Abstract
In eukaryotes, mitochondria carry out numerous functions central to cellular and organismal health. How mitochondrial activities are regulated in response to differing environmental conditions, such as variations in diet, remains an important unsolved question in biology. Here we review emerging evidence suggesting that reversible acetylation of mitochondrial proteins on lysine residues represents a key mechanism by which mitochondrial functions are adjusted to meet environmental demands. In mammals, three members of the sirtuin class of NAD+-dependent deacetylases – SIRT3, SIRT4, and SIRT5 – localize to mitochondria and regulate targets involved in a diverse array of biochemical pathways. The importance of this activity is highlighted by recent studies of SIRT3 indicating that this protein suppresses the emergence of diverse age-related pathologies: hearing loss, cardiac fibrosis, and malignancy. Together, these findings argue that mitochondrial protein acetylation represents a central means by which mammals regulate mitochondrial functions to maintain cellular and organismal homeostasis.
Keywords: Sir2, acetylation, respiration, reactive oxygen species, β-oxidation, ketone body, apoptosis, metabolism, mitochondria, SIRT3, SIRT4, SIRT5, urea cycle, GDH, CPS1, electron transport, glutathione, SOD2, LCAD, IDH2, hearing loss, cardiac hypertrophy, cancer, HMGCS2, AceCS2
1. Introduction: mitochondrial regulation in health and disease
Mitochondria are cytoplasmic organelles that carry out numerous functions critical to cellular and organismal homeostasis (Wallace 2005). They generate the majority of cellular ATP via the Krebs cycle, oxidation of fatty acids, and oxidative phosphorylation. These activities make them the principal source of reactive oxygen species (ROS) within the cell. They also carry out amino acid degradation along with a portion of the urea cycle, and are the site of ketone body formation and heme biosynthesis. They represent a major storage site for cellular Ca2+. In brown adipocytes, they function in heat generation. Finally, they are key mediators of apoptosis. Structurally, mitochondria consist of a matrix space enclosed by an impermeable inner mitochondrial membrane (IMM), in turn surrounded by a permeable outer mitochondrial membrane (OMM). Embedded in the IMM are electron transport chain complexes I–IV that extrude protons from the matrix, generating an electrochemical gradient across the IMM. At complex V (ATP synthase), protons flow back into the matrix with this gradient, coupled to ATP synthesis. Mitochondria possess circular genomes encoding 13 electron transport chain subunits plus some tRNA and rRNA genes. Thus, the great majority of proteins required for respiratory function and other mitochondrial activities (~1500) are encoded in the nucleus. Inherited mutations in the mitochondrial genome cause a variety of syndromes of varied severity and age of onset, whereas acquired mitochondrial dysfunction may contribute to the degenerative manifestations of aging, as well as age-associated diseases such as type 2 diabetes, neurodegeneration, and malignancy (Wallace 2005).
1.1 The challenge of mitochondrial regulation
Coordination of mitochondrial processes with those occurring in other parts of the cell represents a formidable regulatory challenge. This chapter focuses on emerging roles for acetylation of mitochondrial proteins in regulating functions of this organelle, and the involvement of sirtuin-family deacetylases in this process. However, it is important to point out that many other pathways play roles in regulating mitochondrial number and function, a broader topic that is the subject of a number of excellent recent reviews (Finley and Haigis 2009; Ryan and Hoogenraad 2007; Scarpulla 2008). Briefly, factors that induce mitochondrial biogenesis – exercise, electrical stimulation, cold challenge, nitric oxide, thyroid hormone, and glucocorticoids – induce a coordinated transcriptional response from the mitochondrial and nuclear genomes (Butow and Avadhani 2004). Nuclear proteins involved in regulating expression of nuclear-encoded mitochondrial genes include transcription factors (NRF-1 and -2, PPARα and γ, ERRα, and Sp1, among others) and members of the PGC-1 coactivator family (PGC-1α and -1β, and PRC) (Butow and Avadhani 2004; Scarpulla 2008). Other proteins with crucial roles in regulating mitochondrial functions include the deacetylase SIRT1 via its role in activating PGC-1α (Gerhart-Hines et al. 2007; Rodgers et al. 2008), as well as the AMP-activated protein kinase (AMPK) and the mammalian target of rapamycin (mTOR) kinase (Finley and Haigis 2009; Schieke et al. 2006). Mitochondrial dysfunction is signaled to the nucleus by a variety of mechanisms, in yeast collectively termed the retrograde response, that remain poorly understood in mammals. A specialized system involving the nuclear transcription factors CHOP and C/EBPβ exists to indicate the presence of unfolded proteins in mitochondria (Ryan and Hoogenraad 2007).
1.2 Calorie restriction-induced alterations in mitochondrial functions
Variations in diet represent a challenge to mitochondrial function. Mitochondrial adaptation to altered diet is thought to be particularly significant in the context of calorie restriction (CR) – i.e. reduced caloric intake without malnutrition – an intervention that robustly extends lifespan in organisms ranging from budding yeast to rodents, and potentially primates as well (Fontana 2009). In rodent models, CR extends lifespan and delays the onset of a host of age-associated pathologies, including type 2 diabetes, cardiovascular disease, renal failure, cancer, and neurodegeneration (Fontana 2009). Many CR-associated health benefits have been observed in studies of non-human primates, notably dramatic reductions in cancer, cardiovascular disease, brain atrophy, sarcopenia, and type 2 diabetes (Colman et al. 2009; Colman et al. 2008). In humans, CR is associated with greatly improved metabolic and cardiovascular function (Fontana 2009). Thus, pharmacologic mimics of CR would likely have far-reaching health benefits in humans.
Mechanisms of longevity extension by CR remain incompletely understood; however metabolic alterations occurring in the adaptation to CR are likely in part responsible for the beneficial effects of this intervention. These changes implicate alterations in mitochondrial functions as an integral component of the CR response (Anderson et al. 2008b). In budding yeast, increased mitochondrial respiration is required for longevity extension by some (Lin et al. 2002) but not all (Kaeberlein et al. 2005) CR regimens. In C. elegans, CR or genetic and pharmacologic CR mimetics also induce increased mitochondrial respiration, which is required for CR-induced longevity (Bishop and Guarente 2007; Houthoofd et al. 2002a, b; Schulz et al. 2007). In mammals, CR induces mitochondrial biogenesis in a tissue-specific manner, at least in part through increased expression and activity of endothelial nitric oxide synthase and PGC-1α (Anderson et al. 2008a; Civitarese et al. 2007; Lambert et al. 2004; Lopez-Lluch et al. 2006; Nisoli et al. 2005). Conflicting data have been published regarding effects of CR on mitochondrial respiration in mammals (Hunt et al. 2006; Lambert et al. 2004; Nisoli et al. 2005; Weindruch et al. 1980). Numerous reports indicate that CR reduces mitochondrial ROS generation, a source of chronic cellular injury, and concomitantly reduces accumulation of macromolecular oxidative damage (Gredilla and Barja 2005). Several mechanisms may account for this decline in ROS during CR. The recruitment of a larger complement of mitochondria to generate ATP during CR could in and of itself reduce ROS generation (Guarente 2008); as could increased mitochondrial turnover occurring during CR, via removal of damaged mitochondria that would otherwise produce excessive ROS (Miwa et al. 2008). Increased mitochondrial uncoupling observed under CR conditions could also attenuate ROS generation (Brand 2000). As described below, new data suggests that the SIRT3 deacetylase plays a key role in bolstering mitochondrial anti-oxidant defenses during CR (Qiu et al. 2010; Someya et al. 2010). Overall, CR leads to increased mitochondrial biogenesis and decreased ROS generation in mammals.
In addition to these impacts, CR entails a shift from glucose utilization to the use of alternative sources of energy such as amino acids, ketones, and fatty acids (Spindler and Dhahbi 2007). This transition necessitates adaptation of numerous mitochondrial metabolic pathways. Indeed, within mitochondria, activities of a wide variety of enzymes have been shown to be altered in response to CR; much of this work has focused on the liver (Dhahbi et al. 2001; Hagopian et al. 2005; Hagopian et al. 2003, 2004; Tillman et al. 1996). Transcriptional changes occurring during CR have been extensively characterized (Anderson and Weindruch 2007). However, modulation of gene expression does not account for all of these activity changes (Spindler and Dhahbi 2007). This implies that non-transcriptional mechanisms to regulate mitochondrial functions during CR must exist.
2. Protein acetylation is a conserved mechanism of metabolic regulation
Reversible acetylation on the ε-amino group of internal lysine residues (hereinafter referred to as acetylation) has emerged as a post-translational modification with a crucial role in regulating target protein function, akin to phosphorylation. This modification is distinct, chemically and functionally, from acetylation of the α-amino groups of N-terminal residues; the latter occurs during translation and is irreversible (Polevoda and Sherman 2002). Although lysine acetylation was originally discovered on histones in the context of chromatin regulation, it is now clear that acetylation plays a crucial role in regulating a plethora of non-histone proteins, including transcription factors and metabolic enzymes (Spange et al. 2009). As discussed in depth below, regulated acetylation/deacetylation of proteins within mitochondria, regulated by sirtuin deacetylases, likely represents one mechanism by which mitochondrial functions are tailored to meet the demands of dietary challenges such as CR and other metabolic perturbations.
2.1 Mass spectrometry surveys reveal that acetylation of mitochondrial proteins is widespread
One of the first clues as to the wide-ranging impact of acetylation on diverse cellular functions came from a large-scale proteomic survey to identify acetylated proteins (Kim et al. 2006). Using acetyl-lysine affinity purification coupled with mass spectrometry, this study identified acetylation sites on 195 proteins, including numerous non-histone proteins. Strikingly, 133 acetylated proteins were identified within the mitochondrion, an organelle where acetylation of only a single protein had previously been documented (Hallows et al. 2006; Schwer et al. 2006). Moreover, this report showed that acetylation in liver mitochondria changed in response to fasting, suggesting that this modification might play a role in mitochondrial response to food deprivation. The widespread nature of mitochondrial protein acetylation was confirmed in several subsequent independent mass spectrometry surveys (Choudhary et al. 2009; Kendrick et al. 2010; Schwer et al. 2009; Zhao et al. 2010). Work by many laboratories suggests a central role for acetylation of mitochondrial proteins in the regulation of individual enzyme activities and overall metabolism (Zhao et al. 2010). Acetylation of proteins involved in virtually every mitochondrial function has been reported. Much of the work examining the role of acetylation in regulating mitochondrial protein activities has been performed in the context of studies of sirtuin deacetylases and is discussed below. This role for acetylation in regulating metabolic functions is ancient and conserved, and exists even in bacteria, where it is controlled by the opposing activities the Pat acetyltransferase and the sirtuin CobB (Starai et al. 2002; Wang et al. 2010; Zhang et al. 2009).
