopolar extension to bipolar extension.
- Adherens junctions
Cellular adhesions that connect epithelial cells to form a polarized epithelium. Made up of homotypic cadherin interactions and associated intracellular proteins.
- ROBO4
(Roundabout homologue 4). Acts as a receptor for Slit2 protein and regulates vascular integrity.
- Podosome
An adhesive, ring-like, actin-rich structure that is formed on the ventral surface of cells.
- BBSome
A complex of proteins that facilitates membrane traffic into the cilium. Mutant forms of several BBS components have been identified as causative agents for various ciliopathies.
- Phagocytosis
A cellular endocytic process for engulfing large particles, such as bacteria, and bringing them inside the cell.
- Long-term depression
(LTD). A reduction in the efficacy or strength of neuronal synapses that is linked to learning and memory formation.
Biographies
Julie G. Donaldson obtained her Ph.D. in cell biology from the University of Maryland, College Park, USA, and followed this with postdoctoral studies in the laboratory of Richard Klausner at the National Institute of Child Health and Human Development, Rockville, Maryland, USA. She is currently a senior investigator at the National Heart, Lung and Blood Institute at the US National Institutes of Health in Bethesda, Maryland, USA, where her laboratory studies clathrin-independent forms of endocytosis and the role of ADP-ribosylation factor (ARF) family G proteins in the regulation of secretory and endocytic membrane traffic.
Catherine L. Jackson is currently a Centre National de la Recherche Scientifique (CNRS) research director at the Institut Jacques Monod, Paris, France. She carried out doctoral studies in genetics at the University of Washington, Seattle, USA, with Leland H. Hartwell, and postdoctoral studies in the department of André Sentenac at the Commissariat à l'énergie atomique et aux énergies alternatives (CEA), Saclay, France, in collaboration with the laboratory of Marc Chabre, Institut de Pharmacologie Moléculaire et Cellulaire (IPMC), Nice, France. Her laboratory studies the molecular mechanisms regulating membrane trafficking and organelle structure in yeast and mammalian cells.
Related links
FURTHER INFORMATION
Competing interests
The authors declare no competing financial interests.
Contributor Information
Julie G. Donaldson, Email: jdonalds@helix.nih.gov
Catherine L. Jackson, Email: jackson@ijm.univ-paris-diderot.fr
References
- 1.D'Souza-Schorey C, Chavrier P. ARF proteins: roles in membrane traffic and beyond. Nature Rev. Mol. Cell Biol. 2006;7:347–358. doi: 10.1038/nrm1910. [DOI] [PubMed] [Google Scholar]
- 2.Gillingham AK, Munro S. The small G proteins of the Arf family and their regulators. Annu. Rev. Cell Dev. Biol. 2007;23:579–611. doi: 10.1146/annurev.cellbio.23.090506.123209. [DOI] [PubMed] [Google Scholar]
- 3.Antonny B, Beraud-Dufour S, Chardin P, Chabre M. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 1997;36:4675–4684. doi: 10.1021/bi962252b. [DOI] [PubMed] [Google Scholar]
- 4.Chavrier P, Menetrey J. Toward a structural understanding of arf family:effector specificity. Structure. 2010;18:1552–1558. doi: 10.1016/j.str.2010.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Hofmann I, Munro S. An N-terminally acetylated Arf-like GTPase is localised to lysosomes and affects their motility. J. Cell Sci. 2006;119:1494–1503. doi: 10.1242/jcs.02958. [DOI] [PubMed] [Google Scholar]
- 6.Lee MC, et al. Sar1p N-terminal helix initiates membrane curvature and completes the fission of a COPII vesicle. Cell. 2005;122:605–617. doi: 10.1016/j.cell.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 7.Duijsings D, et al. Differential membrane association properties and regulation of class I and class II Arfs. Traffic. 2009;10:316–323. doi: 10.1111/j.1600-0854.2008.00868.x. [DOI] [PubMed] [Google Scholar]
- 8.Chun J, Shapovalova Z, Dejgaard SY, Presley JF, Melancon P. Characterization of class I and II ADP-ribosylation factors (Arfs) in live cells: GDP-bound class II Arfs associate with the ER–Golgi intermediate compartment independently of GBF1. Mol. Biol. Cell. 2008;19:3488–3500. doi: 10.1091/mbc.E08-04-0373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klassen MP, et al. An Arf-like small G protein, ARL-8, promotes the axonal transport of presynaptic cargoes by suppressing vesicle aggregation. Neuron. 2010;66:710–723. doi: 10.1016/j.neuron.2010.04.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen KY, et al. Syt1p promotes activation of Arl1p at the late Golgi to recruit Imh1p. J. Cell Sci. 2010;123:3478–3489. doi: 10.1242/jcs.074237. [DOI] [PubMed] [Google Scholar]
- 11.Beck R, Ravet M, Wieland FT, Cassel D. The COPI system: molecular mechanisms and function. FEBS Lett. 2009;583:2701–2709. doi: 10.1016/j.febslet.2009.07.032. [DOI] [PubMed] [Google Scholar]
- 12.Bonifacino JS, Lippincott-Schwartz J. Coat proteins: shaping membrane transport. Nature Rev. Mol. Cell Biol. 2003;4:409–414. doi: 10.1038/nrm1099. [DOI] [PubMed] [Google Scholar]
- 13.Hong JX, et al. Phospholipid- and GTP-dependent activation of cholera toxin and phospholipase D by human ADP-ribosylation factor-like protein 1 (HARL1) J. Biol. Chem. 1998;273:15872–15876. doi: 10.1074/jbc.273.25.15872. [DOI] [PubMed] [Google Scholar]
- 14.De Matteis MA, Godi A. Protein-lipid interactions in membrane trafficking at the Golgi complex. Biochim. Biophys. Acta. 2004;1666:264–274. doi: 10.1016/j.bbamem.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 15.Volpicelli-Daley LA, Li Y, Zhang CJ, Kahn RA. Isoform-selective effects of the depletion of ADP-ribosylation factors 1–5 on membrane traffic. Mol. Biol. Cell. 2005;16:4495–4508. doi: 10.1091/mbc.E04-12-1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ben-Tekaya H, Kahn RA, Hauri HP. ADP ribosylation factors 1 and 4 and group VIA phospholipase A regulate morphology and intraorganellar traffic in the endoplasmic reticulum–Golgi intermediate compartment. Mol. Biol. Cell. 2010;21:4130–4140. doi: 10.1091/mbc.E10-01-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Honda A, Al-Awar OS, Hay JC, Donaldson JG. Targeting of Arf-1 to the early Golgi by membrin, an ER–Golgi SNARE. J. Cell Biol. 2005;168:1039–1051. doi: 10.1083/jcb.200409138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manolea F, et al. Arf3 is activated uniquely at the trans-Golgi network by brefeldin A-inhibited guanine nucleotide exchange factors. Mol. Biol. Cell. 2010;21:1836–1849. doi: 10.1091/mbc.E10-01-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deretic D, et al. Rhodopsin C terminus, the site of mutations causing retinal disease, regulates trafficking by binding to ADP-ribosylation factor 4 (ARF4) Proc. Natl Acad. Sci. USA. 2005;102:3301–3306. doi: 10.1073/pnas.0500095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mazelova J, et al. Ciliary targeting motif VxPx directs assembly of a trafficking module through Arf4. EMBO J. 2009;28:183–192. doi: 10.1038/emboj.2008.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sadakata T, et al. Interaction of calcium-dependent activator protein for secretion 1 (CAPS1) with the class II ADP-ribosylation factor small GTPases is required for dense-core vesicle trafficking in the trans-Golgi network. J. Biol. Chem. 2010;285:38710–38719. doi: 10.1074/jbc.M110.137414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D'Angelo G, et al. Glycosphingolipid synthesis requires FAPP2 transfer of glucosylceramide. Nature. 2007;449:62–67. doi: 10.1038/nature06097. [DOI] [PubMed] [Google Scholar]
- 23.Guo Y, et al. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453:657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bartz R, et al. Dynamic activity of lipid droplets: protein phosphorylation and GTP-mediated protein translocation. J. Proteome Res. 2007;6:3256–3265. doi: 10.1021/pr070158j. [DOI] [PubMed] [Google Scholar]