2.2 Mitochondrial protein acetylation is altered in response to calorie restriction and other dietary interventions
Work from our group has implicated alterations in mitochondrial protein acetylation specifically in the response to CR (Schwer et al. 2009). Global mitochondrial acetylation is altered in a tissue-specific manner during CR; these changes are particularly striking in liver and in brown adipose tissue, where global mitochondrial acetylation rises and falls, respectively, in response to this diet. Mass spectrometry analysis revealed that acetylation of at least 72 hepatic mitochondrial proteins increases during CR, involving essentially all mitochondrial metabolic pathways. The functional impact of these acetylation changes at the level of individual enzyme activities has been elucidated for only a very few targets (Ahn et al. 2008; Cimen et al. 2010; Nakagawa et al. 2009). One target of CR-associated acetylation changes identified in this study, the E1α subunit of the pyruvate dehydrogenase complex (PDC) (Schwer et al. 2009), is of particular interest, in that this enzyme performs the rate-limiting, final step in glycolysis: conversion of pyruvate to acetyl-CoA for use in the Krebs cycle or biosynthetic processes. Potential alterations in PDC activity mediated by acetylation could have far-reaching metabolic consequences in the cell. In this regard, in budding yeast PDC is required for CR-induced lifespan extension, and overexpression of a PDC subunit extends replicative lifespan in this organism (Easlon et al. 2007), suggesting that regulation of PDC function may represent a conserved component of the response of mitochondria to CR. The actual impact of altered acetylation on PDC remains to be elucidated.
Aside from CR, acetylation of many mitochondrial and non-mitochondrial proteins in liver also rises in response to high-fat diet (HFD) and chronic ethanol ingestion, conditions associated with diminished mitochondrial respiratory function (Kendrick et al. 2010; Picklo 2008; Shepard et al. 2010; Shulga and Pastorino 2010). Whether or not these acetylation changes occurring during CR and other dietary stresses represent regulated events, and whether overlapping or distinct sets of protein substrates and individual lysine residues are targeted under these different conditions, remain unsolved but important questions. Overall, acetylation of mitochondrial proteins has emerged as a widespread modification that plays a crucial role in regulating mitochondrial functions. The critical question is how this modification is regulated at the level of individual substrates in response to varied environmental conditions.
3. Mitochondrial sirtuins in metabolic regulation
Conserved from bacteria to mammals, the sirtuins are a protein family involved in regulating many biological processes, including stress responses, metabolism, development, and longevity (Haigis and Sinclair 2010). Sirtuins modify target proteins by means of their lysine deacetylase and ADP-ribosyltransferase activities; both require NAD+ as an obligate cofactor. Since NAD+ levels rise in response to reduced energy status and/or altered redox, sirtuins provide a means by which cells sense and respond to their environment (Guan and Xiong 2010). Sirtuin activity can be repressed by NADH and by a product of the sirtuin deacetylase reaction, nicotinamide (NAM) (Bitterman et al. 2002; Lin et al. 2004). Mammals possess seven sirtuins, SIRT1-SIRT7; these are a diverse protein family, with varied tissue expression, subcellular localization patterns, activity profiles, and targets (see chapter by S. Imai and colleagues, this volume, for a general overview of sirtuin biology). Three mammalian sirtuins (SIRT3, SIRT4, and SIRT5) are mitochondrial. These sirtuins are thus ideally positioned to regulate mitochondrial functions via modification of proteins within this organelle. Characterization of mitochondrial sirtuin functions has been aided immensely by the availability of mouse strains with targeted mutations in these genes (Haigis et al. 2006; Lombard et al. 2007). SIRT3 is a potent deacetylase with many mitochondrial targets, whereas SIRT5 is a more selective deacetylase; in contrast the only function characterized for SIRT4 to date is as is an ADP-ribosyltransferase (Ahuja et al. 2007; Haigis et al. 2006; Lombard et al. 2007; Nakagawa et al. 2009; North et al. 2003). Although HDAC7, a non-sirtuin deacetylase, has been reported to localize to mitochondria (Bakin and Jung 2004), no mitochondrial substrates have been reported for this enzyme. By contrast, a large amount of data exists supporting a crucial role for mitochondrial sirtuins in the regulation of mitochondrial functions.
4. SIRT3 is a master regulator of mitochondrial functions and suppresses age-associated phenotypes
Among the mitochondrial sirtuins, SIRT3 functions have been characterized in the greatest detail. Initial studies of SIRT3-deficient mice indicated that loss of SIRT3, but not SIRT4 or SIRT5, led to dramatic protein hyperacetylation within mitochondria, suggesting that SIRT3 deacetylates numerous targets in this organelle and is the major mitochondrial deacetylase activity (Lombard et al. 2007). The mitochondrial localization of SIRT3 has been extensively demonstrated by multiple independent laboratories (Cooper et al. 2009; Lombard et al. 2007; Onyango et al. 2002; Schwer et al. 2002). In humans, full length SIRT3 is a 44 kilodalton (kD) protein with an N-terminal mitochondrial targeting sequence that is an enzymatically inactive in vitro. It is proteolytically processed in mitochondria to a mature 28 kD catalytically active deacetylase (Onyango et al. 2002; Schwer et al. 2002). The first mouse SIRT3 cDNA sequence identified encoded a 28 kD protein lacking the N-terminal mitochondrial targeting sequence (Yang et al. 2000). However, several recent studies have identified a longer isoform of murine SIRT3 encoding a 37 kD protein that can be imported into mitochondria and processed into the mature 28 kD protein (Bao et al. 2010a; Cooper et al. 2009; Jin et al. 2009; Yang et al. 2010b).
Whether or not an active fraction of SIRT3 exists outside mitochondria and modifies extra-mitochondrial proteins is a controversial topic. There is one report that human SIRT3 localizes to nuclei, where it deacetylates histones, and is imported into mitochondria upon cellular stress such as genotoxic insult (Scher et al. 2007). Co-overexpression of SIRT5 along with SIRT3 is reported to drive SIRT3 to the nucleus (Nakamura et al. 2008). Rat SIRT3 was detected not only in mitochondria but also in the nucleus and cytoplasm of cardiomyocytes (Sundaresan et al. 2008). Unfortunately, current data supporting the presence of active extra-mitochondrial SIRT3 is based on overexpression and/or knockdown approaches rather than analysis of cells and tissues derived from SIRT3-null mutants. While it is clear that SIRT3 deficiency impacts cellular physiology outside mitochondria, this could occur due to retrograde signaling occurring in the context of mitochondrial dysfunction induced by lack of SIRT3. Conversely, it remains a possibility that an active fraction of SIRT3 might exist outside mitochondria in specific tissues and cell types (e.g. cardiomyocytes) (Sundaresan et al. 2008). The role of SIRT3 in regulating extra-mitochondrial signaling is reviewed below; we now turn to a discussion of mitochondrial processes targeted by SIRT3.
4.1 SIRT3 regulates numerous metabolic pathways within mitochondria
Expression studies of SIRT3 have shown that SIRT3 levels rise in adipose tissue, skeletal muscle, and liver during CR or fasting (Hirschey et al. 2010; Palacios et al. 2009; Schwer et al. 2009; Shi et al. 2005), and conversely decline in insulin-resistant states (Yechoor et al. 2004) or in response to high-fat feeding (Bao et al. 2010b; Kendrick et al. 2010; Palacios et al. 2009). These expression data suggest that SIRT3 might play a role in the response to caloric deprivation. The first mitochondrial SIRT3 substrate identified was Acetyl-CoA Synthetase 2 (AceCS2) (Hallows et al. 2006; Schwer et al. 2006; Shimazu et al. 2010b). AceCS2 converts free acetate, produced from endogenous catabolic reactions or absorbed from the gut, into the active metabolite acetyl-CoA for energy production in the Krebs cycle. In mammals, two independent studies showed that SIRT3 interacts with and deacetylates AceCS2 at the active site lysine to promote AceCS2 activity (Hallows et al. 2006; Schwer et al. 2006). Interestingly, in Salmonella the homolog of AceCS2 is also activated by the sirtuin CobB (Starai et al. 2002; Starai and Escalante-Semerena 2004), an activity that is crucial for bacterial growth on acetate. In mammals under fed conditions, the majority of acetyl-CoA is generated through metabolism of pyruvate by PDC and by fatty acid β–oxidation, largely bypassing the need for AceCS2. In this regard, studies of AceCS2-deficient mice revealed that AceCS2 is specifically required for metabolic homeostasis in the context of a low carbohydrate/high fat diet (LC/HFD); AceCS2-deficient animals are essentially normal on a chow diet but show poor weight gain, hypothermia, hypoglycemia, and impaired survival on a LC/HFD (Sakakibara et al. 2009). It will be of interest to assess the impact of a LC/HFD on SIRT3-deficient animals to determine whether they show similar defects in vivo. Presumably the role of SIRT3 in regulating AceCS2 could also be important during fasting, when acetate can be used as a source of energy in extrahepatic tissues (Hirschey et al. 2010). In this context, SIRT3 has recently been shown to deacetylate and activate 3-hydroxy-3-methylglutaryl-CoA synthase 2 (HMGCS2), a mitochondrial enzyme that converts acetyl-CoA into ketone bodies (acetoacetate, β-hydroxybutyrate, and acetone) in the liver under fasting conditions, which can in turn be used as a source of energy in certain tissues such as the brain (Shimazu et al. 2010a). SIRT3-deficient mice are unable to produce normal levels of ketone bodies upon fasting. Thus SIRT3 promotes multiple aspects of the response to fasting, and, as we shall see, CR.
Emerging data from several laboratories has shown that one major function of SIRT3 is regulation of mitochondrial electron transport chain activity to maintain energy homeostasis. Initial analysis of SIRT3-deficient animals demonstrated that these mice were metabolically unremarkable with respect to overall respiration, fuel utilization, activity, and cold tolerance (Lombard et al. 2007). However, subsequent detailed studies revealed that SIRT3-deficient liver, heart, kidney, and fibroblasts all show reduced basal ATP levels (Ahn et al. 2008). Moreover, SIRT3 interacts with the mitochondrial complex I component NDUFA9, and SIRT3 deficiency is associated with increased complex I acetylation and inhibition of its activity (Ahn et al. 2008; Bao et al. 2010b; Kim et al. 2010). Besides complex I, SIRT3 also regulates other electron transfer chain components, including complex II (Cimen et al. 2010), III (Kendrick et al. 2010; Kim et al. 2010), IV (Kendrick et al. 2010), and V (Bao et al. 2010b). In all of these cases, increased complex acetylation occurring in the context of SIRT3 deficiency or knockdown correlates with decreased complex activity. These findings suggest that SIRT3 regulates many aspects of mitochondrial oxidative phosphorylation. In future studies, it will be of interest to define at a mechanistic level how acetylation on electron transport chain subunits affects ATP generation. For example, acetylation could in principle affect electron transport chain subunit activity, turnover, and/or complex assembly/stability, among other parameters. It is also important to elucidate why it might be desirable under some physiologic conditions to downregulate electron transport chain activity via increased acetylation. As an added wrinkle, SIRT3 negatively regulates translation within mitochondria by deacetylating the ribosomal protein MRPL10, a function proposed to reduce respiration (Yang et al. 2010a).
Another recently identified role for SIRT3 involves its regulation of mitochondrial β–oxidation of fatty acids, the major pathway of fatty acid breakdown in mammals (Hirschey et al. 2010). SIRT3-deficient animals show elevated levels of long-chain fatty acids upon fasting, along with impaired β–oxidation. The authors of this study found that SIRT3 deacetylates and activates the β–oxidation enzyme long-chain specific acyl-CoA dehydrogenase (LCAD). Although young adult (12 week-old) fed SIRT3-deficient animals show no cold sensitivity (Lombard et al. 2007), very young (4-week old) fasted SIRT3-deficient mice are cold-intolerant, much like mice lacking LCAD (Guerra et al. 1998) and other models of defective β–oxidation. As mass spectrometry surveys have revealed acetylation of additional β–oxidation enzymes besides LCAD (Schwer et al. 2009), it remains to be seen whether SIRT3 regulates β–oxidation by deacetylation of additional targets. Independently, it was found that SIRT3-deficient hepatocytes are more susceptible to fatty-acid induced cell death, a phenotype suppressed by anti-oxidants and which the authors attributed to electron transport chain dysfunction leading to elevated ROS production (Bao et al. 2010b). SIRT3 also increases β–oxidation in muscle cells in culture, and reduces lipid accumulation in HepG2 cells, effects ascribed to the role of SIRT3 in promoting AMPK activity (Palacios et al. 2009; Shi et al. 2010). Overall, SIRT3 allows the efficient use of fatty acids as fuel with minimal cellular toxic effects through deacetylation of multiple mitochondrial targets.
4.2 SIRT3 impacts extra-mitochondrial signaling
One important issue that remains to be resolved is how SIRT3 impacts extra-mitochondrial processes. As mentioned above, PGC-1α is a co-activator protein that plays a critical role in promoting mitochondrial biogenesis, adaptive thermogenesis, fatty acid oxidation, and ROS detoxification, among other metabolic processes (Jeninga et al. 2010). PGC-1α stimulates SIRT3 expression by binding to the SIRT3 promoter together with estrogen-related receptor α (ERRα) (Kong et al. 2010). Strikingly, the reciprocal relationship holds as well; SIRT3 is required for normal PGC-1α expression in brown adipocytes and in skeletal muscle (Palacios et al. 2009; Shi et al. 2005). SIRT3 promotes PGC-1α expression by stimulating phosphorylation and activity of factors known to regulate PGC-1α expression, CREB and AMPK (Palacios et al. 2009; Pillai et al. 2010; Shi et al. 2005). SIRT3 is also required for PGC-1α to induce mitochondrial biogenesis and expression of its target genes, particularly genes involved in ROS detoxification, and for PGC-1α to suppress ROS levels (Kong et al. 2010). How SIRT3 impacts PGC-1α is not completely clear, although one study attributes this effect to a role for (presumably extra-mitochondrial) SIRT3 in deacetylating and activating LKB1, a kinase upstream of AMPK (Pillai et al. 2010). Since SIRT3 promotes increased ATP production in many tissues (Ahn et al. 2008), and AMPK is phosphorylated and activated in response to an increased AMP:ATP ratio, SIRT3 might be predicted to suppress rather than stimulate AMPK phosphorylation and activity. These effects may represent functions of extra-mitochondrial SIRT3, or the indirect effects of a retrograde response. In any case, the fact that SIRT3 impacts activities of master metabolic regulators such as PGC-1α and AMPK complicates the interpretation of data generated using SIRT3-deficient mice and cells, since it may be unclear whether a given phenotype results from loss of SIRT3 itself and its role in deacetylating a particular mitochondrial target, or secondary effects on overall cellular physiology associated with loss of SIRT3.
4.3 SIRT3 and cell death
Because mitochondria are central to the intrinsic pathway of cell death, several groups have assessed roles for SIRT3 in modulating cell survival, and have obtained conflicting results. Given that SIRT3 has numerous substrates exerting diverse biological effects (Lombard et al. 2007), it is perhaps not surprising that discrepant results have been obtained in this regard. Experimental differences in the cell type analyzed, the nature of cellular injury, etc. likely explain these disparities. SIRT3 overexpression sensitizes lymphoma cells to kaempferol, a flavonoid that induces ROS and cell death (Marfe et al. 2009). SIRT3-deficient mouse embryonic fibroblasts (MEFs) are resistant to cell death induced by DNA damage (Kim et al. 2010). Similarly, in a variety of tumor cell lines SIRT3 knockdown confers resistance to cell death induced by depletion of Bcl-2 (Allison and Milner 2007). Conversely, SIRT3 protects cardiomyocytes against genotoxin-induced killing, an effect attributed to a role for extra-mitochondrial SIRT3 in deacetylating the Ku70 protein to promote its interaction with the pro-apoptotic protein Bax (Sundaresan et al. 2008). SIRT3 is also required for cell survival in response to the genotoxin methyl methanesulfonate (Yang et al. 2007). Similarly, SIRT3 promotes viability of hepatocytes in response to TNFα exposure via deacetylation and inactivation of cyclophilin D (see below)(Shulga and Pastorino 2010). In bladder cancer cells, SIRT3 also allows continued proliferation following induction of the p53 tumor suppressor, which ordinarily induces senescence (permanent growth arrest) (Li et al. 2010). This function of SIRT3 has been ascribed to the ability of SIRT3 to deacetylate p53 within mitochondria. In vivo, SIRT3 plays an important role in promoting long-term survival of cells in the inner ear to preserve hearing during CR (see below)(Someya et al. 2010), suggesting that in this specific context, the pro-survival function of SIRT3 is dominant.
SIRT3 likely modulates cell death through multiple different mechanisms and targets. Several groups have reported that SIRT3 plays a major role in suppression of intracellular ROS levels (Bao et al. 2010b; Kim et al. 2010; Kong et al. 2010; Qiu et al. 2010; Someya et al. 2010). Since ROS are a potent inducer of apoptosis, this activity of SIRT3 provides a potential means by which SIRT3 could promote cellular survival. Recent studies have revealed that another key mechanism by which SIRT3 suppresses cell death is through deacetylation of cyclophilin D (cypD). CypD is a peptidyl-prolyl isomerase that potentiates activity of the mitochondrial permeability transition pore (MPTP), a non-selective high conductance channel promoting cell death, particularly in the contexts of cardiac and neuronal ischemia (Giorgio et al. 2010). SIRT3 was shown to deacetylate and inactivate cypD to promote mitochondrial respiration in the presence of a non-fermentable carbon source (Shulga et al. 2010). Subsequent elegant studies have revealed that this role of SIRT3 is also important in antagonizing cell death (Shulga and Pastorino 2010). Ethanol treatment of cells suppresses SIRT3 function via decreasing the NAD+:NADH ratio. This in turn promotes hyperacetylation and increased activity of cypD and the MPTP. This effect of ethanol can be prevented by treatment of cells with either the AMPK activator AICAR or acetoacetate, both of which elevate the NAD+:NADH ratio and restore SIRT3 activity. As an important control, introduction of a cypD acetylation site mutant that mimics deacetylation prevents ethanol-induced sensitization to TNFα and bypasses the requirement for SIRT3. Interestingly, although cypD promotes cell death in the context of TNFα treatment, it actually suppresses apoptosis in response to other stimuli (Li et al. 2004; Schubert and Grimm 2004). Thus this single SIRT3 substrate may represent a key target in the context of both pro- and anti-survival roles for SIRT3 described above. Future studies are needed to assess the roles of SIRT3 in regulating cypD in the context of cell death in vivo, particularly in response to medically relevant stressors such as ischemic injury.
4.4 SIRT3 in the regulation of lifespan and age-associated phenotypes
In addition to elucidating its roles in regulating specific biochemical pathways in mitochondria, there is great current interest in testing whether SIRT3 might modulate age-associated phenotypes, or indeed lifespan itself. In this regard, some studies have linked polymorphisms in the SIRT3 genomic locus to human longevity, though others have failed to demonstrate this association (Bellizzi et al. 2007; Bellizzi et al. 2005; Lescai et al. 2009; Rose et al. 2003). A polymorphism associated with decreased SIRT3 mRNA expression was present in cohorts of young but not old men, suggesting that reduced SIRT3 expression may be detrimental to survival in old age (Bellizzi et al. 2009; Bellizzi et al. 2005). In sedentary individuals, SIRT3 protein expression declined with age in skeletal muscle mitochondria, concomitant with a reduction in respiratory function (Lanza et al. 2008).