- 25.Soni KG, et al. Coatomer-dependent protein delivery to lipid droplets. J. Cell Sci. 2009;122:1834–1841. doi: 10.1242/jcs.045849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beller M, et al. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 2008;6:e292. doi: 10.1371/journal.pbio.0060292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hommel A, et al. The ARF-like GTPase ARFRP1 is essential for lipid droplet growth and is involved in the regulation of lipolysis. Mol. Cell. Biol. 2010;30:1231–1242. doi: 10.1128/MCB.01269-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kumari S, Mayor S. ARF1 is directly involved in dynamin-independent endocytosis. Nature Cell Biol. 2008;10:30–41. doi: 10.1038/ncb1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gupta GD, et al. Analysis of endocytic pathways in Drosophila cells reveals a conserved role for GBF1 in internalization via GEECs. PLoS ONE. 2009;4:e6768. doi: 10.1371/journal.pone.0006768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki T, et al. Crucial role of the small GTPase ARF6 in hepatic cord formation during liver development. Mol. Cell. Biol. 2006;26:6149–6156. doi: 10.1128/MCB.00298-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Paleotti O, et al. The small G-protein Arf6GTP recruits the AP-2 adaptor complex to membranes. J. Biol. Chem. 2005;280:21661–21666. doi: 10.1074/jbc.M503099200. [DOI] [PubMed] [Google Scholar]
- 32.Poupart ME, Fessart D, Cotton M, Laporte SA, Claing A. ARF6 regulates angiotensin II type 1 receptor endocytosis by controlling the recruitment of AP-2 and clathrin. Cell Signal. 2007;19:2370–2378. doi: 10.1016/j.cellsig.2007.07.015. [DOI] [PubMed] [Google Scholar]
- 33.Montagnac G, et al. Decoupling of activation and effector binding underlies ARF6 priming of fast endocytic recycling. Curr. Biol. 2011;21:574–579. doi: 10.1016/j.cub.2011.02.034. [DOI] [PubMed] [Google Scholar]
- 34.Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nature Rev. Mol. Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balasubramanian N, Scott DW, Castle JD, Casanova JE, Schwartz MA. Arf6 and microtubules in adhesion-dependent trafficking of lipid rafts. Nature Cell Biol. 2007;9:1381–1391. doi: 10.1038/ncb1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Osmani N, Peglion F, Chavrier P, Etienne-Manneville S. Cdc42 localization and cell polarity depend on membrane traffic. J. Cell Biol. 2010;191:1261–1269. doi: 10.1083/jcb.201003091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smaczynska-de Rooij II, Costa R, Ayscough KR. Yeast Arf3p modulates plasma membrane PtdIns(4,5)P2 levels to facilitate endocytosis. Traffic. 2008;9:559–573. doi: 10.1111/j.1600-0854.2008.00708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang CF, Liu YW, Tung L, Lin CH, Lee FJ. Role for Arf3p in development of polarity, but not endocytosis, in Saccharomyces cerevisiae. Mol. Biol. Cell. 2003;14:3834–3847. doi: 10.1091/mbc.E03-01-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fujita A. ADP-ribosylation factor Arf6p may function as a molecular switch of new end take off in fission yeast. Biochem. Biophys. Res. Commun. 2008;366:193–198. doi: 10.1016/j.bbrc.2007.11.117. [DOI] [PubMed] [Google Scholar]
- 40.Lee SC, Schmidtke SN, Dangott LJ, Shaw BD. Aspergillus nidulans ArfB plays a role in endocytosis and polarized growth. Eukaryot. Cell. 2008;7:1278–1288. doi: 10.1128/EC.00039-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dyer N, et al. Spermatocyte cytokinesis requires rapid membrane addition mediated by ARF6 on central spindle recycling endosomes. Development. 2007;134:4437–4447. doi: 10.1242/dev.010983. [DOI] [PubMed] [Google Scholar]
- 42.Montagnac G, et al. ARF6 interacts with JIP4 to control a motor switch mechanism regulating endosome traffic in cytokinesis. Curr. Biol. 2009;19:184–195. doi: 10.1016/j.cub.2008.12.043. [DOI] [PubMed] [Google Scholar]
- 43.Isabet T, et al. The structural basis of Arf effector specificity: the crystal structure of ARF6 in a complex with JIP4. EMBO J. 2009;28:2835–2845. doi: 10.1038/emboj.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ikenouchi J, Umeda M. FRMD4A regulates epithelial polarity by connecting Arf6 activation with the PAR complex. Proc. Natl Acad. Sci. USA. 2010;107:748–753. doi: 10.1073/pnas.0908423107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hiroi T, Someya A, Thompson W, Moss J, Vaughan M. GEP100/BRAG2: activator of ADP-ribosylation factor 6 for regulation of cell adhesion and actin cytoskeleton via E-cadherin and α-catenin. Proc. Natl Acad. Sci. USA. 2006;103:10672–10677. doi: 10.1073/pnas.0604091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luton F, et al. EFA6, exchange factor for ARF6, regulates the actin cytoskeleton and associated tight junction in response to E-cadherin engagement. Mol. Biol. Cell. 2004;15:1134–1145. doi: 10.1091/mbc.E03-10-0751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tushir JS, D'Souza-Schorey C. ARF6-dependent activation of ERK and Rac1 modulates epithelial tubule development. EMBO J. 2007;26:1806–1819. doi: 10.1038/sj.emboj.7601644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haas AK, et al. Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J. Cell Sci. 2007;120:2997–3010. doi: 10.1242/jcs.014225. [DOI] [PubMed] [Google Scholar]