4.4.1 SIRT3 and cardiac hypertrophy/fibrosis
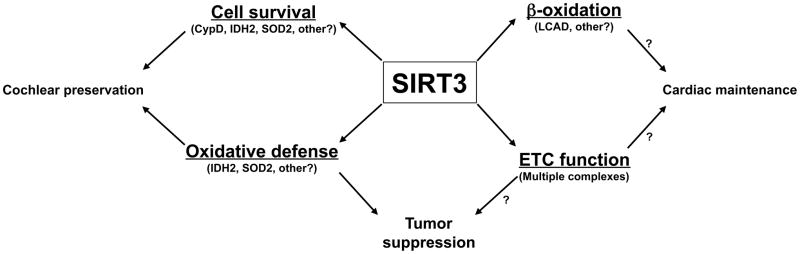
Emerging data from mouse models suggests that SIRT3 may indeed play an important role in delaying the onset of age-associated disorders (Fig. 1). Cardiac hypertrophy is a common age-associated pathology in western societies, where it is most frequently caused by hypertension. While initially an adaptive response, cardiac hypertrophy can lead to various downstream sequelae such as arrhythmias and ischemia. New data suggest a role for SIRT3 in antagonizing the onset of this disease. SIRT3-deficient mice show mild cardiac hypertrophy and fibrosis at baseline, and greatly exaggerated hypertrophy response to pharmacologic stimuli; conversely a SIRT3 overexpressor is protected (Sundaresan et al. 2009). The authors find that SIRT3 negatively modulates intracellular signaling pathways known to promote hypertrophy. The authors attribute this effect to an extra-mitochondrial role for SIRT3 in suppressing ROS levels via deacetylation of the FoxO3A forkhead transcription factor, inducing FoxO3A nuclear localization and increased expression of expression of the oxidative defense proteins catalase and superoxide dismutase 2 (SOD2). FoxO3A activity is required for SIRT3 to induce its anti-hypertrophic effects in a tissue culture model system. The same group has found that exogenous NAD+ can block cardiomyocyte hypertrophy in vivo and in tissue culture in a SIRT3-dependent manner. The authors attribute this role of SIRT3 both to its aforementioned role in suppressing ROS levels, as well as to a role for SIRT3 in deacetylating and inactivating the LKB1 kinase (Pillai et al. 2010). This work reveals important roles for SIRT3 in preventing age-associated cardiac pathology; given the many mitochondrial substrates for SIRT3, it is difficult to know whether extra-mitochondrial FoxO3A and/or LKB1 represent the relevant substrates for SIRT3 in this process in vivo. In particular, some human patients with mitochondrial genome mutations develop cardiac hypertrophy, suggesting that impaired respiratory function occurring in SIRT3 deficiency per se could conceivably contribute to this phenotype (Vydt et al. 2007). Significantly, mice deficient in LCAD, a known SIRT3 substrate, also develop cardiac hypertrophy (Cox et al. 2009; Kurtz et al. 1998); thus SIRT3 could promote cardiac health by multiple mechanisms.
Figure 1. SIRT3 suppresses the onset of diverse age-associated pathologies.
Mitochondrial and cellular impacts of SIRT3 are underlined, along with proposed mitochondrial substrates in parentheses. Some postulated substrate-phenotype relationships represent speculation by the authors; these are designated with question marks. Note that some roles of SIRT3 (e.g., promoting ketone body synthesis and suppressing mitochondrial translation) are omitted for clarity’s sake. See text for details.
4.4.2 SIRT3 and mammary cancer
In most mammals, an increased incidence of malignancy is a prominent feature of aging (Lombard et al. 2005). A recent ground-breaking study has revealed SIRT3 to be a tumor suppressor (Kim et al. 2010). SIRT3-deficient MEFs show higher levels of ROS in response to various forms of stress, aberrations in nuclear chromosome number, and decreased mitochondrial genome integrity, which is also observed in the livers of SIRT3-deficient animals with age. Moreover, SIRT3-deficient MEFs are more easily transformed, and resist apoptosis in response to DNA damage. Many of these cellular phenotypes associated with SIRT3 deficiency can be rescued by expression of exogenous SOD2, suggesting that mitochondrial superoxide plays a causative role in these defects. Most strikingly, a significant fraction of SIRT3-deficient mice develop mammary carcinoma after one year of age. Expression of SIRT3 is decreased in human breast cancer, suggesting that SIRT3 may play a similar tumor suppressor role in humans.
This work reveals a critical novel aspect of SIRT3 function, and raises important mechanistic questions regarding how SIRT3 suppresses mammary tumorigenesis. The authors suggest that SIRT3 might promote SOD2 expression via regulation of FoxO3A activity. However, while SIRT3-deficient mice develop mammary tumors, mice with reduced SOD2 levels develop primarily lymphomas (Van Remmen et al. 2003). Such a discrepancy could in principle result from differences in mouse strain background used in the two studies; alternatively regulation of SOD2 by SIRT3 could be crucial in mammary epithelium but less important in the hematopoietic system. The aneuploidy observed in SIRT3-deficient cells is also notable, in that oxidative stress is most closely associated with chromatid and chromosome breaks leading to genomic rearrangements, rather than ploidy alterations. It would be of interest to test whether SIRT3 deficiency leads to these types of DNA lesions, in addition to altered chromosomal number. In this regard, elevated ROS levels could promote growth of malignant cells by mechanisms other than increased genomic instability. In particular, many phosphatases that negatively regulate growth factor signaling can be inactivated by oxidative modification (Finkel 2003), suggesting that chronically increased ROS in SIRT3 deficiency might provide a trophic stimulus to developing tumors. The authors note that SIRT3-deficient cells generate a greater fraction of their ATP via glycolysis (Kim et al. 2010). This is a phenotype observed in many cancer cells (aerobic glycolysis; the Warburg effect) that promotes aspects of the malignant phenotype (Hsu and Sabatini 2008). Thus, lack of SIRT3 might promote oncogenesis through impaired respiratory function, in turn permitting Warburg-like metabolism. Similarly, germline mutations in genes encoding enzymes of the Krebs cycle, succinate dehydrogenase (SDH) and fumarate hydratase (FH), lead to tumor susceptibility in humans (King et al. 2006). SIRT3 deacetylates SDH to promote its activity in the context of electron transport; FH is also acetylated, although no role for SIRT3 in regulating FH has been identified (Cimen et al. 2010; Kim et al. 2006; Schwer et al. 2009; Zhao et al. 2010). Thus SIRT3 could conceivably suppress tumorigenesis via effects on the Krebs cycle, in addition to the roles discussed above.
4.4.3 SIRT3 and age-related hearing loss
Age-related hearing loss (ARHL), or presbycusis, is a common and vexing problem in the elderly, occurring secondary to cell loss and other degenerative changes in the cochlea (Liu and Yan 2007). A recent elegant study has firmly established a role for SIRT3 in antagonizing ARHL (Someya et al. 2010). Previously, it was known that CR or suppression of oxidative damage prevents cochlear cell loss and ARHL (Someya et al. 2009; Someya et al. 2007). The protective effects of CR on ARHL are SIRT3-dependent (Someya et al. 2010). One mechanism by which SIRT3 mediates this effect is via deacetylation of isocitrate dehydrogenase 2 (IDH2) (Schlicker et al. 2008; Someya et al. 2010), which converts isocitrate to alpha-ketoglutarate concomitant with reduction of NADP+. NADPH in turn allows regeneration of reduced glutathione to promote mitochondrial oxidative defense. In response to CR, wild-type mice, but not SIRT3-deficient animals, show increased NADPH levels, increased reduced glutathione in mitochondria, and decreased DNA damage in the cochlea and in other tissues. In tissue culture cells, overexpression of SIRT3 or IDH2 is protective against oxidative stress-induced cell death, and the two proteins together have a synergistic pro-survival effect. These results do not rule out the possibility that SIRT3 might modify other substrates to prevent AHRL during CR aside from IDH2. Similarly, Qiu and colleagues reported that SIRT3-deficient mice fail to suppress ROS levels and macromolecular damage during CR (Qiu et al. 2010). They find that SIRT3 directly deacetylates SOD2 to increase its activity during CR, whereas SIRT3-deficient mice do not show SOD2 deacetylation in response to this diet (Qiu et al. 2010). Overall, these papers point to crucial role for SIRT3 in suppressing oxidative damage and its negative sequelae during CR. It remains to be seen how SIRT3, or the other mitochondrial sirtuins, might impact other phenotypes of aging and/or effects of CR. In this context, the reduction of serum insulin and triglycerides normally occurring during CR are not observed in SIRT3-deficient mice (Someya et al. 2010), implying that SIRT3 plays additional, uncharacterized roles in the adaptation to this dietary regimen.
5. SIRT4 regulates the urea cycle, insulin secretion, fatty acid oxidation, and respiration
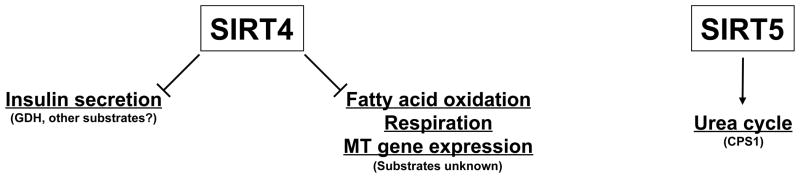
In contrast to numerous substrates and pathways regulated by SIRT3, comparatively little is known regarding the other two mitochondrial sirtuins, SIRT4 and SIRT5 (Fig. 2). SIRT4 is localized in the mitochondrial matrix in both mouse and human cells (Ahuja et al. 2007; Haigis et al. 2006; Michishita et al. 2005) and is broadly expressed, with high SIRT4 levels present in kidney, heart, brain, liver and pancreatic β-cells (Ahuja et al. 2007; Haigis et al. 2006). In liver, SIRT4 expression declines slightly during CR and increases in genetic models of diabetes (Haigis et al. 2006; Nasrin et al. 2010; Schwer et al. 2009). Thus far, no deacetylase function of SIRT4 has been detected, although it is possible that SIRT4 may possess very specific deacetylase activity on as-yet unidentified substrates (Ahuja et al. 2007; Black et al. 2008; Haigis et al. 2006; North et al. 2003). Instead, using SIRT4-deficient mice, it was shown that SIRT4 ADP-ribosylates and inactivates glutamate dehydrogenase (GDH) (Haigis et al. 2006). GDH converts glutamate to α-ketoglutarate in mitochondria, and its inhibition by SIRT4 results in repression of amino acid-stimulated insulin secretion in pancreatic β-cells (Haigis et al. 2006). Reduced SIRT4 activity allows coupling of insulin secretion to serum amino acid levels during CR, when amino acids serve as an important carbon source. It is of interest that SIRT3 deacetylates and activates GDH (Lombard et al. 2007; Schlicker et al. 2008), implying possible coordinated control of GDH by SIRT3 and SIRT4. Indeed, since SIRT5 regulates an additional urea cycle enzyme (CPS1) (Nakagawa et al. 2009), this pathway is the target of regulation by all three mitochondrial sirtuins. In an independent study, SIRT4 was shown to repress insulin secretion in response to glucose in insulinoma cells and to interact with insulin degrading enzyme (IDE) and the ADP/ATP carrier proteins ANT2 and ANT3; the impact of these interactions is not known (Ahuja et al. 2007).
Figure 2. Metabolic functions of SIRT4 and SIRT5.
Mitochondrial and cellular process are underlined and the relevant substrate is indicated in parentheses. MT, mitochondrial. See text for details.
A recent study investigating additional functions of SIRT4 showed that SIRT4 knockdown in tissue culture cells or in mouse liver in vivo results in increased expression of mitochondrial and fatty acid metabolism enzymes (Nasrin et al. 2010), as well as of SIRT1 and SIRT3. SIRT4 knockdown also leads to increased fatty acid oxidation, respiration, and AMPK phosphorylation (Nasrin et al. 2010). The effects of SIRT4 knockdown on fatty acid oxidation require SIRT1. Many of these phenotypes are the opposite of those observed in SIRT3 deficiency, suggesting once again that SIRT3 and SIRT4 activities may antagonize one another in some contexts. Whether SIRT4 mediates these effects via ADP-ribosylation or deacetylation of mitochondrial targets, or alternatively whether an extra-mitochondrial fraction of SIRT4 might play a role in direct regulation of nuclear gene expression, are important unanswered questions raised by this work.
6. SIRT5 regulates the urea cycle
SIRT5 is localized to the mitochondrial matrix and is broadly expressed, with highest levels in brain, heart, liver and kidney (Michishita et al. 2005; Nakagawa et al. 2009; Schlicker et al. 2008). Unlike SIRT3 or SIRT4, hepatic SIRT5 levels are unchanged during CR (Nakagawa et al. 2009; Schwer et al. 2009). SIRT5-deficient mice do not show any gross phenotypes and do not display the global increase in hepatic mitochondrial acetylation observed in SIRT3-deficient animals (Lombard et al. 2007). Also unlike SIRT3, SIRT5 has minimal deacetylase activity on canonical sirtuin substrates such as histones (Black et al. 2008; North et al. 2003; Scher et al. 2007; Schlicker et al. 2008). In vitro, SIRT5 deacetylates purified cytochrome c, although the biological significance of this activity has not yet been determined (Schlicker et al. 2008). More recently, SIRT5 was shown to interact with and deacetylate carbamoyl phosphate synthetase I (CPS1), which catalyzes the first, rate-limiting step of the urea cycle for ammonia detoxification and disposal (Nakagawa et al. 2009). Deacetylation of CPS1 by SIRT5 results in increased enzyme activity, and SIRT5-deficient mice fail to upregulate CPS1 activity and show elevated blood ammonia during a prolonged fast. In the same study it was shown that CPS1 is deacetylated during CR and that CPS1 activity increases on this diet, although the impact of CR on SIRT5-deficient mice was not directly tested (Nakagawa et al. 2009). The ability of SIRT5 to deacetylate and activate CPS1 was independently verified in transgenic mice overexpressing SIRT5 (Ogura et al. 2010). However, we identified CPS1 as a hepatic protein increasing in acetylation during CR (Schwer et al. 2009). The resolution to this discrepancy is not clear, but it may be that CPS1 acetylation and activity are regulated by proteins other than SIRT5 during CR. Alternatively, different techniques of assessing CPS1 acetylation might preferentially uncover different acetylation sites, which could be regulated in opposing ways during CR. In this regard, CPS1 possesses over 20 acetylation sites, implying that acetylation-mediated regulation of CPS1 activity may be quite complex (Schwer et al. 2009).
It is likely that other SIRT5 substrates and functions remain to be identified. In this context, a recent study reported that the presence of a SIRT5 promoter polymorphism correlates with reduced SIRT5 expression and an “older” pattern of gene expression in the human brain (Glorioso et al. 2010). Many of the transcripts altered in the presence of this polymorphism encode mitochondrial proteins, including Parkinson’s disease genes; the authors suggest that SIRT5 polymorphisms may represent a risk factor for diseases related to mitochondrial dysfunction. It will be of great interest to test this hypothesis in human cohorts and using cells and mice with altered levels of SIRT5.
7. Mitochondrial protein acetylation and sirtuins: unresolved questions
Acetylation of mitochondrial proteins plays a major role in regulating functions of this organelle. Despite the rapid progress in this area, there are still many outstanding questions that will no doubt provide fruitful avenues for research for years to come. In particular, how mitochondrial proteins are acetylated in the first place is currently unknown. The identity of putative mitochondrial acetyltransferases remains elusive; identification of such proteins would represent a major step forward in this field. Alternatively or in addition to enzymatic acetylation within mitochondria, mitochondrial proteins could in principle be acetylated outside this organelle, prior to or concomitant with mitochondrial import; or even be acetylated non-enzymatically. These latter models would not permit rapid cycles of acetylation/deacetylation of mitochondrial proteins to regulate target protein function in response to varied environmental challenges. Instead, following deacetylation, restoration of acetylation status would require new protein synthesis. Such models could be distinguished through pulse-chase experiments assessing acetylation of newly synthesized mitochondrial proteins prior to and following mitochondrial import. The fact that a mitochondrially-encoded subunit of complex V is acetylated means that some mechanism of acetylating proteins within this organelle must exist (Huang et al. 2010).
Similarly, how sirtuin activity is regulated in the mitochondria is incompletely understood. Sirtuin require NAD+, and therefore mitochondrial NAD+ levels play a critically important role in governing mitochondrial sirtuin function. Increased NADH generation from NAD+ in the context of HFD leading to reduced sirtuin function may explain the increased global mitochondrial protein acetylation observed during this diet, as could increased acetyl-CoA levels, the substrate for acetyltransferases (Kendrick et al. 2010). It has been reported that NAD+ levels in mouse liver mitochondria rise during CR, which would be predicted to increase sirtuin activity (Nakagawa et al. 2009), and moreover SIRT3 protein levels rise in this tissue in response to this diet (Schwer et al. 2009). However, these observations are at odds with the observation that global mitochondrial acetylation rises dramatically in liver during CR (Schwer et al. 2009). This overall increased acetylation may represent the net effect of increased acetyltransferase activity superimposed upon elevated sirtuin function; alternatively, some protein species hyperacetylated during CR or other conditions may not be substrates for mitochondrial sirtuins. The activity of SIRT3 and other mitochondrial sirtuins might be governed by other influences besides NAD+ levels, such as post-translational modification or interactions with regulatory proteins. It is now clear that SIRT1 activity is tightly regulated by both mechanisms. Surprisingly, no comprehensive quantitative assessment of mitochondrial NAD+ and its metabolites has yet been performed under varied dietary/environmental conditions in different tissues.
While several proteomic studies have provided a detailed snapshot of the suite of acetylated mitochondrial proteins, how this modification changes at individual lysines on various targets in response to different environmental conditions is a topic that is only beginning to be explored. This effort will require the use of mass spectrometry-based approaches that allow quantitative comparison of acetylation on a given peptide between different samples, such as label-free quantitation (Schwer et al. 2009), stable labeling by amino acids in culture (SILAC) (Choudhary et al. 2009), isobaric tagging for relative and absolute quantification (iTRAQ) (Meany et al. 2007), or stable isotope dimethyl labeling (Boersema et al. 2009). It is currently unclear whether interventions that impact acetylation of many mitochondrial proteins – SIRT3 deficiency, CR, HFD, and chronic ethanol ingestion – lead to modification of common sets of proteins on the same lysine sites, or whether this response is tailored to different environmental perturbations. Similarly, it remains unclear whether the mitochondrial sirtuins share targets and/or functions in common. This question could be addressed in mice or cells with compound mitochondrial sirtuin deficiencies or knockdowns. As noted above, whereas SIRT3 deacetylates GDH to modestly stimulate its activity (Lombard et al. 2007; Schlicker et al. 2008), SIRT4 ADP-ribosylates this protein to suppress function (Haigis et al. 2006). This observation raises the possibility that other proteins might be common substrates for multiple sirtuins. Given that SIRT3 deacetylates many proteins in mitochondria as well as suppresses some age-associated phenotypes, it will be of interest to test whether acetylation of mitochondrial proteins changes with age, either individually or globally, and whether prevention of this effect might have a beneficial effect on healthspan or even lifespan.
In addition, whereas the functional impact of acetylation on a few individual protein targets is clear, a global understanding of how sirtuins impact the activity of metabolic pathways at the level of mitochondria, cells, tissues, and the organism overall is still lacking. Metabolic flux analysis, which has already been successfully applied to bacterial cells with altered protein acetylation, can be used to address this question (Wang et al. 2010). The answers to these and related questions regarding sirtuins and mitochondrial protein acetylation will no doubt reveal novel aspects of mitochondrial biology, and perhaps ultimately provide the basis for novel therapeutic strategies for a variety of disorders.
Acknowledgments
The authors gratefully acknowledge the comments of Drs. Rozalyn Anderson, Marcia Haigis and Brian North on the manuscript, and Dr. Wilhelm Haas for helpful discussions. We apologize to those whose work was not cited due to space constraints. D.X.T. is supported by an NIA training grant (T32-AG000114). D.B.L. is a New Scholar in Aging of the Ellison Medical Foundation (AG-NS-0583-09). Work in the Lombard laboratory is supported by the Ellison Medical Foundation as well as research grants from the Elsa U. Pardee Foundation and the American Foundation for Aging Research; pilot and feasibility grants from the Michigan Diabetes Research and Training Center (5P60-DK020572; NIDDK), the Cancer Research Committee of the Michigan Comprehensive Care Center, the Michigan Metabolomics and Obesity Center, the Nathan Shock Center, and the Claude D. Pepper Older American’s Independence Center (5P30-AG024824-05; NIA); and startup funds from the University of Michigan Department of Pathology and the Institute of Gerontology.