- 49.Falace A, et al. TBC1D24, an ARF6-interacting protein, is mutated in familial infantile myoclonic epilepsy. Am. J. Hum. Genet. 2010;87:365–370. doi: 10.1016/j.ajhg.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Martinu L, et al. The TBC (Tre-2/Bub2/Cdc16) domain protein TRE17 regulates plasma membrane-endosomal trafficking through activation of Arf6. Mol. Cell. Biol. 2004;24:9752–9762. doi: 10.1128/MCB.24.22.9752-9762.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Koo TH, Eipper BA, Donaldson JG. Arf6 recruits the Rac GEF Kalirin to the plasma membrane facilitating Rac activation. BMC Cell Biol. 2007;8:29. doi: 10.1186/1471-2121-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eyster CA, et al. Discovery of new cargo proteins that enter cells through clathrin-independent endocytosis. Traffic. 2009;10:590–599. doi: 10.1111/j.1600-0854.2009.00894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Choi W, Karim ZA, Whiteheart SW. Arf6 plays an early role in platelet activation by collagen and convulxin. Blood. 2006;107:3145–3152. doi: 10.1182/blood-2005-09-3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tushir JS, et al. Unregulated ARF6 activation in epithelial cysts generates hyperactive signaling endosomes and disrupts morphogenesis. Mol. Biol. Cell. 2010;21:2355–2366. doi: 10.1091/mbc.E09-09-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jones CA, et al. Slit2–Robo4 signalling promotes vascular stability by blocking Arf6 activity. Nature Cell Biol. 2009;11:1325–1331. doi: 10.1038/ncb1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heckel T, et al. Src-dependent repression of ARF6 is required to maintain podosome-rich sealing zones in bone-digesting osteoclasts. Proc. Natl Acad. Sci. USA. 2009;106:1451–1456. doi: 10.1073/pnas.0804464106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yano H, et al. Fbx8 makes Arf6 refractory to function via ubiquitination. Mol. Biol. Cell. 2008;19:822–832. doi: 10.1091/mbc.E07-08-0763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sabe H, et al. The EGFR-GEP100-Arf6-AMAP1 signaling pathway specific to breast cancer invasion and metastasis. Traffic. 2009;10:982–993. doi: 10.1111/j.1600-0854.2009.00917.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Man Z, et al. Arfaptins are localized to the trans-Golgi by interaction with Arl1, but not Arfs. J. Biol. Chem. 2011;286:11569–11578. doi: 10.1074/jbc.M110.201442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhou C, Cunningham L, Marcus AI, Li Y, Kahn RA. Arl2 and Arl3 regulate different microtubule-dependent processes. Mol. Biol. Cell. 2006;17:2476–2487. doi: 10.1091/mbc.E05-10-0929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bowzard JB, Cheng D, Peng J, Kahn RA. ELMOD2 is an Arl2 GTPase-activating protein that also acts on Arfs. J. Biol. Chem. 2007;282:17568–17580. doi: 10.1074/jbc.M701347200. [DOI] [PubMed] [Google Scholar]
- 62.Nachury MV, Seeley ES, Jin H. Trafficking to the ciliary membrane: how to get across the periciliary diffusion barrier? Annu. Rev. Cell Dev. Biol. 2010;26:59–87. doi: 10.1146/annurev.cellbio.042308.113337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin H, et al. The conserved Bardet-Biedl syndrome proteins assemble a coat that traffics membrane proteins to cilia. Cell. 2010;141:1208–1219. doi: 10.1016/j.cell.2010.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wiens CJ, et al. Bardet-Biedl syndrome-associated small GTPase ARL6 (BBS3) functions at or near the ciliary gate and modulates Wnt signaling. J. Biol. Chem. 2010;285:16218–16230. doi: 10.1074/jbc.M109.070953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Li Y, Wei Q, Zhang Y, Ling K, Hu J. The small GTPases ARL-13 and ARL-3 coordinate intraflagellar transport and ciliogenesis. J. Cell Biol. 2010;189:1039–1051. doi: 10.1083/jcb.200912001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cevik S, et al. Joubert syndrome Arl13b functions at ciliary membranes and stabilizes protein transport in Caenorhabditis elegans. J. Cell Biol. 2010;188:953–969. doi: 10.1083/jcb.200908133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Veltel S, Gasper R, Eisenacher E, Wittinghofer A. The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3. Nature Struct. Mol. Biol. 2008;15:373–380. doi: 10.1038/nsmb.1396. [DOI] [PubMed] [Google Scholar]
- 68.Schrick JJ, Vogel P, Abuin A, Hampton B, Rice DS. ADP-ribosylation factor-like 3 is involved in kidney and photoreceptor development. Am. J. Pathol. 2006;168:1288–1298. doi: 10.2353/ajpath.2006.050941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Evans RJ, et al. The retinitis pigmentosa protein RP2 links pericentriolar vesicle transport between the Golgi and the primary cilium. Hum. Mol. Genet. 2010;19:1358–1367. doi: 10.1093/hmg/ddq012. [DOI] [PubMed] [Google Scholar]
- 70.Jiang L, Rogers SL, Crews ST. The Drosophila Dead end Arf-like3 GTPase controls vesicle trafficking during tracheal fusion cell morphogenesis. Dev. Biol. 2007;311:487–499. doi: 10.1016/j.ydbio.2007.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nakae I, et al. The arf-like GTPase Arl8 mediates delivery of endocytosed macromolecules to lysosomes in Caenorhabditis elegans. Mol. Biol. Cell. 2010;21:2434–2442. doi: 10.1091/mbc.E09-12-1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ishizaki R, Shin HW, Mitsuhashi H, Nakayama K. Redundant roles of BIG2 and BIG1, guanine-nucleotide exchange factors for ADP-ribosylation factors in membrane traffic between the trans-Golgi network and endosomes. Mol. Biol. Cell. 2008;19:2650–2660. doi: 10.1091/mbc.E07-10-1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Boal F, Stephens DJ. Specific functions of BIG1 and BIG2 in endomembrane organization. PLoS ONE. 2010;5:e9898. doi: 10.1371/journal.pone.0009898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao X, Lasell TK, Melancon P. Localization of large ADP-ribosylation factor-guanine nucleotide exchange factors to different Golgi compartments: evidence for distinct functions in protein traffic. Mol. Biol. Cell. 2002;13:119–133. doi: 10.1091/mbc.01-08-0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Szul T, et al. Dissecting the role of the ARF guanine nucleotide exchange factor GBF1 in Golgi biogenesis and protein trafficking. J. Cell Sci. 2007;120:3929–3940. doi: 10.1242/jcs.010769. [DOI] [PubMed] [Google Scholar]
- 76.Puxeddu E, et al. Interaction of phosphodiesterase 3A with brefeldin A-inhibited guanine nucleotide-exchange proteins BIG1 and BIG2 and effect on ARF1 activity. Proc. Natl Acad. Sci. USA. 2009;106:6158–6163. doi: 10.1073/pnas.0901558106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Monetta P, Slavin I, Romero N, Alvarez C. Rab1b interacts with GBF1 and modulates both ARF1 dynamics and COPI association. Mol. Biol. Cell. 2007;18:2400–2410. doi: 10.1091/mbc.E06-11-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dumaresq-Doiron K, Savard MF, Akam S, Costantino S, Lefrancois S. The phosphatidylinositol 4-kinase PI4KIIIα is required for the recruitment of GBF1 to Golgi membranes. J. Cell Sci. 2010;123:2273–2280. doi: 10.1242/jcs.055798. [DOI] [PubMed] [Google Scholar]
- 79.Natarajan P, et al. Regulation of a Golgi flippase by phosphoinositides and an ARFGEF. Nature Cell Biol. 2009;11:1421–1426. doi: 10.1038/ncb1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hsu NY, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Belov GA, Feng Q, Nikovics K, Jackson CL, Ehrenfeld E. A critical role of a cellular membrane traffic protein in poliovirus RNA replication. PLoS Pathog. 2008;4:e1000216. doi: 10.1371/journal.ppat.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Verheije MH, et al. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 2008;4:e1000088. doi: 10.1371/journal.ppat.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lanke KH, et al. GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication. J. Virol. 