References
- Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci U S A. 2008;105:14447–52. doi: 10.1073/pnas.0803790105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, Verdin E. Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem. 2007;282:33583–92. doi: 10.1074/jbc.M705488200. [DOI] [PubMed] [Google Scholar]
- Allison SJ, Milner J. SIRT3 is pro-apoptotic and participates in distinct basal apoptotic pathways. Cell Cycle. 2007;6:2669–77. doi: 10.4161/cc.6.21.4866. [DOI] [PubMed] [Google Scholar]
- Anderson RM, Barger JL, Edwards MG, Braun KH, O’Connor CE, Prolla TA, Weindruch R. Dynamic regulation of PGC-1alpha localization and turnover implicates mitochondrial adaptation in calorie restriction and the stress response. Aging Cell. 2008a;7:101–11. doi: 10.1111/j.1474-9726.2007.00357.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Shanmuganayagan D, Weindruch R. Caloric Restriction and Aging: Studies in Mice and Monkeys. Toxicol Pathol. 2008b;37:47–51. doi: 10.1177/0192623308329476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Weindruch R. Metabolic reprogramming in dietary restriction. Interdiscip Top Gerontol. 2007;35:18–38. doi: 10.1159/000096554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakin RE, Jung MO. Cytoplasmic sequestration of HDAC7 from mitochondrial and nuclear compartments upon initiation of apoptosis. J Biol Chem. 2004;279:51218–25. doi: 10.1074/jbc.M409271200. [DOI] [PubMed] [Google Scholar]
- Bao J, Lu Z, Joseph JJ, Carabenciov D, Dimond CC, Pang L, Samsel L, McCoy JP, Jr, Leclerc J, Nguyen P, Gius D, Sack MN. Characterization of the murine SIRT3 mitochondrial localization sequence and comparison of mitochondrial enrichment and deacetylase activity of long and short SIRT3 isoforms. J Cell Biochem. 2010a;110:238–47. doi: 10.1002/jcb.22531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao J, Scott I, Lu Z, Pang L, Dimond CC, Gius D, Sack MN. SIRT3 is regulated by nutrient excess and modulates hepatic susceptibility to lipotoxicity. Free Radic Biol Med. 2010b;49:1230–7. doi: 10.1016/j.freeradbiomed.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellizzi D, Covello G, Di Cianni F, Tong Q, De Benedictis G. Identification of GATA2 and AP-1 Activator elements within the enhancer VNTR occurring in intron 5 of the human SIRT3 gene. Mol Cells. 2009;28:87–92. doi: 10.1007/s10059-009-0110-3. [DOI] [PubMed] [Google Scholar]
- Bellizzi D, Dato S, Cavalcante P, Covello G, Di Cianni F, Passarino G, Rose G, De Benedictis G. Characterization of a bidirectional promoter shared between two human genes related to aging: SIRT3 and PSMD13. Genomics. 2007;89:143–50. doi: 10.1016/j.ygeno.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Bellizzi D, Rose G, Cavalcante P, Covello G, Dato S, De Rango F, Greco V, Maggiolini M, Feraco E, Mari V, Franceschi C, Passarino G, De Benedictis G. A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics. 2005;85:258–63. doi: 10.1016/j.ygeno.2004.11.003. [DOI] [PubMed] [Google Scholar]
- Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–9. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem. 2002;277:45099–107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Black JC, Mosley A, Kitada T, Washburn M, Carey M. The SIRT2 deacetylase regulates autoacetylation of p300. Mol Cell. 2008;32:449–55. doi: 10.1016/j.molcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boersema PJ, Raijmakers R, Lemeer S, Mohammed S, Heck AJ. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat Protoc. 2009;4:484–94. doi: 10.1038/nprot.2009.21. [DOI] [PubMed] [Google Scholar]
- Brand MD. Uncoupling to survive? The role of mitochondrial inefficiency in ageing. Exp Gerontol. 2000;35:811–20. doi: 10.1016/s0531-5565(00)00135-2. [DOI] [PubMed] [Google Scholar]
- Butow RA, Avadhani NG. Mitochondrial signaling: the retrograde response. Mol Cell. 2004;14:1–15. doi: 10.1016/s1097-2765(04)00179-0. [DOI] [PubMed] [Google Scholar]
- Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–40. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- Cimen H, Han MJ, Yang Y, Tong Q, Koc H, Koc EC. Regulation of succinate dehydrogenase activity by SIRT3 in mammalian mitochondria. Biochemistry. 2010;49:304–11. doi: 10.1021/bi901627u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E. Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4:e76. doi: 10.1371/journal.pmed.0040076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Anderson RM, Johnson SC, Kastman EK, Kosmatka KJ, Beasley TM, Allison DB, Cruzen C, Simmons HA, Kemnitz JW, Weindruch R. Caloric restriction delays disease onset and mortality in rhesus monkeys. Science. 2009;325:201–4. doi: 10.1126/science.1173635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman RJ, Beasley TM, Allison DB, Weindruch R. Attenuation of sarcopenia by dietary restriction in rhesus monkeys. J Gerontol A Biol Sci Med Sci. 2008;63:556–9. doi: 10.1093/gerona/63.6.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper HM, Huang JY, Verdin E, Spelbrink JN. A new splice variant of the mouse SIRT3 gene encodes the mitochondrial precursor protein. PLoS ONE. 2009;4:e4986. doi: 10.1371/journal.pone.0004986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KB, Liu J, Tian L, Barnes S, Yang Q, Wood PA. Cardiac hypertrophy in mice with long-chain acyl-CoA dehydrogenase or very long-chain acyl-CoA dehydrogenase deficiency. Lab Invest. 2009;89:1348–54. doi: 10.1038/labinvest.2009.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhahbi JM, Mote PL, Wingo J, Rowley BC, Cao SX, Walford RL, Spindler SR. Caloric restriction alters the feeding response of key metabolic enzyme genes. Mech Ageing Dev. 2001;122:1033–48. doi: 10.1016/s0047-6374(01)00230-5. [DOI] [PubMed] [Google Scholar]
- Easlon E, Tsang F, Dilova I, Wang C, Lu SP, Skinner C, Lin SJ. The dihydrolipoamide acetyltransferase is a novel metabolic longevity factor and is required for calorie restriction-mediated life span extension. J Biol Chem. 2007;282:6161–71. doi: 10.1074/jbc.M607661200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel T. Oxidant signals and oxidative stress. Curr Opin Cell Biol. 2003;15:247–54. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- Finley LW, Haigis MC. The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Ageing Res Rev. 2009;8:173–88. doi: 10.1016/j.arr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L. The scientific basis of caloric restriction leading to longer life. Curr Opin Gastroenterol. 2009;25:144–50. doi: 10.1097/MOG.0b013e32831ef1ba. [DOI] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P. Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1alpha. EMBO J. 2007;26:1913–23. doi: 10.1038/sj.emboj.7601633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgio V, Soriano ME, Basso E, Bisetto E, Lippe G, Forte MA, Bernardi P. Cyclophilin D in mitochondrial pathophysiology. Biochim Biophys Acta. 2010;1797:1113–8. doi: 10.1016/j.bbabio.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glorioso C, Oh S, Douillard GG, Sibille E. Brain molecular aging, promotion of neurological disease and modulation by Sirtuin5 longevity gene polymorphism. Neurobiol Dis. 2010 doi: 10.1016/j.nbd.2010.09.016. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gredilla R, Barja G. Minireview: the role of oxidative stress in relation to caloric restriction and longevity. Endocrinology. 2005;146:3713–7. doi: 10.1210/en.2005-0378. [DOI] [PubMed] [Google Scholar]
- Guan KL, Xiong Y. Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci. 2010 doi: 10.1016/j.tibs.2010.09.003. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Mitochondria--a nexus for aging, calorie restriction, and sirtuins? Cell. 2008;132:171–6. doi: 10.1016/j.cell.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerra C, Koza RA, Walsh K, Kurtz DM, Wood PA, Kozak LP. Abnormal nonshivering thermogenesis in mice with inherited defects of fatty acid oxidation. J Clin Invest. 1998;102:1724–31. doi: 10.1172/JCI4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagopian K, Harper ME, Ram JJ, Humble SJ, Weindruch R, Ramsey JJ. Long-term calorie restriction reduces proton leak and hydrogen peroxide production in liver mitochondria. Am J Physiol Endocrinol Metab. 2005;288:E674–84. doi: 10.1152/ajpendo.00382.2004. [DOI] [PubMed] [Google Scholar]
- Hagopian K, Ramsey JJ, Weindruch R. Caloric restriction increases gluconeogenic and transaminase enzyme activities in mouse liver. Exp Gerontol. 2003;38:267–78. doi: 10.1016/s0531-5565(02)00202-4. [DOI] [PubMed] [Google Scholar]
- Hagopian K, Ramsey JJ, Weindruch R. Krebs cycle enzymes from livers of old mice are differentially regulated by caloric restriction. Exp Gerontol. 2004;39:1145–54. doi: 10.1016/j.exger.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L. SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic beta cells. Cell. 2006;126:941–54. doi: 10.1016/j.cell.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Haigis MC, Sinclair DA. Mammalian sirtuins: biological insights and disease relevance. Annu Rev Pathol. 2010;5:253–95. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM. Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci U S A. 2006;103:10230–5. doi: 10.1073/pnas.0604392103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, Stevens RD, Li Y, Saha AK, Ruderman NB, Bain JR, Newgard CB, Farese RV, Jr, Alt FW, Kahn CR, Verdin E. SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature. 2010;464:121–5. doi: 10.1038/nature08778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houthoofd K, Braeckman BP, Lenaerts I, Brys K, De Vreese A, Van Eygen S, Vanfleteren JR. Axenic growth up-regulates mass-specific metabolic rate, stress resistance, and extends life span in Caenorhabditis elegans. Exp Gerontol. 2002a;37:1371–8. doi: 10.1016/s0531-5565(02)00173-0. [DOI] [PubMed] [Google Scholar]
- Houthoofd K, Braeckman BP, Lenaerts I, Brys K, De Vreese A, Van Eygen S, Vanfleteren JR. No reduction of metabolic rate in food restricted Caenorhabditis elegans. Exp Gerontol. 2002b;37:1359–69. doi: 10.1016/s0531-5565(02)00172-9. [DOI] [PubMed] [Google Scholar]
- Hsu PP, Sabatini DM. Cancer cell metabolism: Warburg and beyond. Cell. 2008;134:703–7. 3. doi: 10.1016/j.cell.2008.08.021. [DOI] [PubMed] [Google Scholar]
- Huang JY, Hirschey MD, Shimazu T, Ho L, Verdin E. Mitochondrial sirtuins. Biochim Biophys Acta. 2010;1804:1645–51. doi: 10.1016/j.bbapap.2009.12.021. [DOI] [PubMed] [Google Scholar]
- Hunt ND, Hyun DH, Allard JS, Minor RK, Mattson MP, Ingram DK, de Cabo R. Bioenergetics of aging and calorie restriction. Ageing Res Rev. 2006;5:125–43. doi: 10.1016/j.arr.2006.03.006. [DOI] [PubMed] [Google Scholar]
- Jeninga EH, Schoonjans K, Auwerx J. Reversible acetylation of PGC-1: connecting energy sensors and effectors to guarantee metabolic flexibility. Oncogene. 2010;29:4617–24. doi: 10.1038/onc.2010.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Galonek H, Israelian K, Choy W, Morrison M, Xia Y, Wang X, Xu Y, Yang Y, Smith JJ, Hoffmann E, Carney DP, Perni RB, Jirousek MR, Bemis JE, Milne JC, Sinclair DA, Westphal CH. Biochemical characterization, localization, and tissue distribution of the longer form of mouse SIRT3. Protein Sci. 2009;18:514–25. doi: 10.1002/pro.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaeberlein M, Hu D, Kerr EO, Tsuchiya M, Westman EA, Dang N, Fields S, Kennedy BK. Increased life span due to calorie restriction in respiratory-deficient yeast. PLoS Genet. 2005;1:e69. doi: 10.1371/journal.pgen.0010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendrick AA, Choudhury M, Rahman SM, McCurdy CE, Friederich M, Vanhove JL, Watson PA, Birdsey N, Bao J, Gius D, Sack MN, Jing E, Kahn CR, Friedman JE, Jonscher KR. Fatty liver is associated with reduced SIRT3 activity and mitochondrial protein hyperacetylation. Biochem J. 2010 doi: 10.1042/BJ20100791. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-S, Patel K, Muldoon-Jacobs K, Bisht KS, Aykin-Burns N, Pennington JD, van der Meer R, Nguyen P, Savage J, Owens KM, Vassiliopolous O, Ozden O, Park S-H, Singh KK, Abdulkadir SA, Spitz DR, Deng C, Gius D. SIRT3 Is a Mitochondria-Localized Tumor Suppressor Required for Maintenance of Mitochondrial Integrity and Metabolism during Stress. Cancer Cell. 2010;17:41–52. doi: 10.1016/j.ccr.2009.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol Cell. 2006;23:607–18. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- King A, Selak MA, Gottlieb E. Succinate dehydrogenase and fumarate hydratase: linking mitochondrial dysfunction and cancer. Oncogene. 2006;25:4675–82. doi: 10.1038/sj.onc.1209594. [DOI] [PubMed] [Google Scholar]
- Kong X, Wang R, Xue Y, Liu X, Zhang H, Chen Y, Fang F, Chang Y. Sirtuin 3, a new target of PGC-1alpha, plays an important role in the suppression of ROS and mitochondrial biogenesis. PLoS ONE. 2010;5:e11707. doi: 10.1371/journal.pone.0011707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz DM, Rinaldo P, Rhead WJ, Tian L, Millington DS, Vockley J, Hamm DA, Brix AE, Lindsey JR, Pinkert CA, O’Brien WE, Wood PA. Targeted disruption of mouse long-chain acyl-CoA dehydrogenase gene reveals crucial roles for fatty acid oxidation. Proc Natl Acad Sci U S A. 1998;95:15592–7. doi: 10.1073/pnas.95.26.15592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert AJ, Wang B, Yardley J, Edwards J, Merry BJ. The effect of aging and caloric restriction on mitochondrial protein density and oxygen consumption. Exp Gerontol. 2004;39:289–95. doi: 10.1016/j.exger.2003.12.009. [DOI] [PubMed] [Google Scholar]
- Lanza IR, Short DK, Short KR, Raghavakaimal S, Basu R, Joyner MJ, McConnell JP, Nair KS. Endurance exercise as a countermeasure for aging. Diabetes. 2008;57:2933–42. doi: 10.2337/db08-0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lescai F, Blanche H, Nebel A, Beekman M, Sahbatou M, Flachsbart F, Slagboom E, Schreiber S, Sorbi S, Passarino G, Franceschi C. Human longevity and 11p15.5: a study in 1321 centenarians. Eur J Hum Genet. 2009;17:1515–9. doi: 10.1038/ejhg.2009.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Banck M, Mujtaba S, Zhou MM, Sugrue MM, Walsh MJ. p53-induced growth arrest is regulated by the mitochondrial SirT3 deacetylase. PLoS ONE. 2010;5:e10486. doi: 10.1371/journal.pone.0010486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Johnson N, Capano M, Edwards M, Crompton M. Cyclophilin-D promotes the mitochondrial permeability transition but has opposite effects on apoptosis and necrosis. Biochem J. 2004;383:101–9. doi: 10.1042/BJ20040669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Ford E, Haigis M, Liszt G, Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–6. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L. Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature. 2002;418:344–8. doi: 10.1038/nature00829. [DOI] [PubMed] [Google Scholar]
- Liu XZ, Yan D. Ageing and hearing loss. J Pathol. 2007;211:188–97. doi: 10.1002/path.2102. [DOI] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese RV, Jr, Weissman S, Verdin E, Schwer B. Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol. 2007;27:8807–14. doi: 10.1128/MCB.01636-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Chua KF, Mostoslavsky R, Franco S, Gostissa M, Alt FW. DNA repair, genome stability, and aging. Cell. 2005;120:497–512. doi: 10.1016/j.cell.2005.01.028. [DOI] [PubMed] [Google Scholar]
- Lopez-Lluch G, Hunt N, Jones B, Zhu M, Jamieson H, Hilmer S, Cascajo MV, Allard J, Ingram DK, Navas P, de Cabo R. Calorie restriction induces mitochondrial biogenesis and bioenergetic efficiency. Proc Natl Acad Sci U S A. 2006;103:1768–73. doi: 10.1073/pnas.0510452103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marfe G, Tafani M, Indelicato M, Sinibaldi-Salimei P, Reali V, Pucci B, Fini M, Russo MA. Kaempferol induces apoptosis in two different cell lines via Akt inactivation, Bax and SIRT3 activation, and mitochondrial dysfunction. J Cell Biochem. 2009;106:643–50. doi: 10.1002/jcb.22044. [DOI] [PubMed] [Google Scholar]
- Meany DL, Xie H, Thompson LV, Arriaga EA, Griffin TJ. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics. 2007;7:1150–63. doi: 10.1002/pmic.200600450. [DOI] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily Conserved and Nonconserved Cellular Localizations and Functions of Human SIRT Proteins. Mol Biol Cell. 2005;16:4623–35. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miwa S, Lawless C, von Zglinicki T. Mitochondrial turnover in liver is fast in vivo and is accelerated by dietary restriction: application of a simple dynamic model. Aging Cell. 2008;7:920–3. doi: 10.1111/j.1474-9726.2008.00426.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L. SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell. 2009;137:560–70. doi: 10.1016/j.cell.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Ogura M, Tanaka D, Inagaki N. Localization of mouse mitochondrial SIRT proteins: shift of SIRT3 to nucleus by co-expression with SIRT5. Biochem Biophys Res Commun. 2008;366:174–9. doi: 10.1016/j.bbrc.2007.11.122. [DOI] [PubMed] [Google Scholar]
- Nasrin N, Wu X, Fortier E, Feng Y, Bare OC, Chen S, Ren X, Wu Z, Streeper RS, Bordone L. SIRT4 regulates fatty acid oxidation and mitochondrial gene expression in liver and muscle cells. J Biol Chem. 2010;285:31995–2002. doi: 10.1074/jbc.M110.124164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba MO. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science. 2005;310:314–7. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell. 2003;11:437–44. doi: 10.1016/s1097-2765(03)00038-8. [DOI] [PubMed] [Google Scholar]
- Ogura M, Nakamura Y, Tanaka D, Zhuang X, Fujita Y, Obara A, Hamasaki A, Hosokawa M, Inagaki N. Overexpression of SIRT5 confirms its involvement in deacetylation and activation of carbamoyl phosphate synthetase 1. Biochem Biophys Res Commun. 2010;393:73–8. doi: 10.1016/j.bbrc.2010.01.081. [DOI] [PubMed] [Google Scholar]
- Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP. SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci U S A. 2002;99:13653–8. doi: 10.1073/pnas.222538099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacios OM, Carmona JJ, Michan S, Chen KY, Manabe Y, Ward JL, 3rd, Goodyear LJ, Tong Q. Diet and exercise signals regulate SIRT3 and activate AMPK and PGC-1alpha in skeletal muscle. Aging (Albany NY) 2009;1:771–83. doi: 10.18632/aging.100075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picklo MJ., Sr Ethanol intoxication increases hepatic N-lysyl protein acetylation. Biochem Biophys Res Commun. 2008;376:615–9. doi: 10.1016/j.bbrc.2008.09.039. [DOI] [PubMed] [Google Scholar]
- Pillai VB, Sundaresan NR, Kim G, Gupta M, Rajamohan SB, Pillai JB, Samant S, Ravindra PV, Isbatan A, Gupta MP. Exogenous NAD blocks cardiac hypertrophic response via activation of the SIRT3-LKB1-AMP-activated kinase pathway. J Biol Chem. 2010;285:3133–44. doi: 10.1074/jbc.M109.077271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polevoda B, Sherman F. The diversity of acetylated proteins. Genome Biol. 2002;3:reviews0006. doi: 10.1186/gb-2002-3-5-reviews0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu X, Brown K, Hirschey MD, Verdin E, Chen D. Calorie restriction reduces oxidative stress by SIRT3-mediated SOD2 activation. Cell Metab. 2010;12:662–7. doi: 10.1016/j.cmet.2010.11.015. [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Gerhart-Hines Z, Puigserver P. Metabolic adaptations through the PGC-1 alpha and SIRT1 pathways. FEBS Lett. 2008;582:46–53. doi: 10.1016/j.febslet.2007.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose G, Dato S, Altomare K, Bellizzi D, Garasto S, Greco V, Passarino G, Feraco E, Mari V, Barbi C, BonaFe M, Franceschi C, Tan Q, Boiko S, Yashin AI, De Benedictis G. Variability of the SIRT3 gene, human silent information regulator Sir2 homologue, and survivorship in the elderly. Exp Gerontol. 2003;38:1065–70. doi: 10.1016/s0531-5565(03)00209-2. [DOI] [PubMed] [Google Scholar]