2009;83:11940–11949. doi: 10.1128/JVI.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Goueslain L, et al. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J. Virol. 2010;84:773–787. doi: 10.1128/JVI.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gillingham AK, Munro S. Identification of a guanine nucleotide exchange factor for Arf3, the yeast orthologue of mammalian Arf6. PLoS ONE. 2007;2:e842. doi: 10.1371/journal.pone.0000842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Morishige M, et al. GEP100 links epidermal growth factor receptor signalling to Arf6 activation to induce breast cancer invasion. Nature Cell Biol. 2008;10:85–92. doi: 10.1038/ncb1672. [DOI] [PubMed] [Google Scholar]
- 87.Casanova JE. Regulation of ARF activation: the Sec7 family of guanine nucleotide exchange factors. Traffic. 2007;8:1476–1485. doi: 10.1111/j.1600-0854.2007.00634.x. [DOI] [PubMed] [Google Scholar]
- 88.Kolanus W. Guanine nucleotide exchange factors of the cytohesin family and their roles in signal transduction. Immunol. Rev. 2007;218:102–113. doi: 10.1111/j.1600-065X.2007.00542.x. [DOI] [PubMed] [Google Scholar]
- 89.Cohen LA, et al. Active Arf6 recruits ARNO/cytohesin GEFs to the PM by binding their PH domains. Mol. Biol. Cell. 2007;18:2244–2253. doi: 10.1091/mbc.E06-11-0998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hofmann I, Thompson A, Sanderson CM, Munro S. The Arl4 family of small G proteins can recruit the cytohesin Arf6 exchange factors to the plasma membrane. Curr. Biol. 2007;17:711–716. doi: 10.1016/j.cub.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 91.Li CC, et al. ARL4D recruits cytohesin-2/ARNO to modulate actin remodeling. Mol. Biol. Cell. 2007;18:4420–4437. doi: 10.1091/mbc.E07-02-0149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.DiNitto JP, et al. Structural basis and mechanism of autoregulation in 3-phosphoinositide-dependent Grp1 family Arf GTPase exchange factors. Mol. Cell. 2007;28:569–583. doi: 10.1016/j.molcel.2007.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Stalder D, et al. Kinetic studies of the Arf activator Arno on model membranes in the presence of Arf effectors suggest control by a positive feedback loop. J. Biol. Chem. 2011;286:3873–3883. doi: 10.1074/jbc.M110.145532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Beemiller P, Hoppe AD, Swanson JA. A phosphatidylinositol-3-kinase-dependent signal transition regulates ARF1 and ARF6 during Fcγ receptor-mediated phagocytosis. PLoS Biol. 2006;4:e162. doi: 10.1371/journal.pbio.0040162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lim J, Zhou M, Veenstra TD, Morrison DK. The CNK1 scaffold binds cytohesins and promotes insulin pathway signaling. Genes Dev. 2010;24:1496–1506. doi: 10.1101/gad.1904610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bill A, et al. Cytohesins are cytoplasmic ErbB receptor activators. Cell. 2010;143:201–211. doi: 10.1016/j.cell.2010.09.011. [DOI] [PubMed] [Google Scholar]
- 97.Fuss B, Becker T, Zinke I, Hoch M. The cytohesin Steppke is essential for insulin signalling in Drosophila. Nature. 2006;444:945–948. doi: 10.1038/nature05412. [DOI] [PubMed] [Google Scholar]
- 98.Hafner M, et al. Inhibition of cytohesins by SecinH3 leads to hepatic insulin resistance. Nature. 2006;444:941–944. doi: 10.1038/nature05415. [DOI] [PubMed] [Google Scholar]
- 99.White DT, McShea KM, Attar MA, Santy LC. GRASP and IPCEF promote ARF-to-Rac signaling and cell migration by coordinating the association of ARNO/cytohesin 2 with Dock180. Mol. Biol. Cell. 2010;21:562–571. doi: 10.1091/mbc.E09-03-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Quast T, et al. Cytohesin-1 controls the activation of RhoA and modulates integrin-dependent adhesion and migration of dendritic cells. Blood. 2009;113:5801–5810. doi: 10.1182/blood-2008-08-176123. [DOI] [PubMed] [Google Scholar]
- 101.Sakagami H, et al. Distinct spatiotemporal expression of EFA6D, a guanine nucleotide exchange factor for ARF6, among the EFA6 family in mouse brain. Brain Res. 2006;1093:1–11. doi: 10.1016/j.brainres.2006.02.058. [DOI] [PubMed] [Google Scholar]
- 102.Hernandez-Deviez DJ, Casanova JE, Wilson JM. Regulation of dendritic development by the ARF exchange factor ARNO. Nature Neurosci. 2002;5:623–624. doi: 10.1038/nn865. [DOI] [PubMed] [Google Scholar]