- Ryan MT, Hoogenraad NJ. Mitochondrial-nuclear communications. Annu Rev Biochem. 2007;76:701–22. doi: 10.1146/annurev.biochem.76.052305.091720. [DOI] [PubMed] [Google Scholar]
- Sakakibara I, Fujino T, Ishii M, Tanaka T, Shimosawa T, Miura S, Zhang W, Tokutake Y, Yamamoto J, Awano M, Iwasaki S, Motoike T, Okamura M, Inagaki T, Kita K, Ezaki O, Naito M, Kuwaki T, Chohnan S, Yamamoto TT, Hammer RE, Kodama T, Yanagisawa M, Sakai J. Fasting-Induced Hypothermia and Reduced Energy Production in Mice Lacking Acetyl-CoA Synthetase 2. Cell Metab. 2009;9:191–202. doi: 10.1016/j.cmet.2008.12.008. [DOI] [PubMed] [Google Scholar]
- Scarpulla RC. Transcriptional paradigms in mammalian mitochondrial biogenesis and function. Physiol Rev. 2008;88:611–38. doi: 10.1152/physrev.00025.2007. [DOI] [PubMed] [Google Scholar]
- Scher MB, Vaquero A, Reinberg D. SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007;21:920–8. doi: 10.1101/gad.1527307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schieke SM, Phillips D, McCoy JP, Jr, Aponte AM, Shen RF, Balaban RS, Finkel T. The mammalian target of rapamycin (mTOR) pathway regulates mitochondrial oxygen consumption and oxidative capacity. J Biol Chem. 2006;281:27643–52. doi: 10.1074/jbc.M603536200. [DOI] [PubMed] [Google Scholar]
- Schlicker C, Gertz M, Papatheodorou P, Kachholz B, Becker CF, Steegborn C. Substrates and regulation mechanisms for the human mitochondrial sirtuins Sirt3 and Sirt5. J Mol Biol. 2008;382:790–801. doi: 10.1016/j.jmb.2008.07.048. [DOI] [PubMed] [Google Scholar]
- Schubert A, Grimm S. Cyclophilin D, a component of the permeability transition-pore, is an apoptosis repressor. Cancer Res. 2004;64:85–93. doi: 10.1158/0008-5472.can-03-0476. [DOI] [PubMed] [Google Scholar]
- Schulz TJ, Zarse K, Voigt A, Urban N, Birringer M, Ristow M. Glucose restriction extends Caenorhabditis elegans life span by inducing mitochondrial respiration and increasing oxidative stress. Cell Metab. 2007;6:280–93. doi: 10.1016/j.cmet.2007.08.011. [DOI] [PubMed] [Google Scholar]
- Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E. Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci U S A. 2006;103:10224–9. doi: 10.1073/pnas.0603968103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Eckersdorff M, Li Y, Silva JC, Fermin D, Kurtev MV, Giallourakis C, Comb MJ, Alt FW, Lombard DB. Calorie restriction alters mitochondrial protein acetylation. Aging Cell. 2009;8:604–6. doi: 10.1111/j.1474-9726.2009.00503.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, North BJ, Frye RA, Ott M, Verdin E. The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol. 2002;158:647–57. doi: 10.1083/jcb.200205057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepard BD, Tuma DJ, Tuma PL. Chronic ethanol consumption induces global hepatic protein hyperacetylation. Alcohol Clin Exp Res. 2010;34:280–91. doi: 10.1111/j.1530-0277.2009.01091.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi T, Fan GQ, Xiao SD. SIRT3 reduces lipid accumulation via AMPK activation in human hepatic cells. J Dig Dis. 2010;11:55–62. doi: 10.1111/j.1751-2980.2009.00416.x. [DOI] [PubMed] [Google Scholar]
- Shi T, Wang F, Stieren E, Tong Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem. 2005;280:13560–7. doi: 10.1074/jbc.M414670200. [DOI] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Hua L, Dittenhafer-Reed KE, Schwer B, Lombard DB, Li Y, Bunkenborg J, Alt FW, Denu JM, Jacobsen MP, Verdin E. SIRT3 deacetylates mitochondrial 3-Hydroxy-3-Methylglutaryl CoA Synthase 2 and regulates ketone body production. Cell Metab. 2010a;12:654–61. doi: 10.1016/j.cmet.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimazu T, Hirschey MD, Huang JY, Ho LT, Verdin E. Acetate metabolism and aging: An emerging connection. Mech Ageing Dev. 2010b;131:511–6. doi: 10.1016/j.mad.2010.05.001. [DOI] [PubMed] [Google Scholar]
- Shulga N, Pastorino JG. Ethanol sensitizes mitochondria to the permeability transition by inhibiting deacetylation of cyclophilin-D mediated by sirtuin-3. J Cell Sci. 2010;123:4117–27. doi: 10.1242/jcs.073502. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Shulga N, Wilson-Smith R, Pastorino JG. Sirtuin-3 deacetylation of cyclophilin D induces dissociation of hexokinase II from the mitochondria. J Cell Sci. 2010;123:894–902. doi: 10.1242/jcs.061846. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Someya S, Xu J, Kondo K, Ding D, Salvi RJ, Yamasoba T, Rabinovitch PS, Weindruch R, Leeuwenburgh C, Tanokura M, Prolla TA. Age-related hearing loss in C57BL/6J mice is mediated by Bak-dependent mitochondrial apoptosis. Proc Natl Acad Sci U S A. 2009;106:19432–7. doi: 10.1073/pnas.0908786106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Someya S, Yamasoba T, Weindruch R, Prolla TA, Tanokura M. Caloric restriction suppresses apoptotic cell death in the mammalian cochlea and leads to prevention of presbycusis. Neurobiol Aging. 2007;28:1613–22. doi: 10.1016/j.neurobiolaging.2006.06.024. [DOI] [PubMed] [Google Scholar]
- Someya S, Yu W, Hallows WC, Xu J, Vann J, Leeuwenburgh C, Tanokura M, Denu JM, Prolla TA. SIRT3 mediates reduction of oxidative damage and prevention of age-related hearing loss under caloric restriction. Cell. 2010;143:802–12. doi: 10.1016/j.cell.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spange S, Wagner T, Heinzel T, Kramer OH. Acetylation of non-histone proteins modulates cellular signalling at multiple levels. Int J Biochem Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- Spindler SR, Dhahbi JM. Conserved and tissue-specific genic and physiologic responses to caloric restriction and altered IGFI signaling in mitotic and postmitotic tissues. Annu Rev Nutr. 2007;27:193–217. doi: 10.1146/annurev.nutr.27.061406.093743. [DOI] [PubMed] [Google Scholar]
- Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- Starai VJ, Escalante-Semerena JC. Identification of the protein acetyltransferase (Pat) enzyme that acetylates acetyl-CoA synthetase in Salmonella enterica. J Mol Biol. 2004;340:1005–12. doi: 10.1016/j.jmb.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Sundaresan NR, Gupta M, Kim G, Rajamohan SB, Isbatan A, Gupta MP. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J Clin Invest. 2009;119:2758–71. doi: 10.1172/JCI39162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 is a stress-responsive deacetylase in cardiomyocytes that protects cells from stress-mediated cell death by deacetylation of Ku70. Mol Cell Biol. 2008;28:6384–401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillman JB, Dhahbi JM, Mote PL, Walford RL, Spindler SR. Dietary calorie restriction in mice induces carbamyl phosphate synthetase I gene transcription tissue specifically. J Biol Chem. 1996;271:3500–6. doi: 10.1074/jbc.271.7.3500. [DOI] [PubMed] [Google Scholar]
- Van Remmen H, Ikeno Y, Hamilton M, Pahlavani M, Wolf N, Thorpe SR, Alderson NL, Baynes JW, Epstein CJ, Huang TT, Nelson J, Strong R, Richardson A. Life-long reduction in MnSOD activity results in increased DNA damage and higher incidence of cancer but does not accelerate aging. Physiol Genomics. 2003;16:29–37. doi: 10.1152/physiolgenomics.00122.2003. [DOI] [PubMed] [Google Scholar]
- Vydt TC, de Coo RF, Soliman OI, Ten Cate FJ, van Geuns RJ, Vletter WB, Schoonderwoerd K, van den Bosch BJ, Smeets HJ, Geleijnse ML. Cardiac involvement in adults with m.3243A>G MELAS gene mutation. Am J Cardiol. 2007;99:264–9. doi: 10.1016/j.amjcard.2006.07.089. [DOI] [PubMed] [Google Scholar]
- Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: a dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning ZB, Zeng R, Xiong Y, Guan KL, Zhao S, Zhao GP. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–7. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weindruch RH, Cheung MK, Verity MA, Walford RL. Modification of mitochondrial respiration by aging and dietary restriction. Mech Ageing Dev. 1980;12:375–92. doi: 10.1016/0047-6374(80)90070-6. [DOI] [PubMed] [Google Scholar]
- Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA. Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell. 2007;130:1095–107. doi: 10.1016/j.cell.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Cimen H, Han MJ, Shi T, Deng JH, Koc H, Palacios OM, Montier L, Bai Y, Tong Q, Koc EC. NAD+-dependent deacetylase SIRT3 regulates mitochondrial protein synthesis by deacetylation of the ribosomal protein MRPL10. J Biol Chem. 2010a;285:7417–29. doi: 10.1074/jbc.M109.053421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Hubbard BP, Sinclair DA, Tong Q. Characterization of murine SIRT3 transcript variants and corresponding protein products. J Cell Biochem. 2010b;111:1051–8. doi: 10.1002/jcb.22795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Chen YH, Zhang CY, Nimmakayalu MA, Ward DC, Weissman S. Cloning and characterization of two mouse genes with homology to the yeast Sir2 gene. Genomics. 2000;69:355–69. doi: 10.1006/geno.2000.6360. [DOI] [PubMed] [Google Scholar]
- Yechoor VK, Patti ME, Ueki K, Laustsen PG, Saccone R, Rauniyar R, Kahn CR. Distinct pathways of insulin-regulated versus diabetes-regulated gene expression: an in vivo analysis in MIRKO mice. Proc Natl Acad Sci U S A. 2004;101:16525–30. doi: 10.1073/pnas.0407574101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol Cell Proteomics. 2009;8:215–25. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–4. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]