- 103.Choi S, et al. ARF6 and EFA6A regulate the development and maintenance of dendritic spines. J. Neurosci. 2006;26:4811–4819. doi: 10.1523/JNEUROSCI.4182-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Sheen VL, et al. Mutations in ARFGEF2 implicate vesicle trafficking in neural progenitor proliferation and migration in the human cerebral cortex. Nature Genet. 2004;36:69–76. doi: 10.1038/ng1276. [DOI] [PubMed] [Google Scholar]
- 105.Ferland RJ, et al. Disruption of neural progenitors along the ventricular and subventricular zones in periventricular heterotopia. Hum. Mol. Genet. 2009;18:497–516. doi: 10.1093/hmg/ddn377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shoubridge C, et al. Mutations in the guanine nucleotide exchange factor gene IQSEC2 cause nonsyndromic intellectual disability. Nature Genet. 2010;42:486–488. doi: 10.1038/ng.588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Scholz R, et al. AMPA receptor signaling through BRAG2 and Arf6 critical for long-term synaptic depression. Neuron. 2010;66:768–780. doi: 10.1016/j.neuron.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 108.O'Rourke SM, Christensen SN, Bowerman B. Caenorhabditis elegans EFA-6 limits microtubule growth at the cell cortex. Nature Cell Biol. 2010;12:1235–1241. doi: 10.1038/ncb2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Belov GA, Kovtunovych G, Jackson CL, Ehrenfeld E. Poliovirus replication requires the N-terminus but not the catalytic Sec7 domain of ArfGEF GBF1. Cell. Microbiol. 2010;12:1463–1479. doi: 10.1111/j.1462-5822.2010.01482.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Cukierman E, Huber I, Rotman M, Cassel D. The ARF1 GTPase-activating protein: zinc finger motif and Golgi complex localization. Science. 1995;270:1999–2002. doi: 10.1126/science.270.5244.1999. [DOI] [PubMed] [Google Scholar]
- 111.Inoue H, Randazzo PA. Arf GAPs and their interacting proteins. Traffic. 2007;8:1465–1475. doi: 10.1111/j.1600-0854.2007.00624.x. [DOI] [PubMed] [Google Scholar]
- 112.Bharti S, et al. Src-dependent phosphorylation of ASAP1 regulates podosomes. Mol. Cell Biol. 2007;27:8271–8283. doi: 10.1128/MCB.01781-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ismail SA, Vetter IR, Sot B, Wittinghofer A. The structure of an Arf-ArfGAP complex reveals a Ca2+ regulatory mechanism. Cell. 2010;141:812–821. doi: 10.1016/j.cell.2010.03.051. [DOI] [PubMed] [Google Scholar]
- 114.Mandiyan V, Andreev J, Schlessinger J, Hubbard SR. Crystal structure of the ARF-GAP domain and ankyrin repeats of PYK2-associated protein β. EMBO J. 1999;18:6890–6898. doi: 10.1093/emboj/18.24.6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jian X, et al. Autoinhibition of Arf GTPase-activating protein activity by the BAR domain in ASAP1. J. Biol. Chem. 2009;284:1652–1663. doi: 10.1074/jbc.M804218200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Inoue H, Ha VL, Prekeris R, Randazzo PA. Arf GTPase-activating protein ASAP1 interacts with Rab11 effector FIP3 and regulates pericentrosomal localization of transferrin receptor-positive recycling endosome. Mol. Biol. Cell. 2008;19:4224–4237. doi: 10.1091/mbc.E08-03-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Nam JM, et al. CIN85, a Cbl-interacting protein, is a component of AMAP1-mediated breast cancer invasion machinery. EMBO J. 2007;26:647–656. doi: 10.1038/sj.emboj.7601534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Cuthbert EJ, Davis KK, Casanova JE. Substrate specificities and activities of AZAP family Arf GAPs in vivo. Am. J. Physiol. Cell Physiol. 2008;294:C263–C270. doi: 10.1152/ajpcell.00292.2007. [DOI] [PubMed] [Google Scholar]
- 119.Moore CA, Milano SK, Benovic JL. Regulation of receptor trafficking by GRKs and arrestins. Annu. Rev. Physiol. 2007;69:451–482. doi: 10.1146/annurev.physiol.69.022405.154712. [DOI] [PubMed] [Google Scholar]
- 120.Bahri SM, Choy JM, Manser E, Lim L, Yang X. The Drosophila homologue of Arf-GAP GIT1, dGIT, is required for proper muscle morphogenesis and guidance during embryogenesis. Dev. Biol. 2009;325:15–23. doi: 10.1016/j.ydbio.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 121.Schmalzigaug R, et al. Impaired fear response in mice lacking GIT1. Neurosci. Lett. 2009;458:79–83. doi: 10.1016/j.neulet.2009.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Menon P, et al. Impaired spine formation and learning in GPCR kinase 2 interacting protein-1 (GIT1) knockout mice. Brain Res. 2010;1317:218–226. doi: 10.1016/j.brainres.2009.11.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hajdo-Milasinovic A, van der Kammen RA, Moneva Z, Collard JG. Rac3 inhibits adhesion and differentiation of neuronal cells by modifying GIT1 downstream signaling. J. Cell Sci. 2009;122:2127–2136. doi: 10.1242/jcs.039958. [DOI] [PubMed] [Google Scholar]
- 124.Selyunin AS, et al. The assembly of a GTPase-kinase signalling complex by a bacterial catalytic scaffold. Nature. 2011;469:107–111. doi: 10.1038/nature09593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vernoud V, Horton AC, Yang Z, Nielsen E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiol. 2003;131:1191–1208. doi: 10.1104/pp.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Matheson LA, Suri SS, Hanton SL, Chatre L, Brandizzi F. Correct targeting of plant ARF GTPases relies on distinct protein domains. Traffic. 2008;9:103–120. doi: 10.1111/j.1600-0854.2007.00671.x. [DOI] [PubMed] [Google Scholar]
- 127.Richter S, et al. Role of the GNOM gene in Arabidopsis apical-basal patterning — from mutant phenotype to cellular mechanism of protein action. Eur. J. Cell Biol. 2010;89:138–144. doi: 10.1016/j.ejcb.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 128.Naramoto S, et al. ADP-ribosylation factor machinery mediates endocytosis in plant cells. Proc. Natl Acad. Sci. USA. 2010;107:21890–21895. doi: 10.1073/pnas.1016260107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Teh OK, Moore I. An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature. 2007;448:493–496. doi: 10.1038/nature06023. [DOI] [PubMed] [Google Scholar]
- 130.Tanaka H, Kitakura S, De Rycke R, De Groodt R, Friml J. Fluorescence imaging-based screen identifies ARF GEF component of early endosomal trafficking. Curr. Biol. 2009;19:391–397. doi: 10.1016/j.cub.2009.01.057. [DOI] [PubMed] [Google Scholar]
- 131.Nomura K, et al. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- 132.Koizumi K, et al. VAN3 ARF-GAP-mediated vesicle transport is involved in leaf vascular network formation. Development. 2005;132:1699–1711. doi: 10.1242/dev.01716. [DOI] [PubMed] [Google Scholar]
- 133.Sieburth LE, et al. SCARFACE encodes an ARF-GAP that is required for normal auxin efflux and vein patterning in Arabidopsis. Plant Cell. 2006;18:1396–1411. doi: 10.1105/tpc.105.039008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Stefano G, et al. AGD5 is a GTPase-activating protein at the trans-Golgi network. Plant J. 2010;64:790–799. doi: 10.1111/j.1365-313X.2010.04369.x. [DOI] [PubMed] [Google Scholar]
- 135.Liljegren SJ, et al. Regulation of membrane trafficking and organ separation by the NEVERSHED ARF-GAP protein. Development. 2009;136:1909–1918. doi: 10.1242/dev.033605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Price HP, Stark M, Smith DF. Trypanosoma brucei ARF1 plays a central role in endocytosis and Golgi–lysosome trafficking. Mol. Biol. Cell. 2007;18:864–873. doi: 10.1091/mbc.E06-08-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Price HP, Peltan A, Stark M, Smith DF. The small GTPase ARL2 is required for cytokinesis in Trypanosoma brucei. Mol. Biochem. Parasitol. 2010;173:123–131. doi: 10.1016/j.molbiopara.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Price HP, Panethymitaki C, Goulding D, Smith DF. Functional analysis of TbARL1, an N-myristoylated Golgi protein essential for viability in bloodstream trypanosomes. J. Cell Sci. 2005;118:831–841. doi: 10.1242/jcs.01624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Frearson JA, et al. N-myristoyltransferase inhibitors as new leads to treat sleeping sickness. Nature. 2010;464:728–732. doi: 10.1038/nature08893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Kahn RA, et al. Consensus nomenclature for the human ArfGAP domain-containing proteins. J. Cell Biol. 2008;182:1039–1044. doi: 10.1083/jcb.200806041. [DOI] [PMC free article] [PubMed] [Google Scholar]


